Abstract
A C-terminal segment of the yeast activator Gal4 manifests two functions: When tethered to DNA, it elicits gene activation, and it binds the inhibitor Gal80. Here we examine the effects on these two functions of cysteine and proline substitutions. We find that, although certain cysteine substitutions diminish interaction with Gal80, those substitutions have little effect on the activating function in vivo and interaction with TATA box-binding protein (TBP) in vitro. Proline substitutions introduced near residues critical for Gal80 binding abolish that interaction but once again have no effect on the activating function. Crosslinking experiments show that a defined position in the activating peptide is in close proximity to TBP and Gal80 in the two separate reactions and show that binding of the inhibitor blocks binding to TBP. Thus, the same stretch of amino acids are involved in two quite different protein–protein interactions: binding to Gal80, which depends on a precise sequence and the formation of a defined secondary structure, or interactions with the transcriptional machinery in vivo, which are not impaired by perturbations of either sequence or structure.
Activating regions are defined as peptides that confer on a DNA binding domain the ability to activate transcription when bound near a gene (1). Several lines of evidence show that activating regions touch targets in the transcriptional machinery (2–10). In several instances, such interactions, provided the activating region is tethered to DNA, help bring the machinery to a nearby promoter, where transcription then initiates (1). The precise compositional and sequence requirements to constitute an activating region are not known. Activating regions evidently are not structured in the absence of their interacting partners (11, 12). Recent experiments suggest, however, that various activating regions are induced to fold into amphipathic alpha helices when bound to their interacting partners (13–17).
The yeast activator Gal4, a protein of 881 amino acids, activates genes whose products are required for the metabolism of galactose (18). When cells are grown under noninducing conditions—in the presence of raffinose, for example—Gal4 binds DNA but is prevented from activating transcription by the inhibitor Gal80. Growth of cells in galactose frees Gal4 from the inhibitory effects of Gal80 (18, 19).
A series of experiments, taken together, indicate that Gal4 residues 855–870, in the context of a slightly larger peptide, play an especially important role in two functions: transcriptional activation and sensitivity to Gal80. Thus, Ma and Ptashne showed that a region comprising the carboxyl 100 residues of Gal4 when fused to a DNA binding domain functions as a powerful activator and is fully sensitive to Gal80 (20). In a more recent study, it was observed that both of these properties are retained by the peptide comprising the 42 carboxyl terminal residues of Gal4 (residues 840–881). Activation by this DNA-tethered fragment also was inhibited by Gal80, and in vitro experiments confirmed the interaction between the two proteins (21). Carboxyl deletions of this 41-aa peptide extending beyond residue 869 destroyed interaction with Gal80, and further deletions beyond residue 854 severely impaired the activation function (21).
The studies described above that emphasize the dual role of residues 855–870 of Gal4 are consistent with studies of Johnston and colleagues (22, 23, 24). They showed that, in the context of intact Gal4, carboxyl deletions that extended up to residue 870, but not one that extended to residue 868, retained the ability to activate and to interact with Gal80 (24, 25). Johnston and colleagues also reported that this region of Gal4 (residues 840–874) was unstructured in solution at physiological pH but formed a β-sheet at pH 5.9 (26). These findings led to the suggestion that the activating region of Gal4 is unstructured in the absence of an interacting partner but that formation of the β-hairpin is required for both interaction with Gal80 and for the activation function (25).
Here, we systematically mutagenize residues 855–870 in the context of Gal4 (1–100) + (840–881) and study the activating function, sensitivity to Gal80, and affinity for the TATA box-binding protein (TBP) of each mutant. We begin with a cysteine scanning mutagenesis of these residues and then introduce proline substitutions at key positions. Alanine scanning mutagenesis often has been used to identify amino acid side chains important in protein–protein interactions (27). We chose cysteine as the substituting residue because it is accommodated readily in α-helices and β-sheets (28–32) and because it is found both on the surfaces and the interiors of proteins, and, like alanine, it has a small side chain. Moreover, the sulfhydryl (thiol) group of cysteine is specifically modifiable by chemical crosslinkers, a property we exploit in certain experiments here. In contrast, proline residues seriously perturb secondary structure (28–35), and we use this property to test the requirement of secondary structure for function.
MATERIALS & METHODS
Strains and Plasmids.
