Abstract
Eukaryotic mRNAs in which a poly(A) sequence precedes the initiation codon are known to exhibit a significantly enhanced cap-independent translation, both in vivo and in cell-free translation systems. Consistent with high expression levels of poxviral mRNAs, they contain poly(A) sequences at their 5′ ends, immediately before the initiation AUG codon. Here we show that poly(A) as a leader sequence in mRNA constructs promotes the recruitment of the 40S ribosomal subunits and the efficient formation of initiation complexes at cognate AUG initiation codons in the absence of two essential translation initiation factors, eIF3 and eIF4F. These factors are known to be indispensable for the cap-dependent (and ATP-dependent) mechanism of translation initiation but are shown here to be not required if an mRNA contains a 5′-proximal poly(A). Thus, the presence of a pre-AUG poly(A) sequence results in an alternative mechanism of translation initiation. It involves the binding of initiating 40S ribosomal subunits within the 5′ UTR and their phaseless, ATP-independent, diffusional movement (“phaseless wandering”) along the leader sequence, with subsequent recognition of the initiation (AUG) codon.
Keywords: cap-independent initiation, eIF3, eIF4F, poxvirus mRNA, toeprinting
In eukaryotes, including mammalian and human cells, expression of genetic information is extensively regulated at the level of translation. Almost all mechanisms of translational control involve the initiation step of translation. The translation initiation machinery is supported by proteins called initiation factors (eIFs), which determine the selectivity of initiation and are targets of translational regulation. In eukaryotes, the most important and universally present are eIF1, eIF1A, eIF2, eIF3 (a large complex consisting of >10 different protein subunits), eIF4A, eIF4B, eIF4F (a complex consisting of three subunits—eIF4A, eIF4E, and eIF4G), eIF5, and eIF5B (reviewed in ref. 1). The initiation process starts with dissociated ribosomal particles. The 40S ribosomal subunit associates with eIF1, eIF1A, eIF2, and eIF3; eIF2 binds GTP and initiator Met-tRNAi. The resultant complex is usually designated as the 43S preinitiation complex. With capped mRNAs, this complex binds to the 5′ end of mRNA, and the cap structure (m7GpppN) normally present at the 5′ end of cellular mRNAs strongly stimulates the binding owing to the presence of eIF4F and its cap-binding eIF4E subunit. eIF4F (its eIF4G subunit) has an affinity for eIF3 and thus interacts with the 43S preinitiation complex. The main step in the subsequent selection of a correct initiation codon on mRNA is the process of unidirectional movement of the preinitiation 43S ribosomal complex along the 5′ UTR of mRNA from its 5′ end, scanning RNA sequence until the first AUG triplet in the proper context is encountered (the scanning model of M. Kozak; see refs. 2 and 3). The recognized AUG triplet becomes the initiation codon. The above movement requires the hydrolysis of ATP coupled with the unwinding of RNA helices. This reaction is catalyzed by eIF4A, which functions as an ATP-dependent helicase or an RNA-dependent ATPase. Both the eIF4A subunit of eIF4F and free eIF4A are involved in the scanning process. Selective recognition of the initiation codon in mRNA and the proper positioning of the ribosomal complex involve the eIF2-bound Met-tRNAi, eIF1, and eIF1A, all bound within the complex. The stalled complex is referred to as the initiation 48S ribosomal complex. To begin translation, the free 60S ribosomal subunit must join the 48S complex. This association requires two more eIFs, eIF5 and eIF5B with GTP, and is accompanied by the hydrolysis of the two complex-bound GTP molecules and the release of all initiation factors from the 80S ribosome.
To inactivate or bypass cellular regulatory mechanisms and exploit protein synthesis to their own needs, many viruses employ modified pathways of translation initiation. The paradigmatic viral strategy is the use of a virus-specific structural module in a viral RNA, the so-called internal ribosome entry site (IRES), which is capable of binding (“recruiting”) ribosomal particles in an infected cell independently of several initiation factors that are essential for the normal cellular translation (reviewed in ref. 4). The initiation of translation on viral RNAs with IRESes often proceeds through a simplified route, sometimes with participation of proteins (“IRES trans-acting factors,” ITAFs) that are not involved in translation in uninfected cells. In the case of poxviruses, however, no well defined IRESes have been described for their RNAs. Instead, highly expressed late mRNAs of poxviruses were shown to be the product of an unusual modification of RNA transcripts that result in poly(A) sequences (mostly 30–40 nt long, but sometimes shorter) placed at the 5′ ends of these mRNAs, immediately before the initiation AUG codon (5–8). A specific mechanism of slippage and transcription reinitiation (reiteration) during the initial steps of poxviral RNA polymerase-mediated RNA synthesis was proposed to account for the above addition of poly(A) to the 5′ ends of viral mRNAs (9–11). Early mRNAs of poxviruses may also contain leading poly(A) sequences and may be formed through a similar mechanism (11). Translation of the poxvirus mRNAs with a poly(A) leader was shown to be at most weakly dependent on the cap-binding complex eIF4F and may be cap-independent (12, 13).
