Abstract
SecA, the translocation ATPase in Escherichia coli, undergoes cycles of conformational changes (insertion/deinsertion) in response to ATP and a preprotein. The membrane-embedded portion of protein translocase, SecYEG, has crucial roles in the SecA-driven preprotein translocation reaction. We previously identified a secY mutation (secY205) that did not allow an ATP- and preprotein-dependent (productive) insertion of SecA as well as secA mutations that suppressed the secY205 translocation defect. One of the suppressor mutations, secA36, also suppressed the cold-sensitive phenotype of the secG deletion mutant. In vitro experiments at 20°C showed that inverted membrane vesicles lacking SecG were almost inactive in combination with the wild-type SecA protein in translocation of proOmpA as well as in the accompanying ATP hydrolysis. In contrast, the SecA36 mutant protein was found to be able to execute the translocation activity fully at this temperature, even in the absence of SecG. A SecG requirement and its alleviation by the SecA36 alteration also were shown for the SecA insertion reaction. The finding that the SecA36 protein no longer requires assistance from SecG in its insertion and in its catalysis of protein translocation agrees with the idea that SecG normally assists in the functioning of SecA. In agreement with this notion, when the intrinsic SecA function was compromised by a lowered ATP concentration, SecG became essential even at 37°C and even for the SecA36 protein. We propose that in the normal translocase, SecG cooperates with SecA to facilitate efficient movement of preprotein in each catalytic cycle of SecA.
Keywords: suppressor mutation
Protein translocation across a biological membrane is facilitated by multiple protein factors. Periplasmic and outer membrane proteins of Escherichia coli are synthesized in the cytosol as precursor forms, which are then chaperoned by SecB to the inner (plasma) membrane (1–3). They are then captured by SecA, an ATPase that drives protein movement into and across the membrane (2, 4, 5). During this catalysis, SecA undergoes conformational changes. After binding of ATP and a preprotein, a substantial portion of SecA inserts into the membrane, followed by ATP hydrolysis-coupled release of the bound precursor and deinsertion of SecA (6, 7). The released precursor may be translocated further by the action of the proton-motive force (pmf) across the membrane (7–9). Several integral membrane proteins also participate in the translocation reactions. They include SecY, SecE, SecG, SecD, and SecF (10, 11). Among them, SecY and SecE are the essential components within the membrane; reconstitution studies showed that proteoliposomes containing these two proteins can support SecA-dependent preprotein translocation (12, 13). However, efficient translocation activity requires SecG as well (14–16).
These membrane-embedded components play multiple roles. It is generally believed that these proteins form a channel-like pathway for transit of a preprotein. SecY and SecE provide the high-affinity binding sites for SecA, and the SecYEG complex supports the translocation ATPase activity of SecA (2, 5, 17). The SecA insertion reaction also depends on SecYEG (7, 18, 19). We previously identified a secY mutant (secY205) with a cold-sensitive protein-export defect. Inverted membrane vesicles (IMV) from the secY205 mutant did not support the SecA translocation ATPase reaction effectively (20), nor did they support the preprotein- and ATP-dependent SecA insertion reaction in vitro (19). This mutation causes an amino acid alteration in the most carboxyl-terminal cytoplasmic domain of SecY (21). A suppressor variant of SecA, named SecA36, which has an amino acid substitution near the high-affinity ATP-binding site, was shown to be able to insert into the mutant IMV and to drive protein translocation into otherwise inactive mutant IMV (19). Tokuda et al. (22) showed that SecG undergoes a remarkable topology inversion concomitant with SecA insertion. They proposed that SecG, having two transmembrane segments, assists in SecA function by changing its orientation in the membrane.
Whereas SecG was identified by biochemical studies (12, 14, 16), its role in vivo was established by subsequent genetic studies. The secG-disrupted mutation causes cold-sensitive growth of cells of certain genetic backgrounds. At low temperature, protein export in the ΔsecG mutant is retarded severely (23). Some secG mutations confer tolerance to the expression of an artificial hybrid protein between certain eukaryotic proteins and alkaline phosphatase (24), whereas others suppress signal sequence mutations (25). These results reinforce the idea that SecG is a component of protein translocase, although it is not absolutely required at 37°C.