The experiments were performed in the yeast strain JPY9 bearing an integrated reporter pRJR227 (21). Gal4 derivatives were expressed by a gal4 promoter from an ARS1-CEN4 plasmid (21). Cysteine and Proline mutants in the activating region were constructed by site-directed PCR mutagenesis (36). The level of expression of each mutant was determined by immunoblot analysis with anti-Gal4 polyclonal antibodies (Santa Cruz Biotechnology) and with electrophoretic mobility shift assays from yeast extracts (21). The bacterial expression plasmids were constructed by cloning the cassette coding for the entire Gal4 derivative into pET16B (Novagen).
β-Galactosidase Assay.
The cells were grown on the SC media, under histidine selection, either in the presence of 2% raffinose or 2% galactose. The β-galactosidase activity was measured as described by Rose et al. (37), and the units of activity were calculated as nmol/min/mg protein (as in ref. 21). The SD for all assays was <20%. All assays were done in quadruplicates at least five independent times.
Protein Purification.
Gal4 derivatives were over-expressed in BL21(DE3) pLysS strain, were purified first by SP-Sepharose (as in ref. 38), and were purified further by gel filtration on a Superdex G-75 column (Pharmacia). Hexa-histidine-tagged yTBP and Gal80 were purified by using Ni(II)-NTA-agarose beads, as described (7, 21). After the purification, the proteins were subjected to gel filtration on a superdex G-200 column (Pharmacia). Glutathione S-transferase (GST)-yTBP was purified according to the published procedure (39).
GST Pull-Down.
35S-labeled Gal4 derivatives were made by using the coupled transcription and translation (TnT) kit from Promega. Pull-down experiments were done by incubating 5 μl of labeled Gal4 derivative with 1 μg of immobilized GST-TBP on glutathione-Sepharose (20 μl) beads (40). The incubations and washes were performed by using published conditions (22). Bound proteins were resolved on 12% Tricine-SDS/PAGE (41) and were visualized by autoradiography and on a Fuji BAS2000 phosphorimager.
Surface Plasmon Resonance Analysis.
Sensor chips with an immobilized double-stranded DNA carrying two consensus Gal4 binding sites were prepared as described in refs. 9 and 21. Protein–protein interactions were measured on BIAcore 2000 (Pharmacia) in buffer A (20 mM Hepes, pH 8.0/150 mM NaCl/0.1 mM EDTA/20 μM ZnSO4) at 25°C at a flow rate of 15 μl/min. Typically, 30 μl of 100 nM Gal4 derivative was passed over the immobilized DNA, which resulted in saturated binding to the DNA binding sites and gave the first increase in resonance response units (RU). Interactions between this Gal4–DNA complex and TBP were determined by flowing 30 μl of 150 nM solution of a sample protein over the preformed Gal4–DNA complex. Any consequent increase in RU was interpreted as binding of TBP to the Gal4–DNA complex. Control experiments with nonspecific DNA and with immobilized streptavidin alone (data not shown) were conducted simultaneously to determine the background level of surface binding of proteins and the bulk increase in RUs caused by the difference in the refractive index of the protein storage buffer. Chips were regenerated between runs by washing with 30 μl of 0.3% SDS solution in HBS (10 mM Hepes, pH 7.5/150 mM NaCl). The ratio of TBP retained by each Gal4 derivative was determined after subtraction of the background raise in RU values in the parallel channels, which do not bear Gal4 binding sites (see ref. 9 for details).
Electrophoretic Mobility Super-Shift Assays.
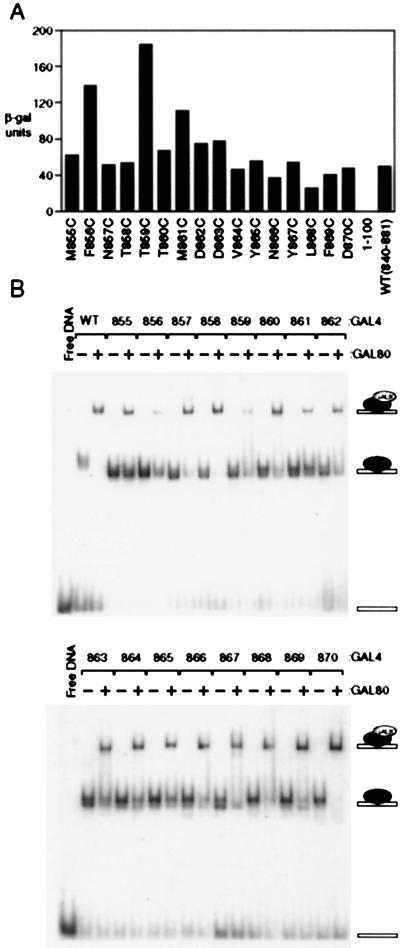
Each cysteine derivative (10 nM) was incubated with a single labeled Gal4 site (100 pM) at room temperature in buffer B (20 mM Hepes, pH 8.0/150 mM NaCl/0.1 mM EDTA/20 μM ZnSO4/100 ng poly dAdT/10% glycerol/3 mM DTT) in the presence or absence of 15 nM Gal80. Under these conditions, Gal4 derivatives are saturating for the DNA binding site (42), but Gal80 is at the threshold of saturated binding (21). The complexes were resolved on a native gel and were visualized by the phosphorimaging; under these conditions, the effects of cysteine mutants on binding of Gal80 are most apparent. At 4-fold higher concentrations of Gal80 (60 nM), all cysteine mutants are saturated, suggesting that Gal80 must have at least a 4-fold decrease in affinity for two most deleterious cysteine mutations (F856C and T859C).