As demonstrated in direct in vitro experiments, when uncapped poly(A) sequences (5, 12, and 25 nt long) were used as mRNA leaders in chimeric mRNA constructs with Luc- or GFP-encoding sequences, they resulted in an efficient translation of such mRNAs in eukaryotic cell-free systems. 5′-Proximal poly(A) sequences could thus be considered as strong translational enhancers comparable, in their efficacy, to the globin mRNA leader and the omega leader of TMV RNA (14). The effect of 5′-proximal poly (A) could not stem simply from an “unstructured nature” of the leader, because another homopolymeric leader, poly(U), which is known to be a largely unstructured homopolymer (see references in Discussion), displayed no enhancing activity.
To determine the requirements of mRNAs with poly(A) leaders for individual eIFs, we analyzed the formation of ribosomal initiation complexes on mRNA where the Luc-coding sequence was preceded by a poly(A) leader sequence. We show that the poly(A) leader mediates the formation of the ribosomal initiation complexes independent of two multifunctional eIFs, eIF3 and eIF4F.
Results
Toeprinting Assay Using Fluorescently Labeled Primers.
To determine the requirements of mRNAs for eIFs we applied the technique called “extension inhibition analysis of translation initiation complexes,” also known as toeprinting (15, 16). The method is based on the observation that specific attachment of a ribosomal particle to mRNA results in blocking the reverse transcriptase (RT) movement along mRNA chain in the upstream (3′–5′) direction when a bound ribosome is encountered. A 5′-labeled oligodeoxyribonucleotide complementary to a region of mRNA downstream (3′-ward) of the initiation codon is annealed to this mRNA and used as a primer for the RT. The product of this reaction (cDNA) is the 5′-labeled elongated polydeoxyribonucleotide (extended primer), the length and the sequence of which correspond to the mRNA section from the annealing site to the upstream site of the bound ribosome. This way, the formation of the ribosomal initiation complex at the proper site of mRNA (initiation AUG codon) can be detected, depending on the presence or absence of individual initiation factors (17). In the present study, a modification of the original technique (where cDNAs were labeled with [32P]phosphate groups, either cotranscriptionally or at 5′ ends of the primers) was used, in which the labeling was by fluorescent dyes and the separation of the fluorescent products of RT reaction was carried out by capillary electrophoresis, as recently described (18). This modification allowed us to quantitate the ribosomal initiation complexes formed on mRNA with different combinations of eIFs and also made it possible to observe the initiation complex formation on different competing mRNAs in a mixture when their cDNAs were labeled with different fluorescent dyes.
The RT is known to terminate the 5′-ward elongation of cDNA on mRNA mostly at the 16th, 17th, and, to a lesser extent, 18th nucleotides downstream from the first nucleotide of the initiation codon on which eukaryotic 48S complex is assembled (19). Therefore, the position of the ribosomal initiation complex relative to the primer position on mRNA chain can be simply deduced from the lengths of terminated polydeoxyribonucleotides, and the amount of the complex formed can be calculated from the total fluorescence of the terminated product.
In figures described below the lengths of dye-labeled products of the AMV RT reaction are plotted against their fluorescent intensity. The first peaks of fluorescence correspond to runoff (full-length) polydeoxyribonucleotide product, i.e., to the 5′ end of mRNA (Fig. 1). A characteristic “trident” of fluorescence (with the third “dent” being minor) appears when reverse transcription is stopped by the initiation 48S complex and thus indicates the position of the initiation AUG triplet; the integral fluorescence of the “trident” reflects the total amount of the initiation complex formed.
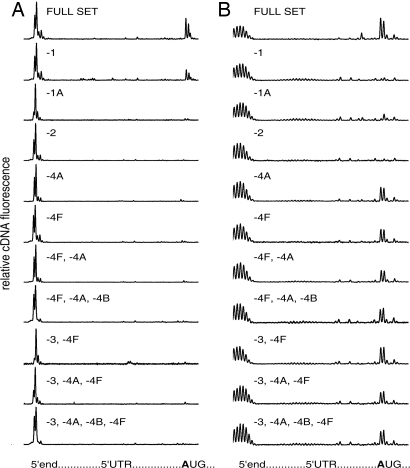
Fig. 1.
Formation of initiation 48S ribosomal complexes on natural capped β-globin mRNA (A) and recombinant noncapped Luc mRNA with poly(A) leader sequence (B). The uppermost plots show the results with the full set of eIFs (eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F) in the complex formation mixtures. The lower plots are the results when one or several eIFs were omitted from the complex formation mixtures. Relative fluorescence intensities of cDNA products generated by RT are plotted versus leader sequences of corresponding mRNAs. The integral fluorescence of the left major peak reflects the amount of the full-length product when mRNA was read out by RT up to the 5′ end without stop. The integral fluorescence of the three major peaks (“trident”) at the initiation site, when it appeared, corresponds to the product of the reversed transcription stopped by initiation 48S ribosomal complex formed at the initiation AUG codon.
