Abstract
Since their discovery, protein tyrosine phosphatases have been speculated to play a role in tumor suppression because of their ability to antagonize the growth-promoting protein tyrosine kinases. Recently, a tumor suppressor from human chromosome 10q23, called PTEN or MMAC1, has been identified that shares homology with the protein tyrosine phosphatase family. Germ-line mutations in PTEN give rise to several related neoplastic disorders, including Cowden disease. A key step in understanding the function of PTEN as a tumor suppressor is to identify its physiological substrates. Here we report that a missense mutation in PTEN, PTEN-G129E, which is observed in two Cowden disease kindreds, specifically ablates the ability of PTEN to recognize inositol phospholipids as a substrate, suggesting that loss of the lipid phosphatase activity is responsible for the etiology of the disease. Furthermore, expression of wild-type or substrate-trapping forms of PTEN in HEK293 cells altered the levels of the phospholipid products of phosphatidylinositol 3-kinase and ectopic expression of the phosphatase in PTEN-deficient tumor cell lines resulted in the inhibition of protein kinase (PK) B/Akt and regulation of cell survival.
Glioblastoma is one of the most common and malignant forms of cancer. It is often characterized by the constitutive activation of epidermal growth factor (EGF)-dependent signaling pathways caused by the amplification of members of the EGF receptor family of protein tyrosine kinases (PTKs). The products of tumor-suppressor genes may attenuate these signaling pathways, and therefore, their loss through deletion or mutation may contribute to tumor progression. The frequent loss of heterozygosity at tumor-suppressor loci is often used to identify tumor-suppressor genes. The 10q23 region of human chromosome 10 is frequently deleted or mutated in a wide variety of tumor types, most frequently in glioblastoma, endometrial cancer, and prostate cancer, indicating the presence of a tumor-suppressor gene at this locus. PTEN was subsequently identified as this tumor suppressor and was found to contain the catalytic signature motif detected in all members of the protein tyrosine phosphatase (PTP) family (1, 2). Importantly, PTEN appears to be lost frequently in advanced cancers, suggesting that its deletion may not be the transforming event but that PTEN may inhibit other cellular functions necessary for tumor progression (3, 4).
Importantly, germ-line transmission of mutations in PTEN was shown to give rise to a related set of disorders, including Cowden disease, that are characterized by numerous small benign tumors and an increased incidence of other malignant growths (5–7). The detection of germ-line mutations in these neoplastic disorders verifies that PTEN is the tumor suppressor residing on human chromosome 10q23. This conclusion is supported by the recent findings that PTEN knockout mice develop tumors similar to those found in Cowden disease (8).
A key step in establishing the cellular function of PTEN is the identification of its physiological substrates. Recently, two groups have identified different candidate substrates for PTEN. Tamura et al. (9) report that overexpression of PTEN results in changes in cell adhesion and spreading and suggest that these effects result from the the dephosphorylation of the PTK focal adhesion kinase (FAK). In contrast, Maehama and Dixon (10) have shown that PTEN recognizes phosphatidylinositol phosphate (PtdInsP) as a substrate. Our initial studies demonstrating catalytic activity in PTEN indicated that it preferred highly acidic or multiply phosphorylated substrates (11). This may explain why, in certain contexts, PTEN will dephosphorylate polyphosphorylated molecules such as FAK or PtdInsP (11, 12). Nevertheless, the important question remains as to which multiply phosphorylated molecules, protein or otherwise, are the physiologically relevant targets of PTEN.
Significantly, we have demonstrated previously that the majority of missense mutations isolated from tumor and Cowden disease samples ablate the protein phosphatase activity of PTEN (11). One exception is a missense mutation that changes a glycine residue in the catalytic signature motif to a glutamate (PTEN-G129E; see ref. 11). Although there was no adverse effect on the protein phosphatase activity of PTEN, this G129E mutation was isolated from two independent Cowden-disease kindreds, indicating that it abolishes the tumor-suppressor activity of PTEN (5, 12). Therefore, this mutation can be used as an important indicator to determine whether a proposed function of PTEN is specific for its role as a tumor suppressor.