Immunoblot Analysis.
Cells were grown to mid-log and were pelleted. The pellet was resuspended in Buffer A and was mixed with an equal volume of glass beads (Sigma). The cells were broken by bead beating immediately after adding 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, and 0.5 mM leupeptin. The supernatant was added to SDS sample buffer and was resolved on 12% Tricine-SDS-polyacrylamide gel. Immunoblotting was performed by standard methods (43). Polyclonal rabbit anti-Gal80 antibodies were used to detect the levels of endogenous Gal80. Gal4 fusions from yeast extract were detected with an anti-Gal4 polyclonal antibody from Santa Cruz Biotechnology. In addition to detecting our 16-kDa Gal4 derivatives, this antibody also cross-reacts with a 90-kDa protein (not endogenous Gal4) in yeast extracts. Horseradish peroxidase conjugated secondary goat anti-rabbit antibodies were used to visualize the proteins by an enhanced chemiluminescence (ECL) kit from Amersham.
Label-Transfer Photoaffinity Crosslinking.
Gal4 (1–100) + (840–881) with an Asn to Cys substitution at residue 857 was reduced with 10 mM DTT for 1 hr at room temperature in buffer A. The reduced protein was incubated for 90 min at room temperature with 125I-APDP (N-[4-(p-azidosalicylamido)butyl]-3′[2′-pyridyldithio]propionamide) as described in ref. 9. Purified radiolabeled Gal4 derivative (0.27 μM) was incubated, in the dark, with TBP or Gal80 (1 μM) in 30 μl of buffer A on ice for 1 hr. Photocrosslinking was activated with long range UV light (320 nm) for 5 min. The reaction was stopped and quenched with 20 mM DTT. This step reduces the disulfide bond between 125I-APDP and Gal4, transferring the radiolabel onto the crosslinked protein (see ref. 9). The proteins were resolved on a 4–20% Tricine-SDS/PAGE and were visualized by using a Fuji BAS2000 phosphorimager.
RESULTS
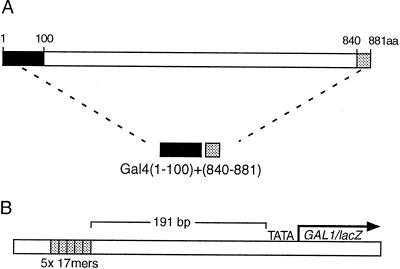
The protein Gal4 (1–100) + (840–881) comprises a DNA binding domain (residues 1–100) attached to a portion of the principal activating region of Gal4, which is found at the carboxyl terminus of the protein (Fig. 1A). This shortened Gal4 derivative activates transcription about half as well as does full length Gal4 as assayed on a reporter bearing five Gal4 binding sites (21), and it, like the full length protein, is inhibited by Gal80 in cells grown in the absence of galactose. In the following studies, we first analyze the effect of sequentially substituting each of the residues spanning the region (855–870) with cysteine residues, and we then analyze various mutants bearing proline substitutions in this region. The following results describe the abilities of these mutants to activate transcription and to interact with TBP and with Gal80.
Figure 1.
Activator and a reporter. (A) A Gal4 derivative comprising the N-terminal, 100-residue DNA binding domain (dark box) fused to the C-terminal, 42-residue activating region (the shaded box). (B) A chromosomally integrated reporter bearing five consensus 17-bp Gal4 binding sites 191 bp upstream of the gal1 promoter, which is fused to the LacZ reporter gene.
Cysteine Substitutions
The Activation Function.