It should be mentioned that poly(A) leader sequence displays some length heterogeneity, as indicated by multiple stop points of the RT at the 5′ end of the poly(A) mRNA (Fig. 1B). This problem is well known and is unavoidable with homopolymers as templates, especially poly(dT). It stems from the fact that most polymerases exhibit a slippage effect, to varying degrees, when transcribing homopolymeric sequences, including the polymerases of phages T3, T7, and SP6 (20–22). Poly(A) leaders of poxviral mRNAs synthesized in vivo are heterogeneous in length for the same reason (6–10, 23). Fortunately, the microheterogeneity observed does not influence the results of the extension inhibition analysis, i.e., the identification of a stop position of a migrating ribosomal subunit at the initiation codon (which is ≈25 nt downstream).
Ribosomal Initiation Complex Formation on mRNA with Poly(A) Leader Does Not Require eIF3, eIF4A, and eIF4F.
Fig. 1A shows that the 48S initiation complex is formed on the natural capped β-globin mRNA at the correct initiation codon in the presence of all canonical eIFs necessary for this stage of translation initiation—eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F (the uppermost plot); the efficiency of its formation can be estimated in terms of the percentage of mRNA involved in the complex, this being ≈30% of input mRNA (Table 1). As expected, the exclusion of any one among the eIFs listed above resulted in either a dramatic decrease (in the case of eIF1) or abolition of the 48S complex formation at the initiation codon. Thus, the lower plots of Fig. 1A demonstrate that the formation of the complex was close to background level in the absence of eIF1A, eIF2, eIF4A, eIF4F, or eIF3 plus eIF4F (see Table 1 for quantitative data). It should be noted that the absence of eIF1 led to an ≈2-fold decrease of the amount of the complex formed but was not critical, whereas the omission of eIF1A was sufficient to prevent the 48S complex formation (see Table 1). These data are in agreement with the results obtained earlier with β-globin mRNA by classical extension inhibition analysis that used radioactively labeled primers (17, 24).
Table 1.
The yield of 48S complex assembly on natural capped β-globin mRNA and chimeric poly(A)-Luc mRNA with different sets of eIFs (distribution of cDNA fluorescence intensity/percentage of input mRNA)
| eIF set | β-Globin mRNA |
poly(A)-Luc mRNA |
||||
|---|---|---|---|---|---|---|
| 5′ end | Nonspecific | AUG | 5′ end | Nonspecific | AUG | |
| Full set | 65 ± 3 | 5 ± 1 | 30 ± 3 | 64 ± 4 | 6 ± 3 | 30 ± 4 |
| -eIF1 | 66 ± 3 | 16 ± 3 | 18 ± 3 | 91 ± 3 | 5 ± 3 | 4 ± 5 |
| -eIF1A | 93 ± 2 | 2 ± 2 | 5 ± 2 | 90 ± 4 | 4 ± 2 | 6 ± 3 |
| -eIF1, -eIF1A | 87 ± 2 | 5 ± 2 | 6 ± 3 | 93 ± 3 | 4 ± 3 | 3 ± 2 |
| -eIF2 | 92 ± 3 | 5 ± 2 | 3 ± 2 | 95 ± 2 | 3 ± 2 | 2 ± 1 |
| -eIF4A | 92 ± 3 | 4 ± 2 | 4 ± 2 | 77 ± 5 | 3 ± 3 | 20 ± 6 |
| -eIF4F | 92 ± 2 | 4 ± 3 | 4 ± 2 | 77 ± 5 | 4 ± 2 | 19 ± 5 |
| -eIF4A, -eIF4F | 93 ± 2 | 4 ± 2 | 3 ± 2 | 78 ± 4 | 3 ± 2 | 19 ± 5 |
| -eIF4A, -eIF4B, -eIF4F | 92 ± 3 | 4 ± 2 | 4 ± 2 | 81 ± 3 | 3 ± 1 | 16 ± 5 |
| -eIF3, -eIF4F | 93 ± 2 | 4 ± 2 | 3 ± 2 | 80 ± 3 | 3 ± 2 | 17 ± 4 |
| -eIF3, -eIF4A, -eIF4F | 93 ± 2 | 3 ± 2 | 4 ± 2 | 80 ± 3 | 2 ± 2 | 18 ± 6 |
| -eIF3, -eIF4A, -eIF4B, -eIF4F | 94 ± 2 | 3 ± 3 | 3 ± 2 | 80 ± 4 | 3 ± 1 | 17 ± 5 |
Data were averaged from several independent experiments. Numbers indicate percent fractions of mRNA in total mRNA, which were (i) not involved in complex formation and correspond to full length mRNA (5′ end column), (ii) nonspecifically spread along mRNA 5′ UTR (Nonspecific column), and (iii) specifically stalled at correct AUG codon by initiation complex (AUG column). The full set included eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F.