We analyzed the PTEN-G129E Cowden disease mutation and found that this mutation specifically inhibits the recognition of PtdInsPs by PTEN. Additionally, we show that expression of PTEN in mammalian cells results in changes in PtdIns(3,4,5)P3 levels, and expression of PTEN in two independent glioblastoma cell lines results in the disruption of signaling downstream of phospahatidylinositol 3-kinase (PI 3-kinase) to PKB/Akt and BAD. Significantly, we also show that expression of PTEN in LnCaP cells, a prostate-tumor cell line, abrogates cell survival and that this effect is inhibited by the expression of a constitutively activated form of PKB. Therefore, we believe that the physiological function of PTEN is to antagonize signaling downstream of PI 3-kinase by dephosphorylating PtdInsPs.
METHODS
PTEN Phosphatase Assays.
Recombinant wild-type and mutant forms of PTEN were expressed in Eschirichia coli and purified by glutathione-affinity chromatography (11). The purified proteins were assayed with polyGluTyr as described (11). Release of 32Pi from radiolabeled PtdInsPs was determined by performing a modified Bligh and Dyer extraction (13). The upper phase (containing inorganic phosphate) was removed, dried, and resuspended in a 1 M trichloroacetic acid/1% ammonium molybdate solution. After extraction with 2 vol of toluene/isobutylalchohol (1:1 vol/vol), the upper phase was removed and counted. Site selectivity was determined by incubating recombinant PTEN or SHIP (a gift from C. Erneux, Free University, Brussels) with radiolabeled PtdIns(3,4,5)P3, and the lipid products of these reactions were analyzed by TLC or HPLC (13). The phospholipids were extracted by performing the modified Bligh and Dyer extraction described, and the resulting lower phases were dried, resuspended in 20 μl of chloroform/methanol (2:1 vol/vol), and applied to an oxalate-activated silica 60 TLC plate. Plates were developed in methanol/chloroform/water/ammonia (100:75:25:15).
Determination of PtdIns[3,4,5]P3 Levels.
HEK293 cells were transfected via calcium phosphate coprecipitation with 20 μg of DNA per 10-cm dish. The calcium phosphate⋅DNA coprecipitate was removed by washing with PBS 16 hr after addition, and the cells were returned to growth medium for 36 hr before harvesting. Cotransfection of PTEN and p110 constructs were performed by using 9.5 μg of each plasmid DNA. Transfection efficiency was determined by including a green fluorescent protein expression plasmid (1 μg) in all transfections and was shown to be 80%. Phospholipids were extracted from the cells exactly as described (14). PtdIns(3,4,5)P3 levels were assayed by using a ligand displacement assay (14) and were normalized to total protein.
Expression of PTEN in Glioblastoma Cell Lines.
PTEN retroviral expression vectors were constructed in pBabePuro (15). After transfection into packaging lines, the viral supernatants were harvested, diluted with growth medium, and incubated with U87MG or U373MG cells for 8 hr at 32°C in the presence of polybrene. Infected cells were selected with puromycin (2 μg/ml), and drug-resistant colonies were expanded to generate clonal cell lines. Cell lysis and immunoblots were performed essentially as described (16). Protein levels were determined by the method of Bradford using BSA as a standard, and equal protein was loaded in each lane. Antibodies to PTEN were generated in rabbits by using a C-terminal peptide of PTEN (ENEPFDEDQHTQITKV) conjugated to keyhole limpet hemocyanin. Antibodies to PKB/AKT, phospho-PKB/Akt (specific to phosphorylated Ser-473) and phospho-BAD were purchased from New England Biolabs.
Expression of PTEN in LnCaP Cells.