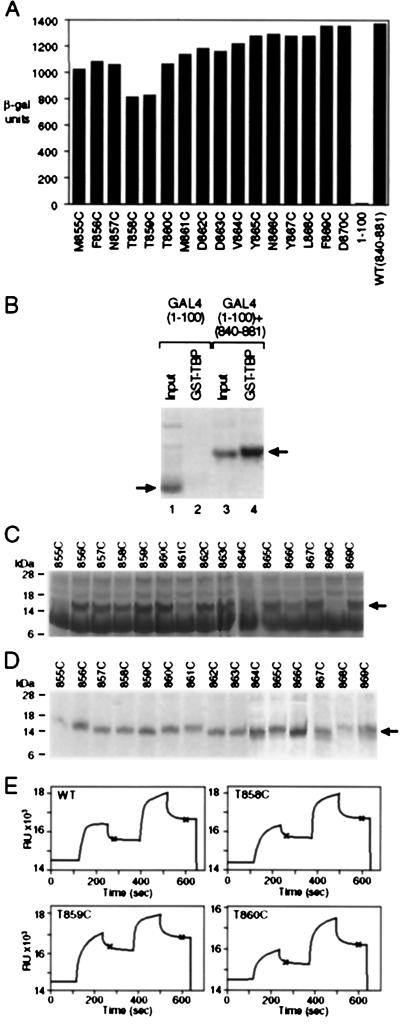
As shown in Fig. 2A, none of the cysteine mutants were severely impaired for activation as assayed in yeast grown under inducing conditions (i.e., in galactose). The least active mutants (T858C and T859C) were ≈60% as active as the parent. In this experiment, activation was measured by using a chromosomally integrated reporter bearing five Gal4 binding sites upstream of a gal1 promoter as shown in Fig. 1B. Each of the derivatives was introduced on a single copy plasmid into cells deleted for endogenous Gal4, and each was expressed from a gal4 promoter. Western blot analysis and electrophoretic mobility-shift assays from yeast extracts confirmed that, as expected, each derivative was expressed at the low levels characteristic of endogenous Gal4 levels in wild-type cells (data not shown).
Figure 2.
The effect of substituting cysteine for wild-type residues on activation and on TBP binding. (A) Reporter gene expression elicited by each of the cysteine substitution mutants as assayed in Gal80+ cells grown in galactose. (B) A GST pull-down experiment. 35S-labeled Gal4 derivatives synthesized in vitro (lanes 1 and 3) and retained by immobilized GST–TBP (lanes 2 and 4). (C) The products of each cysteine derivative synthesized and 35S-labeled in vitro. (D) Binding of Gal4 cysteine derivatives to immobilized GST–TBP. (E) Four surface plasmon resonance sensograms describing binding of immobilized Gal4 derivatives with yeast TBP as measured on a BIAcore machine (9, 21). In each case, the difference in RU values between the three plateaus at the points marked by the asterisks gives a value proportional to the relative binding affinity of the two proteins. The averaged ratio of TBP (ΔRUTBP) retained by each Gal4 (ΔRUGAL4) derivative is: wild-type, 2.02 ± 0.3; T858C, 1.84 ± 0.25; T859C, 1.45 ± 0.3; T860C, 2.06 ± 0.27.
Two separate assays show that the cysteine mutants bind TBP in vitro about as efficiently as does the wild-type parent. The first of these assays was a standard “GST pull-down” experiment in which we measured binding of the Gal4 derivatives to GST–TBP immobilized on beads (40). For this experiment, each mutant Gal4 derivative, as well as the wild-type parent, was synthesized in a cell-free TnT system (Promega). Fig. 2B shows that the Gal4 derivative of Fig. 1A bound GST–TBP whereas the Gal4 DNA binding domain alone did not. Fig. 2C shows the products of the TnT reaction for each cysteine mutant, and Fig. 2D shows that each interacted efficiently with GST–TBP. The gel pattern in Fig. 2C suggests that, in the TnT system, the cysteine derivatives are degraded to various extents and the amount of each cysteine derivative bound by GST-TBP is approximately proportional to the level of the intact product in the TnT reaction.
For the experiment shown in Fig. 2E, we purified each mutant from Escherichia coli, immobilized it on the surface of a sensor chip, and tested its ability to interact with purified TBP by using surface plasmon resonance (9, 21). In the representative examples shown, within experimental error, all of the mutants interacted about as efficiently with TBP as the wild-type parent. The one exception was T859C, which bound slightly less efficiently, and this correlates with the lowered activity of this mutant in vivo.
Sensitivity to Gal80.