Fig. 1B shows a very different factor dependence for the initiation 48S complex formation on an mRNA with an uncapped poly(A) leader sequence. In this case, only eIF2—the eIF responsible for binding and involvement of initiator Met-tRNAi in the initiation process—and the pair of eIF1 and eIF1A were found to be strictly required for the stalling of ribosomal particles in the form of 48S complex at the initiation codon. As to the eIFs of eIF4 group and the largest multifunctional factor eIF3, they proved to be not stringently necessary for the formation of 48S initiation complex on mRNA with poly(A) leader: in the absence of any one of them, and even in the absence of eIF4A+eIF4F, or eIF4A+eIF4B+eIF4F, or eIF4A+eIF4B+eIF4F+eIF3, the efficiency of the 48S complex formation was >50–60% of the control value with a full set of eIFs (see Table 1).
Experiments that examined the initiation complex formation on the mRNA with uncapped poly(A) leader were carried out also in the presence of purified 60S ribosomal subunits, when both subunits (40S and 60S) were present in the incubation mixture from the beginning. The results [see supporting information (SI) Results and Discussion and Fig. S1] did not differ from those in Fig. 1.
The low dependence of the 48S initiation complex formation on the presence of the eIFs required for ATP-dependent unidirectional scanning of leader sequence in the case of mRNA with poly(A) leader suggests that the preinitiation 40S ribosomal subunit binds to the poly(A) sequence at random sites, and the subsequent search for the initiation codon proceeds via energy-independent one-dimensional diffusion (phaseless wandering) along the leader (see Discussion). At the same time, some decrease (≈30%) in the amount of the 48S complexes formed in the absence of ATP-dependent helicases (eIF4A alone, eIF4F alone, or eIF4A and eIF4F together; see Fig. 1B and Table 1) may indicate that the ATP-dependent scanning can also occur on a poly(A) leader sequence and thus additionally contribute to locating the start site.
mRNA with a Poly(A) Leader Outcompetes Capped mRNA in Direct Competition Essays.
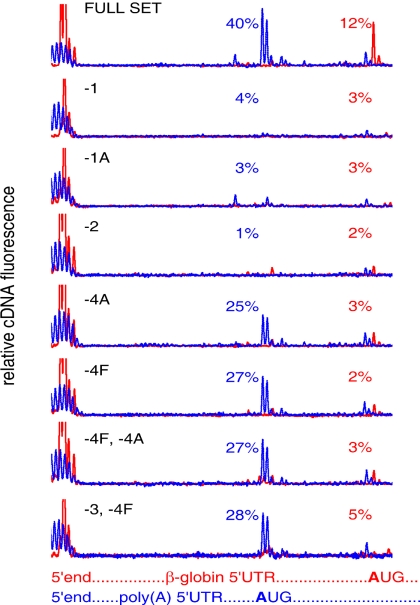
As mentioned above, one of the advantages of toeprinting with fluorescently labeled oligodeoxyribonucleotide primers is the possibility to estimate the efficiency of the initiation complex formation on different mRNAs in the same reaction mixture by using different fluorescent labels. As shown in Fig. 2, in the mixture of capped β-globin mRNA and uncapped A25-Luc mRNA at equal molar concentrations (15 nM each) the initiation 48S complex formation was observed on both mRNAs at correct initiation codons, when all eIFs were present. As expected, the control assay, with the omission of eIF2, showed no significant complex formation in both cases. The absence of only ATP-dependent helicase eIF4A also prevented the formation of the initiation complex on β-globin mRNA, but the complex was well formed at the correct AUG on A25-Luc mRNA (although there was some decrease). The same effect was observed when eIF4F or both eIF4F and eIF4A were omitted. At the same time, as seen also in Fig. 1B, the absence of free helicase (eIF4A), full eIF4F complex, or both eIF4F and eIF3 did not abolish the formation of the initiation 48S complex at the initiation codon of the A25-Luc mRNA, although it somewhat decreased the amount of the complex formed, to the level of ≈60–70% of the control level with the full set of eIFs.
Fig. 2.
Formation of initiation 48S ribosomal complexes in the equimolar mixture of natural capped β-globin mRNA (red line) and recombinant noncapped Luc mRNA with poly(A) leader sequence (blue line). Designations and explanations are the same as in Fig. 1.
The unexpected result of the experiment shown in Fig. 2 was the depression of the 48S complex formation on β-globin mRNA in the presence of equimolar amount of A25-Luc mRNA (the uppermost plot, red line; see Table 1). This seems to be a direct competing effect: it is possible that the A25 leader sequence may possess a higher affinity for 40S ribosomal particles, as compared with a heteronucleotide sequence. On the other hand, some reproducible stimulation of the complex formation on A25-Luc mRNA in the presence of β-globin mRNA (uppermost plot, blue line; see Table 1) can be mentioned (no reasonable explanation of this fact can be proposed by now).