Although the genetic suppression by the secA36 mutation was specific for the secY205 mutation among the various cold-sensitive secY mutations that were examined, we noted that the secA mutation suppressed the cold-sensitivity of the secG-disrupted strain (19). We characterized this suppression in vitro. The SecA36 form of SecA indeed gained the ability to catalyze efficient protein translocation without the aid of SecG. However, we found that SecG was required at lowered concentrations of ATP even when translocation was driven by the SecA36 form of SecA. SecG’s roles in protein translocase are discussed in light of these findings. We suggest that SecG cooperates with SecA to facilitate efficient protein movement coupled with the SecA reaction cycle.
MATERIALS AND METHODS
Materials.
IMVs were prepared from strains GN67 (secG+) and GN68 (ΔsecG∷kan) as described (26) and washed with 6 M urea (20). Western blotting experiments showed that the SecY content of IMV from the ΔsecG mutant was not significantly different from that of the wild-type cells. The wild-type SecA protein and the SecA36 mutant protein were overproduced and purified as described (19). SecB was purified as described (26). The precursor form of OmpA (proOmpA) was purified as described (27, 28). Na[125I]I (17.5 Ci/mg of I, 100 mCi/ml) and [35S]methionine (1,175 Ci/mmol) were purchased from ICN and from American Radiolabeled Chemicals (St. Louis), respectively. ATP, 5′-adenylyl imidodiphosphate, and trypsin (treated with l-1-tosylamido-2-phenylethyl chloromethyl ketone) were purchased from Sigma.
E. coli Strains.
The secG deletion mutant of E. coli, KN370 (ΔsecG∷kan in the background of C600 recD1009) was described by Nishiyama et al. (23). Strains GN68 [MC4100, ΔsecG∷kan ΔompT Δunc(B–C) ilv∷Tn10] and GN67 [MC4100, ΔompT Δunc(B–C) ilv∷Tn10 secG+] were constructed by appropriate P1vir-mediated transductions (29) with strains KN370, MC4100 (30), AD179 (MC4100, ΔompT; ref. 31), and DK8 (Δunc(B–C) ilv∷Tn10; ref. 32) either as donors or recipients.
Measurement of SecA Translocation ATPase.
A reaction mixture (150 μl) contained urea-washed IMV (40 μg of protein per ml), SecA (40 μg/ml), and ATP (2 mM) in buffer containing 50 mM Tris⋅HCl (pH 7.2), 30 mM KCl, 30 mM NH4Cl, and 5 mM magnesium acetate. It was supplemented further with or without 6 μl of proOmpA (4.6 μg in 8 M urea). After incubation at 20°C, 30 μl of sample was removed, and the liberated inorganic phosphate was quantified by the malachite green method (5, 33). To determine translocation ATPase activity, values in the absence of proOmpA were subtracted from those in its presence.
In Vitro Protein Translocation.
35S-labeled proOmpA was synthesized in vitro and subjected to the in vitro translocation reaction as described (19). The reaction mixture contained IMV (150 μg of protein per ml from a specified strain), SecA (150 μg/ml, either wild-type or SecA36), SecB (25 μg/ml), ATP (2 mM, unless otherwise indicated), and an ATP regeneration system (50 mM phosphocreatine and 100 μg/ml creatine kinase) in 50 mM Tris⋅HCl buffer, pH 8.0, supplemented with 50 mM KCl/5 mM MgCl2/200 μg/ml BSA/10 mM 2-mercaptoethanol. After incubation at 37°C or 20°C for indicated lengths of time, samples were chilled on ice and treated with 1 mg/ml trypsin at 0°C for 15 min followed by trichloroacetic acid precipitation and SDS/PAGE. Translocated OmpA was quantitated with a Fuji phosphorimager BAS2000.
Insertion of the 30-kDa Segment of SecA into IMV.
SecA and SecA36 were covalently modified with 125I as described by Economou and Wickner (6) with minor modifications; a 200-μl solution containing 50 mM Tris⋅HCl (pH 8.0), 1 mg of SecA, 0.3 mCi of Na[125I]I, 50 mM KCl, 5 mM MgCl2, and 10% glycerol was added to a tube coated with 100 μg of Iodo-Gen (Pierce) and incubated for 20 min on ice. SecA insertion assay (6, 19) was carried out with IMVs (100 μg of protein per ml), 125I-SecA (100 μg/ml), proOmpA (46 μg/ml), SecB (25 μg/ml), ATP (2 mM), and an ATP regeneration system (50 mM phosphocreatine and 100 μg/ml creatine kinase), in 50 mM Tris⋅HCl buffer, pH 8.0, supplemented with 50 mM KCl/5 mM MgCl2/200 μg/ml BSA/10 mM 2-mercaptoethanol. Samples were treated with 1 mg/ml trypsin at 0°C for 15 min before SDS/PAGE and autoradiography.