Fig. 3A shows that, as assayed in cells grown in noninducing conditions (i.e., in raffinose), two of the cysteine substitution mutants, F856C and T859C, were significantly less sensitive to inhibition by Gal80 than was either the wild-type parent or any of the other cysteine mutants. The mutant M861C was also moderately resistant to Gal80-mediated inhibition. This experiment was performed by expressing each mutant in cells deleted for endogenous Gal4 but bearing intact Gal80. Consistent with these results, Fig. 3B shows that each of the cysteine mutants, with the clear exceptions of F856C and T859C, bound Gal80 as efficiently as did the wild-type parent in an electrophoretic mobility super-shift assay. M861C was evidently also somewhat deficient in this reaction. To perform this experiment, each cysteine mutant was expressed in and purified from E. coli, as was the intact wild-type Gal80. The Gal80 super-shift assays were performed under conditions previously optimized for Gal4-DNA complex formation (38) with a DNA bearing a single consensus Gal4 binding site (42). Taken together, the results presented above suggest that, along the stretch of amino acids comprising residues 855–870, the identities of residues 856 and 859, and perhaps that of 861, are important for Gal80 recognition. In contrast, each residue in this region can be replaced by cysteine without dramatically affecting the activation function. We next explore the role of secondary structure in vivo by introducing the more drastic proline substitutions in key positions as described in the following section.
Figure 3.
The effect of substituting cysteine for wild-type residues on Gal80-mediated inhibition in vivo and interaction with Gal80 in vitro. (A) Reporter gene expression elicited by each of the cysteine substitution mutants of Fig. 1B as assayed in Gal80+ cells grown in raffinose. (B) Electrophoretic mobility super-shift assays performed with recombinant Gal80 and Gal4 derivatives. Gal4 derivatives were bound to a single DNA binding site at saturating concentrations of 10 nM (lanes marked −), and Gal80 was added in adjacent lanes (lanes marked +) at 15 nM concentration. At this inhibitor concentration, the two Gal4 mutants, F856C and T859C, show a 3- to 4-fold decrease in their affinity for the Gal80.
Proline Substitutions
Activation and Sensitivity to Gal80.
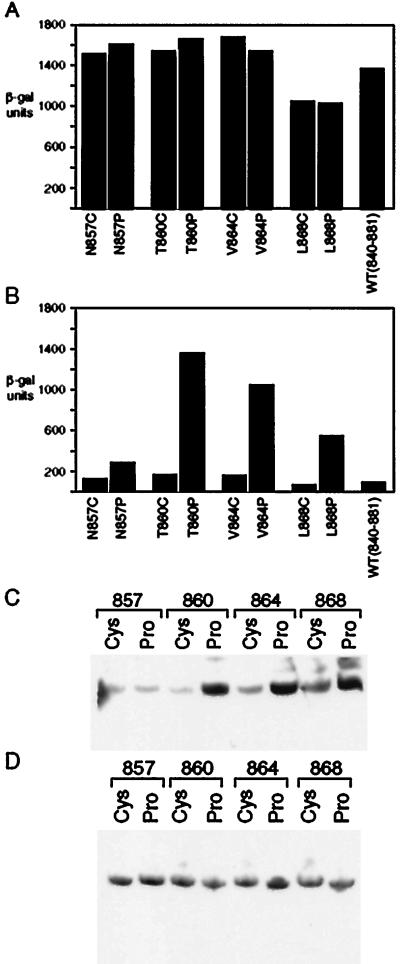
We generated four proline-substitution mutants: N857, T860, V864, and L868. These positions were chosen on the basis of several criteria: first, cysteine substitutions at these positions affect neither Gal80 binding nor activation; second, the first two lie adjacent to residues that, when changed to cysteine, significantly impair interaction with Gal80 and decrease activation slightly; and third, the last two substitutions lie in a region identified as crucial for Gal80 binding by deletion analysis, and that analysis also showed that this region contributes to the activation function (21, 25). Fig. 4A shows that, as assayed in cells grown in galactose, these substitutions had virtually no effect on the activation function.
Figure 4.
The effects of substituting proline for wild-type residues in Gal4 on activation, inhibition by Gal80, and on Gal80 synthesis. This figure shows reporter gene activity elicited by various proline mutants in cells grown in either galactose (A) or raffinose (B). In each case, a mutant bearing a cysteine substitution at the identical position was tested similarly in parallel. Immunoblot studies measured endogenous Gal80 levels in cells expressing either cysteine or proline mutants and grown in raffinose (C) or galactose (D).