Ribosomal Initiation 48S Complex Formed in the Absence of eIF3 and eIF4F Can Join 60S Ribosomal Subunit and Form Initiation 80S Complex.
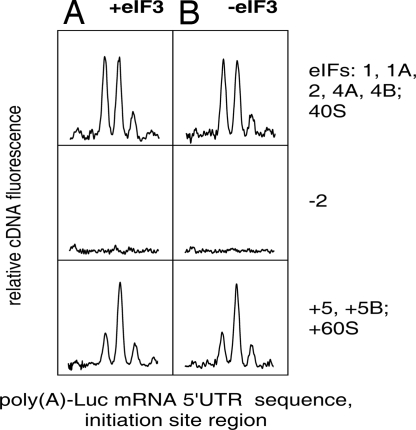
After formation of the 48S complex at the initiation codon, the next step of the initiation process is the eIF5-eIF5B-promoted association with 60S ribosomal subunit and the formation of the initiation 80S complex. Is the 48S complex formed in the absence of eIF3 capable of joining the 60S subunit and thus completing the initiation process? The same toeprinting technique can be applied for detection of the 80S complex at the initiation codon of mRNA. The formation of the 80S complex from the 48S complex is accompanied by a significant change in the pattern of the toeprint: instead of electrophoretic bands reflecting the stop points of RT mainly at the 16th and 17th nucleotide residues from the first nucleotide of the initiation codon, the predominant stop point becomes shifted to the 17th nucleotide (19, 25). We used this observation for the detection of the 80S complex formation on A25-Luc mRNA in the absence of eIF3, in comparison with an otherwise identical assay that contained eIF3 (Fig. 3). The change of the band distribution pattern was the same in both cases, in agreement with the previously described change for eIF3-dependent mRNAs (25). We conclude that the 48S complex formed in the absence of eIF3 is competent for transformation into the 80S complex, and thus eIF3 is not required for the 80S complex formation.
Fig. 3.
Formation of initiation 80S ribosomal complexes from 48S complexes preassembled on noncapped Luc mRNA with poly(A) leader sequence in the presence (A) and absence (B) of eIF3. (Top) The formation of the 48S complex in the presence of required eIFs (eIF1, eIF1A, and eIF2; eIF4A and eIF4B were also present). (Middle) Negative controls where the same factors, except eIF2, were present. (Bottom) The results of addition of eIF5, eIF5B, and 60S ribosomal subunits to the 48S complexes formed under conditions indicated above (see Top). Designations and explanations are the same as in Fig. 1. Only the initiation site region of mRNA is given in the abscissa. The changed trident profiles reflect the formation of 80S ribosomal complexes (see text) in both cases (with and without eIF3).
Discussion
The results of this study emphasize the uniqueness of a poly(A) leader among all known efficient 5′ UTRs of eukaryotic mRNAs. Our findings show that the promotion of translation of a downstream coding sequence by a poly(A) leader reported earlier (14) can be achieved without recruiting several normally essential factors of a eukaryotic host cell. Initiation of translation without eIF3 is especially noteworthy: thus far, no leader sequences (5′ UTRs) that possess the ability to provide initiation in the absence of eIF3 have been known. In addition, most of the known viral IRESes need 40S particles with eIF3 for initiation (4). The known exceptions are the unique intercistronic IRES of cricket paralysis virus RNA (and its relatives), which forms a start complex with 40S ribosomal particle in the absence of eIFs and initiator Met-tRNAi (see ref. 4 for review), and the IRES of hepatitis C virus RNA, which is capable of forming ribosomal initiation complex in the absence of all known eIFs, with only aminoacylated tRNA at an elevated (nonphysiological) Mg2+ concentration (26).
What mechanism underlies an efficacious formation of the initiation 48S complex at the correct initiation codon on an mRNA with a poly(A) leader in the absence of eIF4F and eIF4A (Fig. 1B)? Usually, after the initial binding of the “native” 40S ribosomal subunit, or the 43S preinitiation complex (the complex of 40S subunit with eIF2 and eIF3) to the capped end of mRNA, the monomeric helicase eIF4A and/or the multimeric helicase complex eIF4F provides ATP-dependent catalysis of unidirectional (3′-ward) scanning of 5′ UTR of mRNA by the preinitiation particle; it is the result of this scanning that the ribosomal particle reaches the initiation codon and forms the stalled 48S complex. As Fig. 1B shows, this is not the case for an mRNA with a poly(A) leader. The absence of these eIFs does not prevent the effective formation of the 48S initiation complex at the correct initiation codon. This fact implies that ATP-dependent unidirectional scanning of poly(A) leader is not strictly required for finding the correct initiation codon. At the same time, the length of the leader was shown to be important for its efficiency as a translational enhancer: the enhancing activity of A25 was higher than that of A12 and even higher that that of A5 (14). It is likely that the poly(A) leader sequence can effectively bind preinitiation 40S ribosomal subunits at random internal sites within the poly(A) sequence and may thus allow the particles to perform energy-independent diffusional movement along the 5′ UTR until the particle is fixed at the initiation codon. This mechanism was first proposed by S. Brenner in 1967 (27) for the process of searching for a new initiation codon on polycistronic mRNA by prokaryotic ribosomes after termination of a preceding cistron, and the term “phaseless wandering” was used. Later this mechanism was confirmed and further substantiated for prokaryotes by Adhin and van Duin (28). A similar mechanism of phaseless wandering may be relevant to our case of ATP-independent searching for the initiation codon along poly(A) leader.