Cells were transfected by using cationic lipids (Transfast, Promega) at a DNA/lipid ratio of 1. Cells were incubated with the DNA–lipid complexes for 12 hr, and transfected cells then were identified based on the expression of a cotransfected GFP vector or by performing immunoflourescence by using antibodies to the HA-epitope tag located at the N terminus of PTEN, essentially as described (17).
RESULTS
Activity of PTEN Toward Inositol Phospholipids.
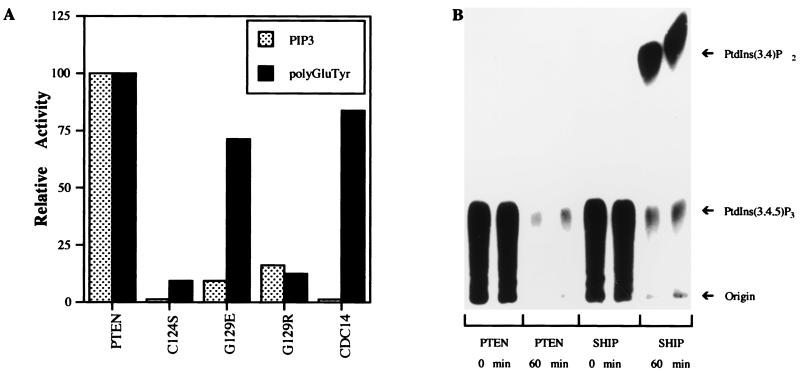
Recombinant PTEN, produced in E. coli (11), was assayed for its ability to release 32Pi from radiolabeled PtdIns(3,4,5)P3 or polyGluTyr (Fig. 1A). As expected, wild-type PTEN catalyzed the dephosphorylation of both substrates (Fig. 1A). Remarkably, the activity of PTEN-G129E toward PtdIns(3,4,5)P3 was reduced by ≈90% relative to the wild-type enzyme while still retaining activity toward polyGluTyr (Fig. 1A). A catalytically inactive mutant of PTEN (PTENC124S) was unable to dephosphorylate polyGluTyr or PtdIns(3,4,5)P3, indicating that the phosphatase activity was not caused by a bacterial contaminant. In addition, cdc14, a dual-specificity phosphatase closely related to PTEN, was also unable to dephosphorylate PtdIns(3,4,5)P3, demonstrating that recognition of this phospholipid substrate is not a general property of other, even closely related, dual-specificity phosphatases. These data, demonstrating that the PTEN-G129E mutation specifically ablates the activity of PTEN toward phosphorylated inositol lipids, demonstrates that the lipid phosphatase, rather than the protein phosphatase, activity of PTEN is required for it to function as a tumor suppressor and that loss of the lipid phosphatase activity results in Cowden disease.
Figure 1.
PTEN is a PtdIns 3-phosphatase. (A) Recombinant PTEN was incubated with radiolabeled polyGluTyr and PtdIns(3,4,5)P3, and the release of 32Pi was measured as described. (B) PtdIns(3,4,5)P3, labeled at the 3 position, was incubated with PTEN or SHIP for 0 or 60 min, and the reaction products were resolved on TLC plates and visualized by autoradiography.
Site Specificity of PTEN Within PtdIns(3,4,5)P3.