In contrast, as shown in Fig. 4B, each of the proline substituted mutants, with the exception of T857P, dramatically reduced sensitivity to Gal80-mediated inhibition as assayed in cells grown in raffinose. This loss of inhibition cannot be attributed to lowered levels of Gal80. The immunoblot analysis of Fig. 4C shows that, in fact, the level of Gal80 was higher in cells expressing each proline mutant—with the notable exception of T857P—than in cells expressing analogous cysteine. This is the expected result: transcription of the Gal80 gene is regulated positively by Gal4 (44), and therefore mutants of Gal4 that retain the activation function, but are not inhibited by Gal80, should cause increased expression of Gal80. Also, as expected from these considerations, cells expressing either the cysteine or the proline mutants at the four positions shown in Fig. 4C, and grown in galactose, all express equal levels of Gal80 (Fig. 4D).
Crosslinking Studies
We performed far-UV CD experiments with peptides comprising residues 840–881 and 850–874 at both pH 7.0 and pH 5.8. At either pH, we obtained spectra characteristic of a random coil with an absorption minima <200 nm (data not shown). These results, taken together, suggest that the same peptide, largely unstructured when free of a binding partner, has quite distinct sequence and structural requirements for interacting with two different partners. One of those partners is Gal80, and the other may include any of several possible targets in the transcriptional machinery, including TBP (21, 22).
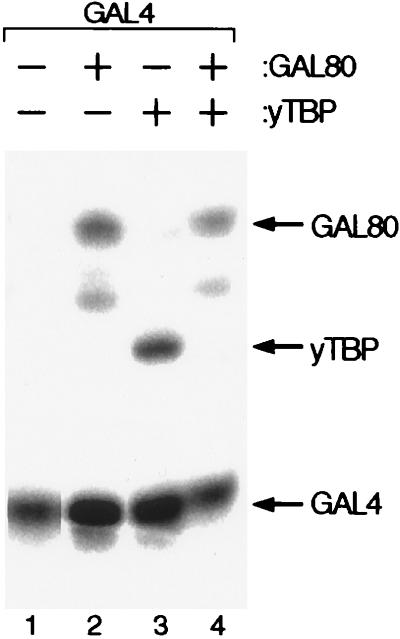
The label-transfer photoaffinity crosslinking experiment of Fig. 5 shows two relevant results. First, as tested in separate reactions, Gal4 residue 857 is in close proximity to TBP and to Gal80, and, thus, it is likely that common Gal4 residues participate in both reactions. Second, binding of the Gal4 activating peptide to Gal80 prevents simultaneous binding to TBP in this region. The experiment was performed by specifically attaching the radiolabeled crosslinking reagent 125I-APDP (9, 45) to the introduced cysteine residue in the N857C mutant of Gal4(1–100) + (840–881). We chose residue 857 for modification because, although neither activation nor Gal80-mediated inhibition are affected by either a proline or a cysteine substitution at this position, 857C lies adjacent to residues that are required for Gal80 binding and that have detectable effects on activation. There are six additional cysteine residues in this protein, but they are complexed with two Zn(II) ions in a Zn2Cys6 binuclear cluster and are unreactive to the crosslinker (46, 47). On interaction of the Gal4 activating region with its partner (in this case Gal80 or TBP), the crosslinker, activated by UV, covalently attached to the interacting partner. Subsequent incubation under reducing conditions reversed the disulfide bond between the crosslinker and the cysteine residue of Gal4, thereby transferring the label to the crosslinked partner. Fig. 5 shows that, when incubated separately with 125I-APDP modified Gal4, both TBP and Gal80 were labeled efficiently, but, when both proteins were mixed with the modified Gal4 at equimolar concentrations, the label was transferred predominantly to Gal80.
Figure 5.
Photoaffinity crosslinking of Gal4 to TBP and to Gal80. 125I-APDP-Gal4 was incubated with purified Gal80 (lane 2), with purified yTBP (lane 3), and with both proteins (lane 4). In each case, after UV-induced crosslinking, the label was transferred by treatment with a reducing agent as described in ref. 9 (the minor band in lanes 2 and 4 is a degradation product of Gal80).