RESULTS
A secA Mutation That Suppresses the Cold-Sensitive Phenotype of the secG Deletion Strain.
Previously, we isolated secA mutations that suppressed the cold-sensitive protein-export defect of the secY205 mutant (19). It was found that one of them, the secA36 mutation causing an Ala-112 to Val substitution in the high-affinity ATP-binding region of SecA, suppressed the cold-sensitivity of strain KN370 as well. In this strain, the secG gene has been replaced by the kan determinant (ΔsecG∷kan), and it is incapable of growing at 20°C at a significant rate (23). The ΔsecG∷kan cells showed markedly slower precursor-to-mature processing rates of maltose-binding protein and OmpA as compared with the isogenic secG+ strain. Introduction of the secA36 mutation into KN370 resulted in the restoration of the processing rates to nearly wild-type levels (ref. 19; data not shown). Bost and Belin (24) reported that the ΔsecG∷kan mutation did not cause cold-sensitive growth in certain different genetic backgrounds. We constructed isogenic derivatives of strain MC4100 carrying either ΔsecG∷kan, secA36, or both. Although none of these strains exhibited clear cold-sensitivity for cell growth, pulse–chase experiments gave results that were qualitatively similar to those obtained in the strains of the KN370 background (data not shown). We used a MC4100-derived ΔsecG∷kan strain that contained additional ΔompT and Δunc mutations for preparation of IMVs.
Wild-Type SecA Cannot Effectively Drive Protein Translocation into the SecG-Lacking IMV at 20°C, but the SecA36 Form of SecA Can Do So.
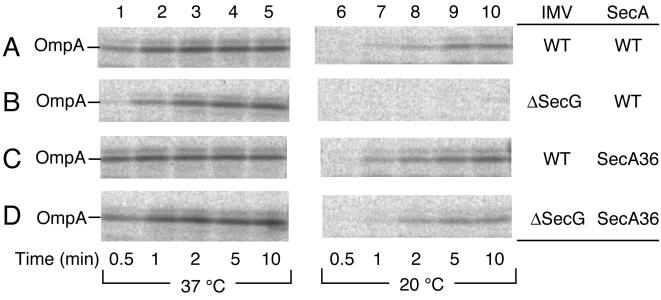
IMVs from secG+ and ΔsecG∷kan cells were washed with 6 M urea to remove and inactivate endogenous SecA. Posttranslational translocation of 35S-labeled proOmpA protein into the urea-washed IMVs was examined in combination with purified SecA+ or SecA36. Translocation was negligible without added SecA and ATP. When proOmpA, wild-type SecA, and excess (2 mM) ATP were incubated with urea-washed wild-type IMV at 37°C, the proportion of translocated proOmpA molecules increased rapidly and reached the maximum level after 1 min of incubation (Fig. 1A, lanes 1–5). This was true also when SecA36 was used instead of SecA+ (Fig. 1C, lanes 1–5). The reaction with ΔsecG IMV in combination with SecA+ was slightly slower than that with the wild-type combination, taking more than 2 min to reach the maximal translocation (Fig. 1B, lanes 1–5). When proOmpA translocation into the ΔsecG IMV was driven by the SecA36 protein, it occurred as rapidly as the standard reaction (Fig. 1D, lanes 1–5).
Figure 1.
SecA36 can drive in vitro translocation of proOmpA into ΔsecG IMV. Urea-washed IMV, either from secG+ strain (GN67; A and C) or the ΔsecG∷kan mutant (GN68; B and D), were combined with wild-type SecA protein (A and B) or the SecA36 mutant protein (C and D) for translocation of 35S-labeled proOmpA. The translocation reaction was allowed at 37°C (lanes 1–5) or at 20°C (lanes 6–10). Samples were withdrawn at 0.5 (lanes 1 and 6), 1 (lanes 2 and 7), 2 (lanes 3 and 8), 5 (lanes 4 and 9), and 10 (lanes 5 and 10) min after initiation of reaction, treated with trypsin, and subjected to SDS/PAGE. Translocated OmpA protein was visualized by phophorimager.