To determine whether PTEN recognizes specific sites in the inositol ring of PtdIns(3,4,5)P3, lipid substrate, labeled exclusively in the 3 position with 32P, was incubated with PTEN or SHIP, a well characterized 5-phosphatase (18). The products of these reactions were analyzed by TLC. Incubation with SHIP yielded a radiolabeled product with the expected mobility of PtdIns(3,4)P2 (Fig. 1B). However, no labeled lipid products were generated following treatment with PTEN under conditions where >50% of the 32P was lost from the substrate (Fig. 1B). The site selectivity of PTEN was confirmed by HPLC analysis of similar reactions by using 3H- and 3-32P-labeled Ins(1,3,4,5)P4 in which the only products detected were 32Pi and [3H]Ins(1,4,5)P3 (data not shown). In addition to PtdIns(3,4,5)P3, PTEN also hydrolyzed the other potential products of PI 3-kinase, PtdIns(3)P and PtdIns(3,4)P2, with the following rank order based on relative percent hydrolysis: PtdIns(3,4,5)P3 = PtdIns(3,4)P2 > PtdIns3P > Ins(1,3,4,5)P4. The specificity of PTEN for 3-phosphorylated inositol lipids indicates that it may function as a negative regulator of PI 3-kinase-mediated signaling and distinguishes PTEN from other known lipid phosphatases.
PTEN Inhibits PI 3-Kinase-Dependent Signaling.
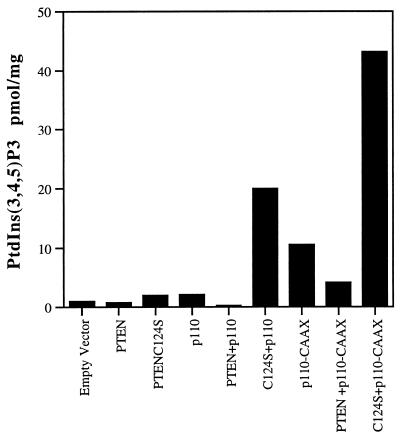
The potential for PTEN to antagonize PI 3-kinase signaling was further investigated in mammalian cells (Fig. 2). Transient expression of PTEN in HEK293 cells lowered the levels of PtdIns(3,4,5)P3 (Fig. 2) from 1 pmol/mg to 0.7 pmol/mg. Importantly, PTEN-C124S, in which the catalytic cysteine has been mutated to serine, resulted in an increase in the levels of PtdIns(3,4,5)P3 (Fig. 2) from 1 pmol/mg to 2 pmol/mg. The accumulation of PtdIns(3,4,5)P3 in response to expression of PTEN-C124S is likely because of the ability of this form of PTEN to behave as a “substrate-trapping” mutant, resulting in a stable complex with the lipid substrate that protects it from dephosphorylation by endogenous phosphatases (19). The accumulation of PtdIns(3,4,5)P3 in the presence of a substrate-trapping mutant confirms that PtdIns(3,4,5)P3 is a physiological target of PTEN (19, 20). Expression of a constitutively activated PI 3-kinase (p110-CAAX) resulted in elevated levels of PtdIns(3,4,5)P3 that were reduced ≈60% following coexpression with PTEN and were increased ≈4.5-fold when coexpressed with PTEN-C124S (Fig. 2). The ability of PTEN to decrease PtdIns(3,4,5)P3 levels produced by p110-CAAX further indicates that this phosphatase exerts its effects on the products of PI 3-kinase rather than by dephosphorylating the tyrosine residues responsible for recruiting the p85-p110 PI 3-kinase complex to the membrane.
Figure 2.
Expression of PTEN-antagonized PI 3-kinase. HEK293 cells were transfected with PTEN or PTENC124S in combination with the p110 subunit of PI 3-kinase or an activated, membrane-bound PI-3 kinase (p110-CAAX). The resulting levels of PtdIns(3,4,5)P3 were determined as described and were normalized to total protein. Data are expressed as pmol of PtdIns(3,4,5)P3 per mg of protein. These data, from a single experiment, are representative of three experiments that yielded similar results.