DISCUSSION
The results presented here show that a peptide encompassing 16 residues—Gal4(855–870)—can participate in two distinct protein–protein interactions with quite different sequence and structural requirements. This peptide, when fused to a protein comprising the Gal4 residues (1–100) + (840–854), confers on the fusion both the ability to activate transcription significantly and to bind, and be inhibited in that function by, Gal80. The transcriptional activating function of this peptide is affected little by the introduction of single cysteine mutations anywhere in the sequence, or by any of four proline substitutions. We find that the various mutants that we have constructed, with one exception (T859C), bind TBP as efficiently as does the wild-type parent. In striking contrast, the second function of this region, interaction with the inhibitor Gal80, is highly sensitive to certain cysteine substitutions (F856C and T859C), and prolines introduced at three nearby sites at which cysteine substitutions have no effect strongly disrupt repression by Gal80 in vivo. The photo-crosslinking experiments we describe show that residue 857 comes in close proximity to both TBP and Gal80 when they are bound separately and that the binding of Gal80 blocks binding of TBP, results reinforcing the notion that a common set of residues is involved in both functions.
Cysteine substitutions are structurally nonobtrusive because they have an equal propensity to be in an α-helix or a β-sheet (28–32) and because thiol groups are nonpolar yet readily polarizable, cysteine residues are found both in the cores and on the surfaces of proteins. The fact that two cysteine substitution mutants—856 and 859—impair interaction with Gal80 suggests that the residues found in the wild-type protein at these positions (Phe and Thr, respectively) contact Gal80. The additional finding that prolines placed near these positions similarly affect Gal80 binding suggests that some structure—an α-helix or a β-sheet—is required for the interaction.
None of the cysteine substitutions significantly affected the activation function or interaction with TBP. Activation domains of a similar or larger size are resistant to deleterious effects of typical point mutations (12, 21). The finding that the F869A substitution has a more deleterious effect on activation than does the F869C mutant suggests that the side chain of cysteine can substitute significantly for the missing phenyl ring but the side chain of alanine, which does not extend as far from the backbone cannot. Furthermore, in contrast to a previously published report (26), our CD experiments with purified peptides comprising residues 840–881 or 850–874 show that, at both pH 7.0 or pH 5.8, the peptides were unstructured (data not shown). Taken together, our results suggest that interaction with Gal80 induces formation of some ordered structure incompatible with the presence of proline residues; in contrast, such a structure is not essential for the activating function. TBP binds our Gal4 derivative at least 10-fold less tightly than does Gal80, and so it is unlikely that TBP but not Gal80 can overcome the constraints placed by a proline residue and force the peptide to adopt an ordered conformation on binding.
Our results argue that it is highly unlikely that a β-hairpin structure forms on interaction with a target in the transcriptional machinery. However, such a structure could be formed on interaction with Gal80 (25). Our results question the physiological relevance of the α-helices that have been reported to form when certain activating regions (e.g., those from VP16, myc, and CREB) are bound to their respective targets in the transcriptional machinery (14, 16, 17). To bring our results into congruence with those findings, we might consider the following possibilities: (i) some activating regions form α-helices on interaction with their targets in the transcriptional machinery and some (e.g., the one examined here) do not; (ii) the α-helices formed by activating regions in such interactions tolerate proline-mediated kinks; or (iii) activating regions form α-helices when bound to their targets in vitro but need not necessarily do so to function in vivo. Whatever the explanation, it may be relevant that shortened variants of certain activating regions (48, 49), including the one studied here (21), are much more sensitive to the effects of point mutations than are their longer parents. This has led to the suggestion that activating regions comprise multiple functional units that can be sampled individually by the target protein, an idea consistent with the fact that reiteration of a short peptide can produce an efficient activator (50–55).
The proposal that acidic activating regions are promiscuous (1, 9, 12, 56), interacting with multiple partners, also is consistent with the idea that binding may depend only on the “stickiness” of the region rather than a very rigidly ordered constellation of functional groups. As a corollary, de novo design of transcriptional activators should be relatively facile, and that, indeed, is the case. In early experiments, eukaryotic activating regions were isolated readily from a screen of random E. coli DNA fused to a DNA binding domain (57). More recently, short peptides that are predominantly hydrophobic and are capable of activating transcription at very high levels when tethered to DNA were isolated in abundance from a yeast genetic screen (X. Lu and M.P., unpublished results).
We know of several instances in which eukaryotic transcriptional activating regions also recognize inhibitors. In one case, that involving the activator p53 and MDM2, the activating region forms an α-helix on interaction with the inhibitor (13). Wherever measured, the inhibitor binds significantly more tightly to the activating region and blocks the binding of the putative targets in the transcriptional machinery (13, 58–60). Evidently, the relative weakness and promiscuity of the activating region–target interactions impose less of a requirement for structure on the activating region (see, for example, ref. 61) than does the stronger interaction with a specific inhibitor.
