Abstract
The biotin-binding site of streptavidin was modified to alter its ligand-binding specificity. In natural streptavidin, the side chains of N23 and S27 make two of the three hydrogen bonds with the ureido oxygen of biotin. These two residues were mutated to severely weaken biotin binding while attempting to maintain the affinity for two biotin analogs, 2-iminobiotin and diaminobiotin. Redesigning of the biotin-binding site used the difference in local electrostatic charge distribution between biotin and these biotin analogs. Free energy calculations predicted that the introduction of a negative charge at the position of S27 plus the mutation N23A should disrupt two of the three hydrogen bonds between natural streptavidin and the ureido oxygen of biotin. In contrast, the imino hydrogen of 2-iminobiotin should form a hydrogen bond with the side chain of an acidic amino acid at position 27. This should reduce the biotin-binding affinity by approximately eight orders of magnitude, while leaving the affinities for these biotin analogs virtually unaffected. In good agreement with these predictions, a streptavidin mutant with the N23A and S27D substitutions binds 2-iminobiotin with an affinity (Ka) of 1 × 106 M−1, two orders of magnitude higher than that for biotin (1 × 104 M−1). In contrast, the binding affinity of this streptavidin mutant for diaminobiotin (2.7 × 104 M−1) was lower than predicted (2.9 × 105 M−1), suggesting the position of the diaminobiotin in the biotin-binding site was not accurately determined by modeling.
Altering the ligand-binding specificity of a protein by redesigning its ligand-binding site requires thorough understanding of the roles of individual amino acid residues in the ligand-binding site. A number of successful redesign studies have been reported (1–10). In most cases, redesign of the ligand (substrate)-binding site of a protein was carried out within a family of related proteins. For example, lactate dehydrogenase to malate dehydrogenase (3) and to phenylpyruvate dehydrogenase (4), aspartate aminotransferase to tyrosine aminotransferase (5), and Δ6-palmytoyl-ACP desaturase to Δ9-stearoyl-ACP desaturase (6). Similar efforts also have been made without specific template proteins (7, 10). However, the availability of homologous proteins with different specificities guided the redesign of ligand-binding sites.
In this work, we attempted to alter the ligand-binding specificity of streptavidin by redesigning its biotin-binding site. Streptavidin has a remarkably high affinity (Ka ≈ 1.8 × 1013 M−1) (11) for its natural ligand, biotin. The structure of its biotin-binding site is quite complex and irregular, highly optimized for the binding to biotin. The streptavidin-biotin complex is used frequently as a model system to study the thermodynamics and structural characteristics of high affinity binding between protein receptors and their ligands (12, 13). The interactions between streptavidin and biotin are well understood at the molecular level (14–19), and this would be of great help in redesigning the biotin-binding site. However, because there is no protein known that has a structure homologous to streptavidin but shows a tight binding affinity for other ligands, redesign of the biotin-binding site of streptavidin must be carried out essentially de novo, unlike the cases in which homologous proteins can be used as templates.
The ligand-binding specificity of streptavidin could be altered by using two strategies. One strategy is to reduce the binding affinity for biotin to a level lower than that for another ligand. The other is to increase the affinity for a weak-binding ligand to a level greater than that for biotin. The latter is quite difficult to achieve because of the extremely high binding affinity of natural streptavidin for biotin. Thus, this work used the first strategy. Structural and thermodynamic data available on streptavidin and its homologue, avidin (15–19), were used for redesign of the biotin-binding site in which the major structural motifs of the natural streptavidin-biotin complex are preserved. This was accomplished by changing amino acid residues deep in the biotin-binding pocket to reduce the biotin-binding affinity considerably without disturbing the binding affinities for two biotin analogs, 2-iminobiotin and diaminobiotin (cis-3, 4-diamino-2-tetrahydrothiophenylvaleric acid) (15).
MATERIALS AND METHODS
Modeling and Protein Design.
Modeling of streptavidin mutants was based on the known three-dimensional structure of the natural streptavidin-biotin complex taken from the Protein Data Bank (18). The coordinates were refined by performing 200 steps of energy minimization using the charmm potential (Version 19) (20) and harmonic constraints on the positions of non-hydrogen atoms (13). Only the polar hydrogens for the protein were included, but all hydrogens for the ligands were considered. After molecular geometries were optimized with AM1 using the mopac 6.0 program (Quantum Chemistry Program Exchange, Program 455), the atomic partial charges of the ligand molecules were determined to reproduce the electrostatic potential obtained by ab initio calculations at the HF/6–31G* level by using the gaussian 94 program package (Revision D.3; Gaussian, Pittsburgh, PA). Only the nonprotonated form of 2-iminobiotin was used in calculations because the protonated form yielded highly unfavorable van der Waals interactions with streptavidin that could not be removed by a reasonable number of minimization steps. This result is in agreement with the crystal structure of the complex between streptavidin and 2-iminobiotin, which contains only the nonprotonated form of the ligand even at pH 4.0 (21).