It was previously shown that a poly(A) sequence placed as an intercistronic insert in a bicistronic mRNA exhibits a high frequency of initiation at the second cistron in both in vivo and in vitro eukaryotic translation systems, thus demonstrating an “IRES-like” effect (29). Recently it has also been reported that internal poly(A) tracts within 5′ UTRs preceding the AUG initiation codon allow a cap-independent translation of mRNAs that are required for starvation-induced differentiation in yeast (30). The above-mentioned possibility of internal binding of preinitiation 40S ribosomal subunits to poly(A) leader is consistent with the finding that poly(A) sequences inside polycistronic mRNA constructs or inside 5′ UTRs may function as IRES-like elements (29, 30). However, typical viral IRESes are characterized by relatively stable tertiary structures and represent compact modules that are capable of specifically binding and positioning the preinitiation 40S ribosomal subunits on mRNA (4). The disposition with poly(A) leaders or poly(A) internal inserts seem to be quite different: they have no fixed tertiary structure and do not position the ribosomal particle at a strictly determined site in the mRNA. It would be therefore appropriate to consider such enhancers of cap-independent initiation of translation as a special case.
The uniqueness of poly(A) as a leader of mRNA may be explained by its conformational peculiarity. In contrast to poly(U), poly(A) is not quite a “random coil” (see refs. 31 and 32). At physiological temperatures and neutral pH, it has a tendency to form a regular single-stranded helix, and its interaction with proteins and/or ribosomal particles may stabilize this structure. It may be that the single-stranded helix of poly(A) possesses a specific affinity for the mRNA-binding site of the small ribosomal subunit. Indeed, the prokaryotic 30S ribosomal subunit was reported to accommodate the ribosome-bound single-stranded mRNA section in the helical A form (33). It is possible that, in the case of heteronucleotide leader sequences, eIF3 is needed to enhance the affinity of the ribosomal subunit for the leader or induce its single-stranded helix conformation—the role that is played by the anti-Shine-Dalgarno sequence of ribosomal RNA in prokaryotes.
It should be emphasized that in all of our experiments that demonstrated high efficiency of poly(A) leader sequence in the process of initiation complex formation no PABP was present. However, this fact is not necessarily in contradiction with the recent article where stimulation of cap-independent translation due to IRES-like activity by internal poly(A) tracts was reported to be mediated by the yeast Pab1 and eIF4G (30). One can presume that the mechanism of the 48S initiation complex formation in the case of an internal poly(A) tract is different from that of poly(A) leader, but it seems to us unlikely. A more probable explanation is the possibility that PABP and eIF4F/eIF4G were required at later stages of cap-independent translation of mRNAs that were required for the invasive growth in yeast. Such a late eIF4F/eIF4G-dependent stage was suggested by a recent study of the translation acceleration effect during translation of uncapped mRNAs and may result from a noncovalent circularization of polysomes in the course of their formation (34).
In summary, we demonstrated that the initiation on homopolymeric poly(A) tract characteristic of poxvirus mRNA leaders requires only the factors analogous (and in some cases homologous) to prokaryotic IF1, IF1A, and IF2. This implies that poxviruses use the most basic (most ancient) elements of translation initiation machinery that are present in both prokaryotes and eukaryotes, a circumstance that may underlie the independence of poxvirus-specific translation from typically eukaryotic factors, such as eIF3 and eIF4F. This feature of poxviruses (it may be a significant contributor to their virulence) and their use of the prokaryotic phaseless wandering mechanism for finding the initiation codon raise the interesting question of the origin and evolution of poxviruses.
Materials and Methods
Plasmids.
Vectors containing recombinant genes of mammalian eF1, eF1A, eF4A, eF4B, eIF5, eIF5B, and Escherichia coli Met-tRNA synthetase for expression in E. coli were generously provided by T. Pestova (State University of New York, Brooklyn, NY). Plasmid pTZA25Luc containing firefly luciferase coding region with GA25CC 5′ UTR under control of the T7 promoter was constructed in our laboratory.
Translation Initiation Factors and Met-tRNA Synthetase.
Natural and recombinant proteins were isolated and purified generally as described (35) with some modifications. Instead of sucrose gradient centrifugation used in the original protocol to separate eIF3 and eIF4F, the eIF3–eIF4F complex was separated from the fraction of eIF3 by affinity chromatography on 7-methyl-GTP Sepharose 4B column (GE Healthcare). The detailed procedures and the results of SDS/PAGE analyses are in SI Results and Discussion and Figs. S2 and S3.