We then examined proOmpA translocation at 20°C. Wild-type IMV in combination with either SecA+ or SecA36 supported proOmpA translocation that reached the maximum level at about 5 min after initiation of the reactions at this temperature (Fig. 1 A and C, lanes 6–10). The ΔsecG IMV did not support any significant translocation when it was combined with SecA+ (Fig. 1B, lanes 6–10). However, when the same IMV preparation was combined with SecA36, proOmpA translocation was restored to a normal level (Fig. 1D, lanes 6–10). Thus, we were able to reproduce in vitro the temperature-dependent defect in protein translocation in the absence of SecG as well as suppression of the defect by the SecA36 mutant form of the SecA ATPase. Experiments with chemically purified proOmpA protein gave similar results (data not shown). The SecA36 alteration makes SecA no longer require SecG for efficient catalysis of protein translocation.
SecG Dependence of the SecA Actions.
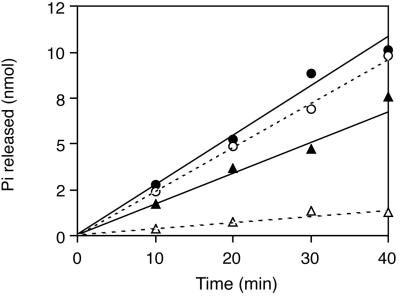
To examine the mutational effects on the SecA functions more directly, we measured the SecA translocation ATPase activity. Wild-type IMV supported proOmpA-dependent ATPase activity at 20°C, which was almost identical in SecA+ (Fig. 2, open circles) and SecA36 (Fig. 2, solid circles). In contrast, ΔsecG IMV did not effectively support the translocation ATPase activity of SecA+ at this temperature (Fig. 2, open triangles), whereas it supported a nearly normal level of the SecA36 activity (Fig. 2, solid triangles).
Figure 2.
Translocation ATPase activity of SecA+ and SecA36 in combination with secG+ IMV or ΔsecG IMV. Translocation ATPase was assayed at 20°C with the wild-type SecA protein (open symbols) or the SecA36 mutant protein (solid symbols) in combination with 6 M urea-washed IMV from the wild-type strain (GN67; circles) or from the ΔsecG∷kan mutant (GN68; triangles). The activity of wild-type SecA at this temperature was about 25% of the activity at 37°C.
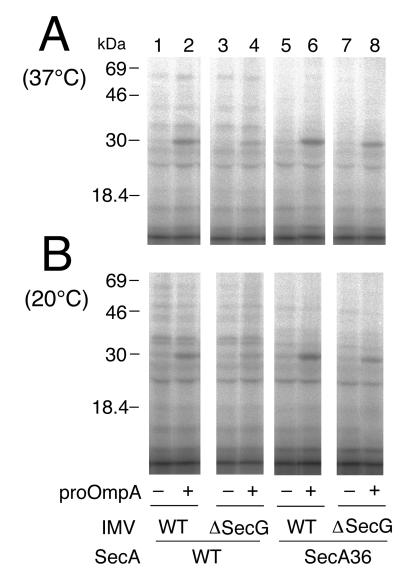
SecA insertion into IMVs was then examined. For this purpose, the SecA+ and the SecA36 proteins were iodinated with 125I and incubated at 37°C or at 20°C with either wild-type IMV or ΔsecG IMV in the presence of ATP. Reactions in the presence and absence of proOmpA were compared. Samples were chilled and treated with trypsin, and 125I-labeled protein fragments were visualized after SDS/PAGE. As shown in Fig. 3 (lanes 2 and 6), reactions with wild-type IMV produced a prominent band with a mass of 30 kDa that appeared only when proOmpA had been included. Although other bands were also seen, they were all preprotein-independent (Fig. 3; compare lanes 1 and 2 as well as lanes 5 and 6). The generation of the 30-kDa band depended also on the presence of ATP (data not shown), indicating its identity with the originally described, inserted fragment of SecA (6).
Figure 3.