The search for the most probable position of each mutated amino acid residue was carried out by using the molecular modeling packages congen (22) and quanta (Molecular Simulations, San Diego, CA). The resulting structure was refined by minimizing the conformational energy of the complexes as in the case of the x-ray structure of the natural streptavidin-biotin complex.
Binding Free Energy Evaluation.
An empirical free energy evaluation model (13) was used to predict the ligand-binding characteristics of streptavidin mutants. The binding free energy ΔG is calculated by a mean field potential of the form ΔG = ΔE + ΔGd − TΔSc + ΔGconst, where ΔE, ΔGd, and ΔSc represent the internal energy change, the desolvation free energy, and the change in conformational entropy, respectively. The ΔGconst term consists of translational, rotational, vibrational, and cratic free energies, and it is considered constant. The energy change ΔE is the sum of electrostatic, van der Waals, and internal energy terms (ΔE = ΔEel + ΔEvdW + ΔEint), where the internal energy change ΔEint is the sum of bond length, bond angle, torsional, and improper energy terms (20). The desolvation term ΔGd is calculated by using an atomic solvation parameter model (23). It is assumed that entropy loss is restricted to side chains. The term TΔSc is calculated by using an empirical entropy scale. The addition of these phenomenological terms to the energy change ΔE results in a mean field free energy potential (13) that has been carefully tested against thermodynamic data (24).
Since no explicit modeling of the solvent is done, the calculation of the solute–solvent van der Waals interactions in ΔE (ΔEvdW) requires an approximation. The most straightforward strategy is to calculate these interactions as part of the desolvation free energy ΔGd. However, this approach has only limited accuracy because the solute–solute and solute–solvent van der Waals terms are based on very different models (25). Thus, we used an alternative approach, used frequently in binding free energy evaluation (13, 26), which assumes that the solute–solute and solute–solvent interfaces are equally well-packed, and hence the van der Waals interactions between a receptor and its ligand in the bound state are balanced by the interactions of the two molecules with the solvent in the free state. This approximation implies that the two contributions nearly cancel, and the van der Waals terms can be removed from both the energy change, ΔE, and the desolvation free energy, ΔGd. The disadvantage of this approach is its insensitivity to imperfect packing, i.e., to steric clashes or cavities. The effect of such packing errors was reduced by a procedure called van der Waals normalization, in which, before free energy evaluation, the complex between a mutant receptor and its ligand is minimized until the complex has the same solute–solute van der Waals energy as its parental receptor-ligand complex (25).
Construction of Expression Vectors.
Expression vectors were constructed by using a bacteriophage M13 mp18 derivative, mpSA-29, which codes for a minimum-sized core streptavidin consisting of amino acids 16 to 133 (27), as the starting material. The following two sets of mutations were introduced into the coding sequence for streptavidin by using an oligonucleotide-directed in vitro mutagenesis system (Amersham) (28): AAC (N23) to GCT (A) and TCG (S27) to GAC (D); and AAC (N23) to GCT (A) and TCG (S27) to GAA (E). The coding sequences containing the desired mutations were cloned into plasmid pET-3a under the control of the bacteriophage T7 Φ10 promoter (29). The resulting expression vectors pTSA-A23D27 and pTSA-A23E27 encode the streptavidin mutants Stv-A23D27 and Stv-A23E27, in which S27 is replaced by D and E, respectively, plus the N23A mutation.
Expression and Purification of Streptavidin Mutants.
Expression of each streptavidin mutant was carried out as previously described (30) by using the Escherichia coli strain BL21(DE3)(pLysE) (29) carrying an expression vector. Insoluble fractions were prepared from the cell lysate and solubilized in 7 M guanidine-HCl (pH 1.5) as described earlier (30). The resulting solution was dialyzed against 0.2 M ammonium acetate (pH 6.0), 0.02% Tween 20, and 0.02% NaN3 to renature streptavidin. Urea, a biotin analog that has a weak binding affinity for natural streptavidin (15), was included in the renaturation solution at a concentration of 10 mM for Stv-A23E27.
Stv-A23D27 and Stv-A23E27 were purified by affinity chromatography by using diaminobiotin-agarose (Sigma). A crude Stv-A23D27 or Stv-A23E27 solution, prepared above, was applied to a diaminobiotin-agarose column equilibrated with 0.5 M NaCl, 0.2 M ammonium acetate (pH 6.0), 0.02% Tween 20, and 0.02% NaN3. After unbound materials were removed by washing the column with the same solution containing 10 mM urea; bound proteins were eluted with 0.02% Tween 20, 10 mM urea and 50 mM Na2CO3 (pH 10.0) for Stv-A23D27, or with 0.02% Tween 20, 10 mM urea, and 50 mM 2-[N-cyclohexylamino]-ethanesulfonic acid (pH 9.3) for Stv-A23E27. Eluted proteins were dialyzed against water, lyophilized, resuspended in 150 mM NaCl and 50 mM Hepes (pH 7.5), and stored at 4°C.