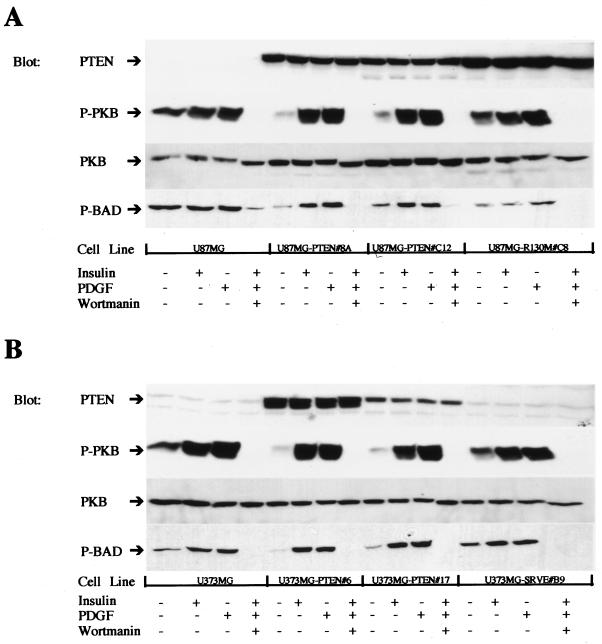
Importantly, we detected defects in PI 3-kinase signaling in tumor cell lines that have lost their endogenous PTEN genes through deletion or mutation. Signaling downstream of PI 3-kinase was assessed in two of these PTEN-deficient cell lines, U87 MG and U373 MG, by determining the phosphorylation status of PKB/Akt, a kinase whose activation depends on PtdIns(3,4,5)P3 or PtdIns(3,4)P2, two products of PI 3-kinase (21). Clonal cell lines were generated in U87 MG and U373 MG in which PTEN expression was restored by infection with recombinant retroviruses (Fig. 3). The parental cell lines exhibited high basal levels of phosphorylated PKB/Akt, whereas in every wild-type PTEN-expressing clonal cell line the basal levels of activated PKB/Akt were significantly lowered (Fig. 3). Expression of a catalytically inactive mutant of PTEN, PTEN-R130M, had no effect on basal PKB/Akt activation (Fig. 3). Similarly, no effect on PKB/Akt activation was observed in a U373 MG (PTEN-SRVE no. B9) clone infected with an unstable mutant of PTEN (Fig. 3). After reconstitution of PTEN expression in the tumor lines, activation of PKB/Akt by insulin or platelet-derived growth factor was unaffected (Fig. 3), suggesting that there are regulatory mechanisms that allow for the generation of PtdIns(3,4,5)P3 following growth factor stimulation. The activation of PKB/Akt was completely inhibited by wortmannin in the tumor-cell lines (Fig. 3), indicating that the increase in PKB/Akt phosphorylation was PI 3-kinase-dependent.
Figure 3.
Expression of PTEN in glioblastoma cell lines decreases the amount of activated PKB/AKT. PTEN expression was reconstituted in U87MG (A) or U373MB glioblastoma (B) cell lines by infection with recombinant retrovirus. Confluent dishes were either left untreated, stimulated with insulin (10 μg/ml for 10 min) or platelet-derived growth factor (50 ng/ml for 10 min), or pretreated with wortmannin (150 nM for 30 min), stimulated with both insulin (10 μg/ml) and platelet-derived growth factor (50 ng/ml) for 10 min, and then lysed. Equal amounts of protein were loaded for each cell line, and immunoblots were probed with antibodies to PTEN, PKB/Akt (PKB), phospho-PKB/Akt (P-PKB), or phosphorylated BAD (P-BAD) and visualized by using enhanced chemiluminescence reagents.
Regulation of Cell Survival.