Insertion of SecA+ and SecA36 into secG+ IMV vs. ΔsecG IMVs. 125I-SecA (100 μg/ml and 1.33 × 105 cpm/μg; lanes 1–4) and 125I-SecA36 (100 μg/ml and 2 × 105 cpm/μg; lanes 5–8) were subjected to incubation with urea-treated IMV (100 μg of protein per ml) from either the wild-type cells (GN67; lanes 1, 2, 5, and 6) or the ΔsecG∷kan cells (GN68; lanes 3, 4, 7, and 8), in the presence of ATP/ATP regeneration system and SecB, and in the presence (lanes 2, 4, 6, and 8) or absence (lanes 1, 3, 5, and 7) of proOmpA. After incubation at 37°C (A) or 20°C (B) for 15 min, samples were treated with trypsin. 125I-labeled protein fragments were separated by SDS/PAGE and visualized by a phosphorimager. The positions of molecular-mass markers are indicated on the left.
Insertion of the wild-type SecA decreased when the ΔsecG IMV was used (Fig. 3 A and B, lane 4). Comparison of the background bands indicated that the reduction seemed somewhat more pronounced at 20°C (Fig. 3B) than at 37°C (Fig. 3A). In contrast, the SecA36 mutant protein was capable of inserting into the ΔsecG IMV with only slightly reduced efficiencies (Fig. 3B, lane 8) as compared with its insertion into wild-type IMV (Fig. 3B, lane 6).
SecG Is Absolutely Required in the Presence of Lowered Concentrations of ATP.
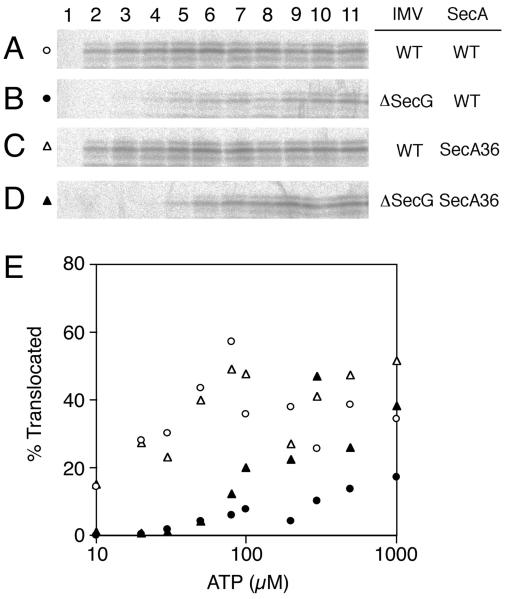
To examine the relationships between the SecG function and the SecA function, we measured the translocation activity of various SecA/IMV combinations at lowered ATP concentrations at 37°C. Because an ATP regeneration system was included in the assays, the initial ATP concentration should have been maintained during the entire reaction. When the wild-type IMV was combined with either SecA+ or SecA36, the proportions of translocated proOmpA varied between ≈20% and 50% (Fig. 4 A and C; Fig. 4E, open symbols). No translocation occurred without any ATP (Fig. 4 A–D, lane 1). In contrast, translocation of proOmpA into the ΔsecG IMV was affected drastically by ATP concentrations (Fig. 4 B and D; Fig. 4E, solid symbols). Essentially no translocation took place into the ΔsecG IMV in the presence of 30 μM or lower concentrations of ATP, irrespective of whether SecA+ or SecA36 was used (Fig. 4E, solid circles and solid triangles, respectively). The translocation yield became significant at 50 μM ATP and increased thereafter, reaching the maximum levels (about 15% translocation with SecA+ and about 40% translocation with SecA36) at a few hundred μM ATP. Thus, whether proOmpA translocation into the ΔsecG IMV was driven by wild-type SecA or by SecA36, the profiles of ATP-dependence were similar in that the activity diminished at lower ATP concentrations. In the above experiments, a translocation intermediate (8) was observed to accumulate when ATP concentration was lowered to 10 μM (with wild-type IMV) or to 30–100 μM (with ΔsecG IMV; data not shown).
Figure 4.