Preparation of 2-Iminobiotin-[U-14C]Glycine.
NHS-2-iminobiotin (25 μg/μl; Sigma), dissolved in dimethylformamide, was mixed with [U-14C]glycine (98 or 104 mCi/mmol; Amersham) in 100 mM sodium borate (pH 8.0) (total reaction volume, 440 μl) at a molar ratio of NHS-2-iminobiotin to glycine of 50. The mixture was incubated at room temperature (≈ 23°C) for 1 hr to allow the NHS group to react with the primary amino group of glycine. The resulting mixture (200 μl) was adjusted to pH 2.5 by the addition of hydrochloric acid, applied to a reversed-phase chromatography column (RPC HR5/5, 0.5 cm × 5 cm; Pharmacia), which had been equilibrated with 100 mM potassium phosphate (pH 2.5) and eluted at room temperature with 100 mM potassium phosphate (pH 2.5) at a flow rate of 0.7 ml/min. Unreacted glycine and 2-iminobiotin-[U-14C]glycine were eluted at retention times 5–13 min and 18–24 min, respectively, followed by the elution of NHS-2-iminobiotin at ≈24 min. Fractions containing 2-iminobiotin-[U-14C]glycine (total volume, 4.2 ml) were pooled, reloaded onto the same column, which had been equilibrated with water, and eluted by using water as the eluant. The compound 2-iminobiotin-[U-14C]glycine was eluted at 20–26 min. Collected fractions were lyophilized, resuspended in 150 mM NaCl and 50 mM Hepes (pH 7.5), and stored at 4°C.
Determination of Ligand-Binding Affinities of Streptavidin Mutants.
The binding affinity of each streptavidin mutant for biotin was determined by using the following procedure. A streptavidin mutant (final subunit concentration; Stv-A23D27, 23.8 μM; Stv-A23E27 5.96 μM) was mixed with various concentrations of d-[8,9-3H]biotin (0.14 Ci/mmol) (5.96–59.6 μM) in 150 mM NaCl and 50 mM Hepes (pH 7.5) (total reaction volume, 200 μl). The mixtures were incubated at 25°C for 18–24 hr. Free, unbound biotin was separated from streptavidin-biotin complexes by using Ultrafree-MC centrifugal filter units (molecular mass cutoff, 10 kDa; Millipore). The amounts of biotin used in each mixture and in the filtrates were measured by ligand scintillation counting to determine the concentrations of total and free biotin, from which the biotin-binding affinity was estimated by using Scatchard plots (31).
The binding affinities between the streptavidin mutants and 2-iminobiotin-[U-14C]glycine were determined by the same procedure as above by using the following conditions: Final subunit concentration of each streptavidin mutant, 1.38 μM; 2-iminobiotin-[U-14C]glycine, 1.38–13.8 μM. The binding affinity of each streptavidin mutant for 2-iminobiotin was estimated by a competition assay in which 2-iminobiotin and 2-iminobiotin-[U-14C]glycine competed for binding to the streptavidin mutant. Each streptavidin mutant (final subunit concentration, 1.38 μM) was mixed with various concentrations of 2-iminobiotin (1.38–13.8 μM) and a constant concentration of 2-iminobiotin-[U-14C]glycine (9.66 μM) in 150 mM NaCl and 50 mM Hepes (pH 7.5) (total reaction volume, 200 μl). The mixtures were incubated at 25°C for 18–24 hr. The amounts of 2-iminobiotin-[U-14C]glycine used in each mixture and in the filtrates were measured by ligand scintillation counting to determine the concentrations of total and free 2-iminobiotin-[U-14C]glycine, from which the 2-iminobiotin-binding affinity was estimated by using Scatchard plots (31).
The affinity constants between streptavidin mutants and diaminobiotin also were estimated by a competition assay between 2-iminobiotin-[U-14C]glycine and diaminobiotin. Each streptavidin mutant (final subunit concentration, 2.3 μM) was mixed with various concentrations of diaminobiotin (2.3–460 μM) and a constant concentration of 2-iminobiotin-[U-14C]glycine (9.66 μM) in 150 mM NaCl and 50 mM Hepes (pH 7.5) (total reaction volume, 200 μl). The mixtures were incubated at 25°C for 18–24 hr. The diaminobiotin-binding affinity was estimated by using the same procedure used for the determination of the binding affinities for 2-iminobiotin, as described above.
Other Methods.
The concentration of each streptavidin mutant was determined from A at 280 nm by using the extinction coefficient for a minimum-sized core streptavidin E0.1% 280 nm = 3.55 (27). SDS/PAGE was carried out by using 15% polyacrylamide gels (32). Proteins were stained with Coomassie Brilliant Blue (Sigma).
RESULTS AND DISCUSSION
Design of Streptavidin Mutants with Altered Ligand-Binding Specificity.