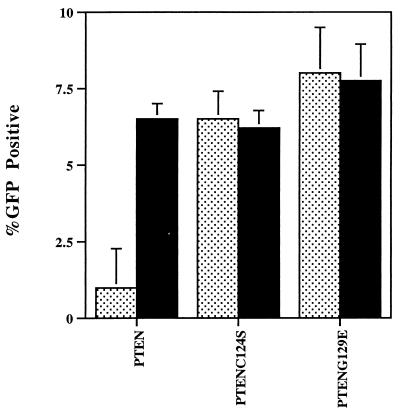
One of the downstream substrates of PKB/Akt is the death-effector protein BAD (22), phosphorylation of which has been linked to the promotion of cell survival (23). Although expression of PTEN in both U87 MG and U373 MG resulted in the reduction of BAD phosphorylation (Fig. 3B), we did not detect evidence of apoptosis in the PTEN-expressing clones. However, expression of wild-type PTEN in LnCaP cells, a PTEN-deficient prostate-cancer cell line, resulted in a decrease in the number of PTEN-positive cells recovered relative to controls expressing the catalytically inactive mutant PTEN-C124S (Fig. 4). In this assay, the Cowden disease mutation PTEN-G129E behaved similarly to PTEN-C124S, consistent with the ablation of the tumor-suppressor activity of PTEN by this mutation (Fig. 4). Significantly, coexpression of a constitutively active, membrane-targeted PKB/Akt (24) completely reverted the PTEN phenotype in that PTEN-positive cells could now be recovered despite the presence of wild-type PTEN (Fig. 4).
Figure 4.
Expression of PTEN inhibits cell survival in LnCaP cells. PTEN, PTENC124S, or PTENG129E were cotransfected with a green fluorescent protein (GFP) expression vector into LnCaP cells. The cotransfections also included either an expression vector for consitutively active PKB/Akt (black bars) or a control empty vector (speckled bars). Transfected cells were identified as GFP positive cells or by immunofluorescence microscopy after staining with antibodies to the transfected PTEN. Transfection efficiency was assessed by determining the percentage of GFP/PTEN-positive cells (total number of cells was determined by counting nuclei stained with 4′,6-diamidino-2-phenylindole). Data are expressed as the mean transfection efficiency (± SD, n = 3).
DISCUSSION
PTEN is a newly defined tumor-suppressor gene that has been implicated in a wide variety of cancers and in a series of related disorders that are characterized by a predispostion to cancer (1, 2, 25–28). Although these studies offer many insights into the biology of PTEN, they do not address the molecular mechanism of PTEN function, which requires the identification of its physiological substrates. However, this has become an area of controversy. Tamura et al. (9) have suggested that PTEN disrupts cell spreading and migration by dephosphorylating FAK. In contrast, Maehema and Dixon (10) have shown that, in certain contexts, PTEN can dephosphorylate the lipid second messenger PtdInsP. However, both of these studies fall short of demonstrating that the ability of PTEN to dephosphorylate FAK or PtdInsP is related to its tumor-suppressive function. In light of the facts that previous studies have shown that PTEN prefers highly acidic or multiply phosphorylated substrates (11) and that FAK and PtdInsP share these properties, one must view these results with caution, as they may result from nonspecific interactions between PTEN and highly charged molecules (12).
Significantly, we have been able to show that a single point mutation in PTEN, PTEN-G129E, specifically ablates the ability of PTEN to recognize phospholipids as substrates. In contrast, Tamura et al. (9) found that the PTEN-G129E point mutation behaves essentially like wild-type PTEN in their assays of cell spreading. Even though this mutation does not alter the ability of PTEN to recognize proteinaceous substrates, the fact that it has been identified in two independent Cowden disease kindreds indicates that this mutation would be expected to impair the tumor-suppressor activity of PTEN (5, 11). Therefore, these findings demonstrate that the lipid phosphatase activity rather than the protein phosphatase activity of PTEN is important for its tumor-suppressive activity and that FAK, at least in this respect, is not a relevant target of the phosphatase.
Characterization of the lipid phosphatase activity of PTEN demonstrates that it shows specificity for phosphatidylinositols phosphorylated at the 3 position. This represents the first phosphatidylinositol phosphatase with specificity for the lipid products of PI 3-kinase and suggests that PTEN may function to antagonize the growth-promoting signals generated by PI 3-kinase. Indeed, overexpression of PTEN in mammalian cells disrupted the PI 3-kinase-dependent production of PtdIns(3,4,5)P3. Furthermore, expression of a catalytically inactive mutant of PTEN, PTEN-C124S, which may function as a substrate trap, results in the accumulation of PtdIns(3,4,5)P3, indicating that PTEN may function in vivo to antagonize PI 3-kinase-dependent signaling.