Ribosomal Subunits.
Isolation and purification of ribosomal subunits were done generally as described (34) with minor modifications (see the detailed procedures and the results of SDS/PAGE analyses of ribosomal subunit proteins in SI Text and Fig. S3).
Natural β-Globin mRNA.
The RNA was isolated from S100 fraction of rabbit reticulocyte lysate by phenol deproteinization and purified by two successive steps of chromatography on an oligo(dT) cellulose (Sigma–Aldrich) column. The details are given in SI Results and Discussion and Fig. S4.
Transcripts.
The transcription reaction mixture contained 80 mM Tris-OAc (pH 7.5), 10 mM KCl, 22.2 mM Mg(OAc)2, 20 mM DTT, 20 mM 2-mercaptoethanol, 2 mM spermidine, 0.01% Triton X-100 (vol/vol), 0.2 mM EDTA, 4 mM ribonucleoside triphosphate (ATP, GTP, CTP, and UTP) each, 80 mg/ml polyethylene glycol 8000, 0.01–0.05 mg/ml DNA template, 0.8 units/ml RNase inhibitor (RiboLock; Fermentas), and 12 units/ml T7 RNA polymerase (Fermentas). The mixture was incubated for 90 min at 37°C, and the reaction was stopped by phenol extraction. The extracted material was then gel-filtered by using either ProbeQuant G-50 micro columns (GE Healthcare) or Superdex 200 HR column (GE Healthcare) and analyzed by denaturing PAGE (Fig. S4).
Aminoacylation of Mammalian tRNAiMet.
The reaction was performed generally as described in ref. 34; detailed procedures can be found in SI Results and Discussion.
Formation of Ribosomal Initiation Complexes.
The assembly of initiation 48S complexes was done as follows: 0.3 pmol of mRNA was added to the ice-cold mixture of 1.5 pmol of 40S ribosomal subunits, 1.2 pmol of eIF2, 1.2 pmol of eIF3, 0.6 pmol of eIF3–eIF4F complex, 15 pmol of eIF1, 15 pmol of eIF1A, 6 pmol of eIF4A, 6 pmol of eIF4B, and 0.6 pmol of Met-tRNAiMet in a buffer containing 40 mM Tris-OAc (pH 7.5), 3.7 mM Mg(OAc)2, 2 mM DTT, 0.25 mM spermidine, 2 mM ATP, 0.2 mM GTP (or guanosine 5′-[β,γ-imido]triphosphate), 0.1 mM EDTA, 120 mM KCl, and 0.3 units/ml RiboLock RNase inhibitor (Fermentas). The reaction mixture volume of 20 ml was incubated at 37°C for 15 min. To form the initiation 80S complex, the above-described mixture was supplemented with 1.2 pmol of eIF5, 0.6 pmol of eIF5B, and 1.5 pmol of 60S ribosomal subunits and further incubated for 15 min at 37°C.
Primer Extension Inhibition Assay (Toeprinting).
To perform primer extension reaction, 0.5 mM dATP, dCTP, dGTP, and dTTP each, 2 pmol/ml DNA primers with fluorescent labels, and 0.3 units/ml AMV RT (Promega) were added to the mixtures with preformed ribosomal initiation complexes. The Mg2+ concentration was adjusted to 7 mM. The reaction mixture was then incubated at 37°C for 45 min. The products of the primer extension reaction were purified with phenol extraction, precipitated in 70% ethanol and 0.7 M NH4OAc, and dissolved in 20 ml of 90% formamide with 89 mM Tris base, 89 mM boric acid, and 2 mM EDTA. Aliquots (0.5 ml) of fluorescent (carboxy-X-rhodamine, CXR) 60- to 400-base DNA size standards (Promega) were added to each sample for capillary electrophoresis. The DNA primers were 5′ [6-carboxyfluorescein]-GGACTCGAAGAACCTCTG 3′ for rabbit β-globin mRNA and 5′ [6-carboxyfluorescein]-GATGTTCACCTCGATATG 3′ or 5′ [6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein, succinimidyl ester]-GATGTTCACCTCGATATG 3′ for A25-Luc mRNA.
Capillary Gel Electrophoresis of Primer Extension Products.
The cDNAs formed in the primer extension reaction were analyzed by capillary gel electrophoresis in an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems) according to the manufacturer's manual. The collected data were processed with GeneMarker 1.5 software (SoftGenetics). Profiles of fluorescent cDNA distribution were attributed to mRNA sequence by using fluorescent (CXR) 60- to 400-base DNA size standards (Promega). Fluorescence intensities corresponding to each cDNA peak were measured to determine the amount of reverse transcription products and thus the amount of ribosomal complexes that resulted in the inhibition of primer extension.
Supplementary Material
Acknowledgments.