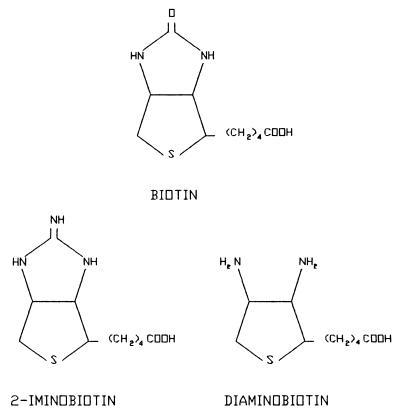
Mutations in the biotin-binding site of streptavidin were designed, by using molecular modeling and binding free energy calculations, to reduce the binding affinity for biotin considerably while maintaining the binding affinities for the biotin analogs 2-iminobiotin and diaminobiotin. These biotin analogs were selected because of their substantial structural similarity to biotin, so that the major structural motifs of the natural streptavidin-biotin complex can be preserved. However, these biotin analogs have electrostatic properties different from those of biotin, particularly at the ureido group of biotin, which is replaced with a guanidino group in 2-iminobiotin and removed in diaminobiotin (Fig. 1). Thus, amino acid mutations introduced into streptavidin were designed that change the local electrostatic charge distribution in the region of the biotin-binding site, which is occupied by the ureido oxygen of biotin that forms hydrogen bonds with the side chains of N23, S27, and Y43 of streptavidin.
Figure 1.
Schematic chemical formulae of biotin and its analogs.
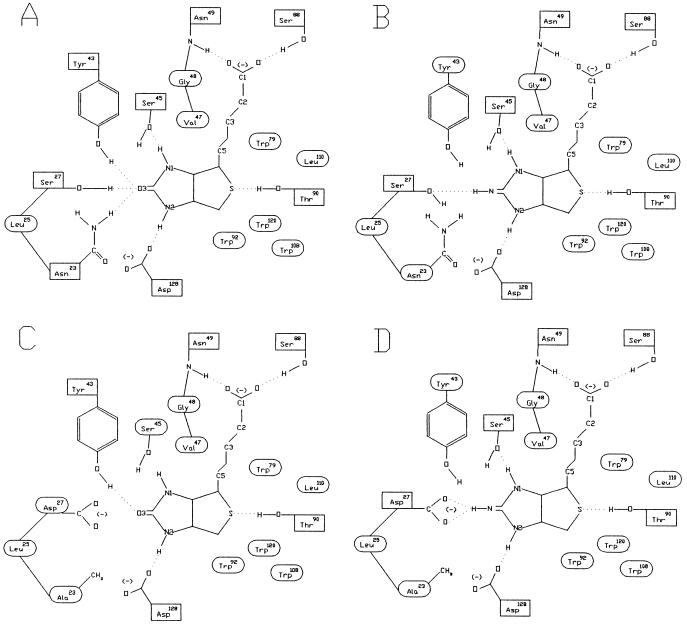
The amino acid residue in the biotin-binding site with the largest electrostatic contribution to the binding free energy is S27 (19). Because the hydroxyl group of S27 is only 1.65 Å away from the ureido oxygen of biotin, which has a partial negative charge, placing an acidic amino acid residue at position 27 is expected to cause electrostatic repulsion between the carboxyl group of the acidic amino acid and the ureido oxygen, thus lowering the biotin-binding affinity. In fact, an avidin with nitrated Y33, equivalent to Y43 in streptavidin, shows pH-dependent biotin-binding affinity, and it has a very low affinity for biotin at high pH due to the ionization of the nitrated Y33 (33). A similar concept also was used successfully to alter the substrate specificity of subtilisin, in which electrostatic interactions between charged substrates and complementary charged amino acid residues in the substrate-binding cleft were engineered (7). Two streptavidin mutants were initially conceived in which S27 is substituted with D or E. Modeling of the resulting streptavidin mutants complexed with biotin showed that the side chains of both Y43 and N23 of these mutants should still make hydrogen bonds with the ureido oxygen of biotin. Thus, the reduction in biotin-binding affinity may not be large enough to shift the ligand-binding specificity toward the biotin analogs. An additional substitution of N23 for A should prevent the formation of one of the two hydrogen bonds with the ureido oxygen, resulting in further reduction of biotin-binding affinity (Fig. 2).
Figure 2.
Schematic representations of the interactions between streptavidins and biotin analogs. (A) Natural streptavidin and biotin. (B) Natural streptavidin and 2-iminobiotin. (C) Stv-A23D27 and biotin. (D) Stv-A23D27 and 2-iminobiotin. Amino acid residues surrounded by a rectangle and an ellipse refer to those involved in hydrogen bonding and van der Waals interactions, respectively, with the ligand. These notations are based on Fig. 3 in ref. 12.