Acknowledgments
We thank A. Jafferally, J. Ruan, and M. Awad for technical assistance; L. Wu for help with CD experiments; Y. Wu and G. Bryant for help with surface plasmon resonance analysis; M. Green for the gift of the GST–yTBP expression plasmid; R. Hellmiss for illustrations; and, particularly, the members of the Ptashne lab for valuable discussions. This study was funded by Grant GM32308 from the National Institutes of Health to M.P., and A.Z.A. was supported by the Helen Hay Whitney Foundation.
ABBREVIATION
- TBP
TATA box-binding protein
- GST
glutathione S-transferase
- TnT
transcription and translation
- RU
resonance response units
- 125I-APDP
N-[4-(p-azidosalicylamido)butyl]-3′[2′-pyridyldithio]propionamide
References
- 1.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 3.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 5.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, et al. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 7.Stargell L A, Struhl K. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 8.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 10.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 11.Sigler P B. Nature (London) 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 12.Triezenberg S J. Curr Opin Gen Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 13.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 14.McEwan I J, Dahlman-Wright K, Ford J, Wright A P. Biochemistry. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- 15.Shen F, Triezenberg S J, Hensley P, Proter D, Knutson J R. J Biol Chem. 1996;271:4827–4837. doi: 10.1074/jbc.271.9.4827. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan I, Perez-Avarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 17.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 18.Johnston M. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohr D, Venkov P, Zlatnova J. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Ptashne M. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 22.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston S A, Salmeron J M, Dincher S S. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 24.Salmeron J M, Leuther K K, Johnston S A. Genetics. 1990;125:21–27. doi: 10.1093/genetics/125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuther K K, Salmeron J M, Johnson S A. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 26.Van Hoy M, Leuther K K, Kodadek T, Johnston S A. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham B C, Wells J A. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 28.Chou P Y, Fasman G D. Biochemistry. 1974;13:211–221. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 29.O’Neil K T, DeGrado W F. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 30.Kim C A, Berg J M. Nature (London) 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- 31.Minor D L, Kim P S. Nature (London) 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 32.Smith C K, Withka J M, Regan L. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- 33.Schulman B A, Kim P S. Nat Struct Biol. 1995;3:682–687. doi: 10.1038/nsb0896-682. [DOI] [PubMed] [Google Scholar]
- 34.Williams K A, Deber C M. Biochemistry. 1991;30:8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]
- 35.Wood S J, Wetzel R, Martin J D, Hurle M R. Biochemistry. 1995;34:724–730. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 36.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic; 1990. [Google Scholar]
- 37.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 38.Reece R J, Ptashne M. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 39.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 40.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 41.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 42.Liang S, Marmorstein R, Harrison S C, Ptashne M. Mol Cell Biol. 1996;356:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 44.Shimada H, Fukasawa T. Gene. 1985;39:1–9. doi: 10.1016/0378-1119(85)90100-3. [DOI] [PubMed] [Google Scholar]
- 45.Weissman J S, Kashi Y, Fenton W A, Horwich A. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 46.Marmorstein R, Carey M, Ptashne M, Harrison S C. Nature (London) 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 47.Pan T, Coleman J E. Biochemistry. 1990;29:3023–3029. [Google Scholar]
- 48.Cress W D, Triezenberg S J. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 49.Hope I A, Mahadevan S, Struhl K. Nature (London) 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 50.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blair W S, Bogerd H P, Madore S J, Cullen B R. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seipel K, Georgiev O, Schaffner W. Biol Chem Hoppe-Seyler. 1994;375:463–470. doi: 10.1515/bchm3.1994.375.7.463. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka M, Herr W. Mol Cell Biol. 1994;14:6056–6067. doi: 10.1128/mcb.14.9.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerber H P, Seipel K, Georgiev O, Hofferer M, Hug M, Russconi S, Schaffner W. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 55.Jackson B, Drysdale C M, Natarajan K, Hinnebusch A G. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drysdale C M, Jackson B, McVeigh R, Klebanow E R, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil A, Hinnebusch A G. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 58.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 59.Hagemeier C, Cook A, Kouzrides T. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weintraub S J, Chow K N B, Luo R X, Zhang H, He S, Dean D. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 61.Nyanguile O, Uesugi M, Austin D J, Verdine G L. Proc Natl Acad Sci USA. 1997;94:13402–13406. doi: 10.1073/pnas.94.25.13402. [DOI] [PMC free article] [PubMed] [Google Scholar]