Importantly, signaling downstream of PI 3-kinase is altered in two different glioblastoma cell lines, U87 MG and U373 MG, which have lost expression of PTEN because of mutation or deletion of both alleles. Both cell lines exhibited elevated levels of active, phosphorylated PKB/Akt, a serine/threonine kinase whose activation requires the production of PtdIns(3,4,5)P3 or PtdIns(3,4)P2 (21, 29). Restoration of PTEN expression by retroviral transduction resulted in the reduction of activated PKB/Akt as is evident from the observed reduction in the phosphorylation of PKB/Akt and in reduced levels of phosphorylated BAD, an apoptosis regulator that is a known in vivo substrate of PKB/Akt (22). Significantly, insulin or platelet-derived growth factor stimulation of these cell lines resulted in the stimulation of PKB/Akt regardless of whether PTEN expression had been reconstituted, suggesting that there are mechanisms in place which, in response to growth factor stimulation, serve to regulate the activity of PTEN and allow the production of PtdIns(3,4,5)P3. This is especially important given that PTEN is widely expressed, including cell lines that produce PtdIns(3,4,5)P3 in response to insulin.
Although the reconstitution of PTEN decreased the levels of phosphorylated BAD, we were unable to detect evidence of apoptosis in these cell lines. PKB/AKT is thought to affect cell survival in many cell types by phosphorylating BAD or inactivating GSK3β (23, 30). Reconstitution of PTEN in LnCaP cells, a PTEN-deficient prostate-cancer cell line, resulted in the inhibition of cell survival. Importantly, expression of the PTEN-G129E Cowden disease point mutation does not result in the abrogation of cell survival. This observation indicates that the lipid phosphatase activity of PTEN is required for these effects on cell survival. The properties of the PTEN-G129E mutant emphasize the requirement of the lipid phosphatase activity for the tumor-suppressor function of PTEN. Furthermore, the PTEN-dependent induction of apoptosis was completely inhibited by coexpressing a constitutively active, membrane-targeted form of PKB/Akt indicating that PTEN, by dephosphorylating phosphatidylinositides, is an upstream regulator of PKB/Akt and, at least in LNCAP cells, serves to regulate survival signals. Thus, in some cell types, PTEN functions as a tumor suppressor to inhibit the PKB/AKT-dependent survival signals that are activated in response to 3-phosphorylated phosphatidylinositols.
The failure of PTEN to regulate cell survival in the glioblastoma cell lines suggests that the generation of phosphatidylinositol 3-phosphates plays other roles during tumor progression. In fact, PI 3-kinase has been implicated in the regulation of a wide variety of cellular processes (31). Therefore, by dephosphorylating the products of PI 3-kinase, PTEN is likely to be involved in the regulation of many of these processes. Significantly, both PI 3-kinase and PKB/Akt have been isolated as transforming oncogenes in retroviruses (32, 33). These observations indicate that PI 3-kinase and PKB/Akt function in a signaling pathway promoting growth and survival. Our data demonstrate that PTEN functions to suppress these growth-promoting and survival signals by dephosphorylating the phospholipid products of PI 3-kinase. Our data provide a molecular basis for the development of Cowden disease, as loss of the lipid phosphatase activity of PTEN leads to the accumulation of the products of PI 3-kinase and results in the neoplastic pathologies characteristic of the disease.