Two streptavidin mutants, Stv-A23D27 and Stv-A23E27 each of which carries the S27D or S27E mutation, respectively, plus the N23A substitution, were analyzed by using an empirical free energy evaluation model (13) (Table 1). Each free energy component for a streptavidin mutant complexed with biotin or its analog was calculated and given as the differential from that for the natural streptavidin-biotin complex. Restricting consideration to differential free energy eliminates the need for determining the terms that depend on translational, rotational, vibrational, and cratic free energies (see Materials and Methods), thus facilitating this analysis.
Table 1.
Binding free energies and binding constants of streptavidin mutants complexed with biotin and its analogs
| Mutations | Ligand | Free energy terms, kcal/mol
|
Ka (M−1)
|
|||||
|---|---|---|---|---|---|---|---|---|
| ΔΔEint | ΔΔEel | ΔΔGd | −TΔΔSc | ΔG* | Predicted† | Measured | ||
| None | Biotin | — | — | — | — | −18.3 | 1.8 × 1013 | 2.5 × 1013 |
| 2-Iminobiotin | 5.0 | 1.8 | 1.0 | 0.0 | −10.5 | 4.0 × 107 | 8.0 × 106 | |
| Diaminobiotin | 2.0 | 4.2 | 3.4 | 0.8 | −7.9 | 4.4 × 105 | nd | |
| A23D27 | Biotin | 3.3 | 10.3 | −0.1 | −2.0 | −6.8 | 8.4 × 104 | 1.4 × 104 |
| 2-Iminobiotin | 2.3 | 6.4 | 1.1 | −2.0 | −10.5 | 4.0 × 107 | 1.0 × 106 | |
| Diaminobiotin | 2.2 | 6.2 | 3.5 | −1.2 | −7.6 | 3.2 × 105 | 2.7 × 104 | |
| A23E27 | Biotin | 4.0 | 7.9 | −0.4 | −1.5 | −8.3 | 1.0 × 106 | 1.4 × 105 |
| 2-Iminobiotin | 5.5 | 5.8 | 0.7 | −1.5 | −7.8 | 4.4 × 105 | 1.2 × 105 | |
| Diaminobiotin | 5.3 | 3.6 | 3.3 | −0.6 | −6.7 | 7.1 × 104 | <5.0 × 103 | |
ΔG = ΔGstv-biotin + ΔΔG, where ΔG of the streptavidin-biotin complex (ΔGstv-biotin) is −18.3 kcal/mol (11) and ΔΔG (=ΔΔEint + ΔΔEel + ΔΔGd − TΔΔSc) is the change in the binding free energy relative to ΔGstv-biotin.
The predicted association constant Ka is calculated by using the equation Ka = exp (−ΔG/RT), where R is the universal gas constant (1.98719 cal/K⋅mol) and T is the temperature (298 K).
nd, not determined.
In natural streptavidin, replacing biotin with 2-iminobiotin or diaminobiotin should introduce some structural strains into the complex as shown by positive ΔΔEint values. This also is seen with Stv-A23D27 and Stv-A23E27. Particularly, Stv-A23E27 has the highest ΔΔEint values because the longer side chain of E27 would be accommodated only with local structural rearrangements. The electrostatic contribution to the binding free energy is most favorable in the streptavidin-biotin complex as shown by positive ΔΔEel values for other complexes. The electrostatic energy is highest for the two streptavidin mutants complexed with biotin (10.31 kcal/mol for Stv-A23D27 and 7.87 kcal/mol for Stv-A23E27), indicating that the electrostatic attraction between these mutants and biotin should be very weak. These mutations have two different effects on the differential desolvation free energy (ΔΔGd). When complexed with biotin, these streptavidin mutants exhibit slightly more favorable desolvation than natural streptavidin due, presumably, to the N23A mutation. However, the ΔΔGd becomes positive when complexed with the two biotin analogs because of their smaller contact surfaces than that between natural streptavidin and biotin. These mutations should decrease the side chain conformation entropy in the unbound state, and thus they should reduce the loss of conformational entropy upon binding (positive ΔΔSc), favorably contributing to the binding free energy.
The A23D27 and A23E27 mutations should reduce the binding affinity for biotin substantially, as seen with the Ka values estimated from the free energies. In these mutations, the N23A mutation on its own should reduce the binding affinity for biotin by two orders of magnitude but only one order of magnitude for the two biotin analogs (data not shown). The estimated biotin-binding affinity of Stv-A23D27 is 8.4 × 104 M−1; this is almost eight orders of magnitude smaller than that of natural streptavidin. In contrast, these mutations should have minimal effects on the binding affinity for 2-iminobiotin and diaminobiotin. Comparing these two streptavidin mutants, Stv-A23D27 should have better binding properties because of its lower affinity for biotin and higher affinity for 2-iminobiotin.
Expression and Purification of Streptavidin Mutants.