We thank Tatyana Pestova and Christopher Hellen for providing one of us (N.E.S.) the possibility to learn the original toeprint analysis technique in their laboratory and for providing us with expression vectors for recombinant mammalian eIFs. We are grateful to Konstantin S. Vassilenko and Vyacheslav A. Kolb for critically reading the manuscript and for useful suggestions. This work would be impossible without the methodological help of Elena Z. Alkalaeva, Lev L. Kisselev, Alexey G. Ryazanov, and Andrey B. Poltaraus. The work was supported by Russian Foundation for Basic Research Grant 06-04-48964-a, by a grant from the President of the Russian Federation (2238.2006.4), and by the Program on Molecular and Cellular Biology of the Russian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804940105/DCSupplemental.
References
- 1.Pestova TV, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–128. [Google Scholar]
- 2.Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna JA, Sarnow P. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 129–153. [Google Scholar]
- 5.Bertholet C, Van Meir E, ten Heggeler-Bordier B, Wittek R. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell. 1987;50:153–162. doi: 10.1016/0092-8674(87)90211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwer B, Visca P, Vos JC, Stunnenberg HG. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell. 1987;50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel DD, Pickup DJ. Messenger RNAs of a strongly-expressed late gene of cowpox virus contain 5′-terminal poly(A) sequences. EMBO J. 1987;6:3787–3794. doi: 10.1002/j.1460-2075.1987.tb02714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright CF, Moss B. In vitro synthesis of vaccinia virus late mRNA containing a 5′ poly(A) leader sequence. Proc Natl Acad Sci USA. 1987;84:8883–8887. doi: 10.1073/pnas.84.24.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwer B, Stunnenberg HG. Vaccinia virus late transcripts generated in vitro have a poly(A) head. EMBO J. 1988;7:1183–1190. doi: 10.1002/j.1460-2075.1988.tb02929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Magistris L, Stunnenberg HG. Cis-acting sequences affecting the length of the poly(A) head of vaccinia virus late transcripts. Nucleic Acids Res. 1988;16:3141–3156. doi: 10.1093/nar/16.8.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ink BS, Pickup DJ. Vaccinia virus directs the synthesis of early mRNAs containing 5′ poly(A) sequences. Proc Natl Acad Sci USA. 1990;87:1536–1540. doi: 10.1073/pnas.87.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulder J, Robertson ME, Seamons RA, Belsham GJ. Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J Virol. 1998;72:8813–8819. doi: 10.1128/jvi.72.11.8813-8819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bablanian R, Goswami SK, Esteban M, Banerjee AK, Merrick WC. Mechanism of selective translation of vaccinia virus mRNAs: Differential role of poly(A) and initiation factors in the translation of viral and cellular mRNAs. J Virol. 1991;65:4449–4460. doi: 10.1128/jvi.65.8.4449-4460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudkov AT, Ozerova MV, Shiryaev VM, Spirin AS. 5′-poly(A) sequence as an effective leader for translation in eukaryotic cell-free systems. Biotechnol Bioeng. 2005;91:468–473. doi: 10.1002/bit.20525. [DOI] [PubMed] [Google Scholar]
- 15.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 16.Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- 17.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould PS, Bird H, Easton AJ. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques. 2005;38:397–400. doi: 10.2144/05383ST02. [DOI] [PubMed] [Google Scholar]
- 19.Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 20.Kassavetis GA, Zentner PG, Geiduschek EP. Transcription at bacteriophage T4 variant late promoter. J Biol Chem. 1986;261:14256–14265. [PubMed] [Google Scholar]
- 21.Cunningham PR, Weitzmann CJ, Ofengand J. SP6 RNA polymerase shutters when initiating from AAA… sequence. Nucleic Acids Res. 1991;17:4669–4673. doi: 10.1093/nar/19.17.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiyama R, Oishi M. In vitro transcription of a poly(dA)·poly(dT)-containing sequences is inhibited by interaction between the template and its transcripts. Nucleic Acids Res. 1996;22:4577–4583. doi: 10.1093/nar/24.22.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn B-Y, Moss B. Capped poly(A) leaders of variable lengths at the 5′ ends of vaccinia virus late mRNAs. J Virol. 1989;63:226–232. doi: 10.1128/jvi.63.1.226-232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 25.Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarabhai A, Brenner S. A mutant which reinitiates the polypeptide chain after chain termination. J Mol Biol. 1967;27:145–162. doi: 10.1016/0022-2836(67)90357-9. [DOI] [PubMed] [Google Scholar]
- 28.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 29.Dorokhov YL, et al. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 31.Cantor CR, Schimmel PR. Biophysical Chemistry. San Francisco: Freeman; 1980. [Google Scholar]
- 32.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 33.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 34.Alekhina OM, Vassilenko KS, Spirin AS. Translation of non-capped mRNAs in a eukaryotic cell-free system: acceleration of initiation rate in the course of polysome formation. Nucleic Acids Res. 2007;35:6547–6559. doi: 10.1093/nar/gkm725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.