Acknowledgments
The authors thank B. Neel and L. Faleiro for invaluable comments. M.P.M. was supported by a National Cancer Institute training grant (5T32 CA09311-18). I.P. was supported by an Biotechnology and Biological Sciences Research Council Cooperative Awards in Science and Engineering studentship. This work was supported by Grants CA53840 and GM55989 to N.K.T. and by a Medical Research Council Program Grant to C.P.D.; M.H.W. is an American Cancer Society Research Professor and is supported by the U.S. Department of the Army (DAMD-17-94-14247), the National Cancer Institute (5R35 CA39829), Amplicon Corporation, and the “1 in 9” breast cancer organization.
ABBREVIATIONS
- EGF
epidermal growth factor
- PK
protein kinase
- PTK
protein tyrosine kinase
- PTP
protein tyrosine phosphatase
- PtdIns
phosphatidylinositol
- PtdInsP
PtdIns phosphate
References
- 1.Steck P A, Perhouse M A, Jasser S A, Yung W K A, Lin H, Ligon A H, Lauren A L, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Milliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1946. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Whang Y E, Wu X, Suzuki H, Reiter R E, Tran C, Vessella R L, Said J W, Isaacs W B, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasheed B K, Stenzel T T, McLendon R E, Parsons R, Friedman A H, Friedman H S, Bigner D D, Bigner S H. Cancer Res. 1997;57:4187–4190. [PubMed] [Google Scholar]
- 5.Liaw D, Marsh D J, Li J, Dahia P L M, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, et al. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 6.Marsh D J, Dahia P L M, Zheng Z, Liaw D, Parsons D, Gorlin R J, Eng C. Nat Genet. 1997;16:334–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 7.Eng C, Murday V, Seal S, Mohammed S, Hodgson S V, Chaudray M A, Fentiman I S, Ponder B A J, Eeles R A. J Med Genet. 1994;31:458–461. doi: 10.1136/jmg.31.6.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 9.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 10.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 11.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers M P, Tonks N K. Am J Hum Genet. 1997;61:1234–1238. doi: 10.1086/301659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batty I H, Carter A N, Challis R A J, Hawthorne J N. In: Neurochemistry: A Practical Approach. 2nd Ed. Turner A J, Bachelard H S, editors. Oxford: IRL; 1997. pp. 229–268. [Google Scholar]
- 14.van der Kaay J, Batty I H, Cross D A, Watt P W, Downes C P. J Biol Chem. 1997;272:5477–5481. doi: 10.1074/jbc.272.9.5477. [DOI] [PubMed] [Google Scholar]
- 15.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S H, Kobayashi R, Graves P R, Piwnica-Worms H, Tonks N K. J Biol Chem. 1997;272:27281–27287. doi: 10.1074/jbc.272.43.27281. [DOI] [PubMed] [Google Scholar]
- 17.Tiganis T, Bennett A M, Ravichandran K S, Tonks N K. Mol Cell Biol. 1998;18:1622–1634. doi: 10.1128/mcb.18.3.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold A, Rohrschneider L R. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Charles C H, Lau L F, Tonks N K. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 20.Garton A J, Flint A J, Tonks N K. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bos J L. Trends Biochem Sci. 1995;20:441–442. doi: 10.1016/s0968-0004(00)89097-0. [DOI] [PubMed] [Google Scholar]
- 22.Marte B M, Downward J. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/s0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 23.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 24.Andjelkovic M, Alessi D R, Meier R, Fernandez A, Lamb N J, Frech M, Cron P, Cohen P, Lucocq J M, Hemmings B A. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 25.Risinger J I, Hayes A K, Berchuck A, Barrett J C. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 26.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman J G, Jen J, Isaacs W B, Bova G S, Sidransky D. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 27.Wang S I, Puc J, Li J, Bruce J N, Cairns P, Sidransky D, Parsons R. Cancer Res. 1997;57:4183–4186. [PubMed] [Google Scholar]
- 28.Tashiro H, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parsons R, Ellenson L H. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 29.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 31.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 32.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 33.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 30.Pap M, Cooper G M. J Biol Chem. 1997;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]