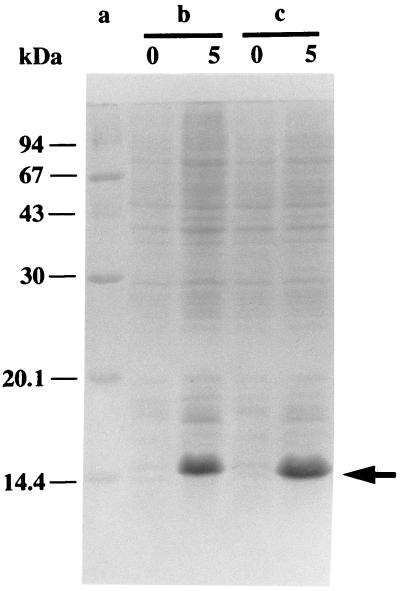
Stv-A23D27 and Stv-A23E27 were expressed efficiently in E. coli (Fig. 3) by using the T7 expression system (29). Attempts were made to purify these streptavidin mutants by using biotin affinity chromatography. However, their biotin-binding affinities were so weak that only a very small fraction of the proteins was bound by immobilized biotin. Similarly, 2-iminobiotin affinity chromatography also failed to purify these streptavidin mutants under the conditions used commonly for the purification of streptavidin (34). To determine the conditions under which these mutants can be bound to 2-iminobiotin and diaminobiotin, each of these proteins was incubated for 10 min with 2-iminobiotin-agarose or diaminobiotin-agarose in a pH range from 2 to 11. After unbound materials were removed, bound proteins were analyzed by SDS/PAGE. Stv-A23D27 and Stv-A23E27 bound to 2-iminobiotin-agarose in a pH range from 7 to 11 and 5 to 11, respectively, with the maximum binding at pH 8. Stv-A23D27 and Stv-A23E27 bound to diaminobiotin-agarose in a pH range from 4 to 9 and from 5 to 8, respectively, with the maximum binding at pH 6.
Figure 3.
Expression of streptavidin mutants Stv-A23D27 and Stv-A23E27. Lane a, molecular mass standard proteins; b, BL21(DE3)(pLysE)(pTSA-A23D27); and c, BL21(DE3)(pLysE)(pTSA-A23E27). Total cell protein at time 0 (at the time of induction; 133 μl of culture) and 5 hr after induction (67 μl of culture) was analyzed by SDS/PAGE. Proteins were stained with Coomassie Brilliant Blue. An arrow indicates the position where the streptavidin mutants migrate.
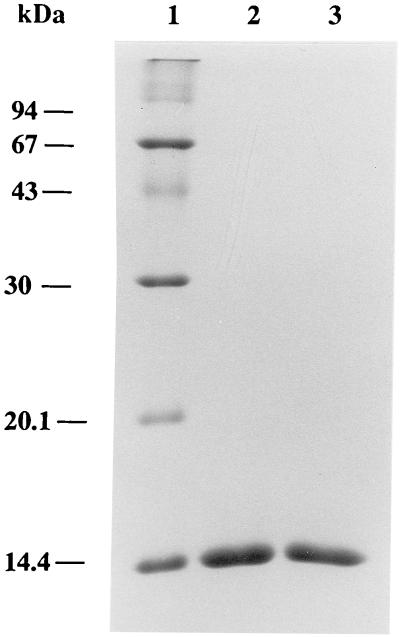
To determine the pH at which each streptavidin mutant is released from 2-iminobiotin-agarose and diaminobiotin-agarose, Stv-A23D27 and Stv-A23E27, bound to 2-iminobiotin-agarose, were incubated for 10 min in a pH range from 1.5 to 4. Similarly, Stv-A23D27 and Stv-A23E27, bound to diaminobiotin-agarose, were incubated for 10 min in a pH range from 2 to 5 and from 8 to 11. Then, the mixtures were centrifuged, and the supernatants were analyzed by SDS/PAGE. Stv-A23D27 and Stv-A23E27 were released from diaminobiotin-agarose at pHs 10 and 9.3, respectively. In contrast, Stv-A23D27 and Stv-A23E27 were not released from 2-iminobiotin agarose under the pH conditions tested. Therefore, both of these proteins were purified by using diaminobiotin-agarose: Stv-A23D27 was bound at pH 6.0 and eluted at pH 10.0, and Stv-A23D27 was bound at pH 6.0 and eluted at pH 9.3. Under these conditions, these streptavidin mutants were purified to homogeneity, as judged by SDS/PAGE (Fig. 4). The final yields of purified Stv-A23D27 and Stv-A23E27 were ≈900 μg and 150 μg, respectively, from 100 ml of culture. SDS/PAGE analysis showed that each of these proteins maintains a tetrameric structure in the presence of SDS, as shown previously for natural core streptavidin (35).
Figure 4.
SDS/PAGE analysis of purified streptavidin mutants Stv-A23D27 and Stv-A23E27. Lane 1, molecular mass standard proteins; 2, Stv-A23D27; and 3, Stv-A23E27. Proteins were stained with Coomassie Brilliant Blue.
Binding Affinities of Streptavidin Mutants for Biotin and Its Analogs.
The biotin-binding affinities (Ka) of Stv-A23D27 and Stv-A23E27 were estimated at 1.4 × 104 M−1 and 1.4 × 105 M−1, respectively. This indicates that the amino acid mutations introduced into the biotin-binding site were sufficient to destabilize the strong biotin binding of streptavidin, leading to a reduction in binding affinity by at least eight orders of magnitude. This is in excellent agreement with molecular modeling predictions (Table 1) that the mutations in Stv-A23D27 and Stv-A23E27 would weaken the biotin-binding affinity by seven and eight orders of magnitude, respectively.
The binding affinities of these streptavidin mutants for 2-iminobiotin were determined by competition assays using 2-iminobiotin-[U-14C]glycine. First, the Ka of both Stv-A23D27 and Stv-A23E27 for 2-iminobiotin-[U-14C]glycine was estimated at 3.3 × 105 M−1. By competition assays in which 2-iminobiotin-[U-14C]glycine and 2-iminobiotin compete for binding to streptavidin, the binding affinities of Stv-A23D27 and Stv-A23E27 for 2-iminobiotin were estimated at 1.0 × 106 M−1 and 1.2 × 105 M−1, respectively. The predicted hydrogen bonding in the complexes of Stv-A23D27 with biotin and 2-iminobiotin suggests that the difference in binding affinity for these ligands can be primarily attributed to the hydrogen bond between the carboxyl oxygen of D27 and the imino hydrogen of 2-iminobiotin that should not exist in the complex with biotin (Fig. 2 C and D).
The binding affinities of the streptavidin mutants for diaminobiotin were estimated by competition assays, in which 2-iminobiotin-[U-14C]glycine and diaminobiotin compete for binding. The Ka between Stv-A23D27 and diaminobiotin was estimated at 3.0 × 104 M−1. It was difficult to determine accurately the binding affinity of Stv-A23E27 for diaminobiotin due to the significant difference in affinity of Stv-A23E27 for 2-iminobiotin-[U-14C]glycine and diaminobiotin. An upper limit for the Ka between Stv-A23D27 and diaminobiotin was estimated at 5 × 103 M−1.
According to previous calculations on protease-protein inhibitor complexes, the uncertainty in calculated binding free energies is approximately ±1.5 kcal/mol (10). Taking this into account, there is good agreement between ligand-binding affinities predicted and those experimentally determined. The prediction had reduced accuracy in the determination of diaminobiotin-binding affinities; this is probably due to a shift in the position of diaminobiotin in the biotin-binding site that has not been fully accounted for by modeling. Compared with the natural streptavidin-biotin pair, each streptavidin mutant-biotin analog complex is somewhat strained structurally, as shown by positive ΔΔEint.
CONCLUSIONS
The ligand-binding specificity of streptavidin has been altered from its natural ligand biotin to its analog, 2-iminobiotin, by decreasing the binding affinity for biotin to a level lower than that for 2-iminobiotin. Molecular modeling and free energy calculations were used to redesign the biotin-binding site. By introducing only two amino acid mutations into the biotin-binding site, the biotin-binding affinity was reduced by eight orders of magnitude without significant effects on the binding affinity for 2-iminobiotin. This resulted in the production of a streptavidin mutant that has a greater binding specificity to 2-iminobiotin than to biotin, with the difference in binding affinity constant of two orders of magnitude. A unique aspect of this work is that redesign of the biotin-binding site of streptavidin was carried out essentially de novo because there is no protein known that has a structure similar to streptavidin but shows greater binding specificity to other ligands than to biotin. This implies that the ligand-binding characteristics of a receptor protein could be tailored by redesigning its ligand-binding site even if the binding site is highly optimized for its natural ligand. This streptavidin mutant and its derivatives could be very useful in biotechnological applications. For example, they could provide an assay system with specific binding pairs, in addition to natural streptavidin-biotin, so that multiple targets could be analyzed in single assays. They could also be useful for in vivo targeting applications, in which the presence of endogenous biotin currently reduces their efficacy (36).
Acknowledgments
We thank Dr. Mark Murcko for his help in calculating the partial charges for 2-iminobiotin and diaminobiotin. This work was supported by Grant DBI-9630188 from the National Science Foundation and Grants DE-FG02-93ER61656 and DE-FG02-96ER62263 from the Department of Energy.
References
- 1.Wells J A, Cunningham B C, Graycar T P, Estell D A. Proc Natl Acad Sci USA. 1987;84:5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahmhsen L, Tom J, Burnier J, Butcher K A, Kosiakoff A, Wells J A. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- 3.Wilks H M, Hart K W, Feeney R, Dunn C R, Muirhead H, Chia W N, Barstow D A, Atkinson T, Clarke A R, Holbrook J J. Science. 1988;242:1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- 4.Dunn C R, Wilks H M, Halsall D J, Atkinson T, Clarke A R, Muirhead H, Holbrook J J. Phil Trans Royal Soc London B. 1991;332:177–184. doi: 10.1098/rstb.1991.0047. [DOI] [PubMed] [Google Scholar]
- 5.Onuffer J J, Kirsch J F. Protein Sci. 1995;4:1750–1757. doi: 10.1002/pro.5560040910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoon E B, Lindquist Y, Schneider G, Shanklin J. Proc Natl Acad Sci USA. 1997;94:4872–4877. doi: 10.1073/pnas.94.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells J A, Powers D B, Bott R R, Graycar T P, Estell D A. Proc Natl Acad Sci USA. 1987;84:1219–1223. doi: 10.1073/pnas.84.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craik C S, Roczniak S, Sprang S, Fletterick R, Rutter W. Science. 1985;228:291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- 9.Perona J J, Tsu C A, McGrath M E, Craik C S, Fletterick R J. J Mol Biol. 1993;230:934–949. doi: 10.1006/jmbi.1993.1211. [DOI] [PubMed] [Google Scholar]
- 10.Caputo A, James M N G, Powers J C, Hudig D, Bleackley R C. Nat Struct Biol. 1994;1:364–367. doi: 10.1038/nsb0694-364. [DOI] [PubMed] [Google Scholar]
- 11.Weber P C, Wendoloski J J, Pantoliano M W, Salemme F R. J Am Chem Soc. 1992;114:3197–3200. [Google Scholar]
- 12.Miyamoto S, Kollman P A. Proteins Struct Funct Genet. 1993;16:226–245. doi: 10.1002/prot.340160303. [DOI] [PubMed] [Google Scholar]
- 13.Vajda S, Weng Z, Rosenfeld R, DeLisi C. Biochemistry. 1994;33:13977–13988. doi: 10.1021/bi00251a004. [DOI] [PubMed] [Google Scholar]
- 14.Chaiet L, Wolf F J. Arch Biochem Biophys. 1964;106:1–5. doi: 10.1016/0003-9861(64)90150-x. [DOI] [PubMed] [Google Scholar]
- 15.Green N M. Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 16.Green N M. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 17.Weber P C, Ohlendorf D H, Wendoloski J J, Salemme F R. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson W A, Pahler A, Smith J A, Satow Y, Merrit E A, Phizackerly R P. Proc Natl Acad Sci USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sano T, Vajda S, Reznik G O, Smith C L, Cantor C R. Ann NY Acad Sci. 1996;799:383–389. doi: 10.1111/j.1749-6632.1996.tb33229.x. [DOI] [PubMed] [Google Scholar]
- 20.Brooks B R, Bruccoleri R B, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 21.Athappilly F K, Hendrickson W A. Protein Sci. 1997;6:1338–1342. doi: 10.1002/pro.5560060623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruccoleri R E. Mol Simul. 1993;10:151–174. [Google Scholar]
- 23.Eisenberg D, McLachlan A D. Nature (London) 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 24.Weng Z, DeLisi C, Vajda S. Protein Sci. 1997;9:1976–1984. doi: 10.1002/pro.5560060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janardhan A, Vajda S. Protein Sci. 1998;7:1772–1780. doi: 10.1002/pro.5560070812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novotny J, Bruccoleri R E, Saul F A. Biochemistry. 1989;28:4735–4749. doi: 10.1021/bi00437a034. [DOI] [PubMed] [Google Scholar]
- 27.Sano T, Pandori M W, Chen X, Smith C L, Cantor C R. J Biol Chem. 1995;270:28204–28209. doi: 10.1074/jbc.270.47.28204. [DOI] [PubMed] [Google Scholar]
- 28.Sayers J R, Schmidt W, Eckstein F. Nucleic Acids Res. 1988;16:791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Sano T, Cantor C R. Proc Natl Acad Sci USA. 1990;87:142–146. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantor C R, Schimmel P R. Biophysical Chemistry, Part III: The Behavior of Biological Macromolecules. San Francisco: Freeman; 1980. pp. 849–886. [Google Scholar]
- 32.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Morag E, Bayer E A, Wilchek M. Biochem J. 1996;316:193–199. doi: 10.1042/bj3160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann K, Wood S W, Brinton C C, Montibeller J A, Finn F M. Proc Natl Acad Sci USA. 1980;77:4666–4668. doi: 10.1073/pnas.77.8.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sano T, Pandori M W, Chen X, Smith C L, Cantor C R. In: Advances in Biomagnetic Separation. Uhlén M, Hornes E, Olsvik O, editors. Natick, MA: Eaton; 1994. pp. 21–29. [Google Scholar]
- 36.Rusckowski M, Fogarasi M, Virzi F, Hnatowich D J. Nucl Med Commun. 1995;16:38–46. doi: 10.1097/00006231-199501000-00009. [DOI] [PubMed] [Google Scholar]