ATP-dependence of in vitro translocation of proOmpA into SecG+ vs. ΔsecG IMVs. (A–D) Translocation of [35S]proOmpA was allowed at 37°C for 15 min, as described for Fig. 1, by using urea-washed IMVs from either the wild-type cells (GN67; A and C) or the ΔsecG cells (GN68; B and D) in combination with either wild-type SecA protein (A and B) or the SecA36 mutant protein (C and D). ATP concentrations were 0 (lane 1), 10 (lane 2), 20 (lane 3), 30 (lane 4), 50 (lane 5), 80 (lane 6), 100 (lane 7), 200 (lane 8), 300 (lane 9), 500 (lane 10), or 1,000 (lane 11) μM. After treatment with trypsin and SDS/PAGE, radioactive OmpA was visualized by phosphorimager. (E) The results in A–D were graphically depicted after quantification of the radioactive full-length mature OmpA by using BAS-1800 phosphorimager. Open symbols, wild-type IMV; solid symbols, ΔsecG IMV; circles, SecA+; triangles, SecA36.
These results indicate that SecA36 does not alleviate the defect of the translocation system lacking SecG when ATP is insufficiently available. It should be noted that no pmf was imposed in these experiments and that the IMVs lacked the F0-F1 ATPase, preventing any exchange between ATP and pmf.
To address whether the inability to function at lower ATP concentrations was specific for the lack of SecG or whether it was some nonspecific consequence of the lowered translocation activity, we examined ATP-dependence of translocation with the secY205 IMV. Although translocation into this IMV, which has the altered SecY protein, was inefficient (19), it was affected only moderately even when the ATP concentration was lowered to 10 μM (data not shown). Thus, there is a clear distinction between the IMV that lacked SecG and the secY205 IMV that retained SecG. Although both mutational defects are suppressible by the SecA36 variant of SecA (19), only the ΔsecG IMV is incapable of supporting protein translocation at the lowered concentrations of ATP. At least in vitro, SecG enables efficient protein translocation in the presence of an insufficient ATP supply.
DISCUSSION
The SecG protein stimulates protein translocation when reconstituted into proteoliposomes together with SecY and SecE (14, 34). Its requirement is especially evident at low temperature and in the absence of pmf across the membrane (35). The SecG requirement in vivo is also evident at 20°C (23). Although the extent of the cold-sensitivity of the secG-disruption mutation varied in different genetic backgrounds (24), retardation of protein export was observed at 20°C even in the strain background that did not yield a clearly cold-sensitive growth phenotype. Nishiyama et al. (22) reported that SecG undergoes an inversion of its topology in the membrane under the conditions that would allow insertion of SecA and a preprotein into the membrane. They proposed that SecG assists in SecA function through the coordinated topology inversion (of SecG) and membrane insertion (of SecA). SecG indeed stimulates the SecA insertion reaction in vitro (17). More recently, it was shown that the secG deletion mutation and a cold-sensitive secA mutation are synthetically lethal (36).
The consequence of the gain-of-ability alteration of SecA provides strong evidence for the SecG’s role in assisting SecA. The SecA36 form of SecA, with an Ala to Val amino acid change near the primary ATP-binding site, alleviates the cold-sensitive protein export and growth defects of the secG-deleted mutant in vivo. Although the mutational alteration is adjacent to the prlD4 alteration (37), the secA36 mutation does not suppress a lamB signal sequence mutation (G.M., unpublished results). Purified SecA36 protein can efficiently drive protein translocation into the SecG-lacking IMV, which does not allow the wild-type SecA protein to function at 20°C. The SecA36-driven protein translocation into the ΔsecG IMV is accompanied by wild-type levels of ATP hydrolysis. Although Duong and Wickner (17) reported that their SecG-lacking IMV was as active as the wild-type IMV in supporting the translocation ATPase (of wild-type SecA) at 37°C, our results indicated clearly that IMV without SecG resulted in severe defects in SecA function at low temperature.
Our assays for the steady-state levels of SecA insertion showed that wild-type SecA inserted into the ΔsecG IMV less efficiently than into the wild-type IMV, especially at 20°C. In contrast, the SecA36 mutant protein inserted into both IMVs with comparable steady-state efficiency. Thus, SecA actions that are stimulated by SecG include the membrane-insertion reaction, in which the SecA36 mutant protein circumvents the SecG requirement. The trypsin-resistant 30-kDa fragment of SecA is observable in the presence of both ATP and proOmpA but not of either alone (6, 7, 19). It has been noted that a similar conformational change in SecA can take place in the absence of a preprotein (7, 19) or even in the absence of membranes (38), provided that a nonhydrolyzable ATP analog is used instead of ATP. However, conformational changes observed under the latter condition should be regarded as distinct from the productive SecA insertion reaction that requires both ATP and a preprotein (39). We previously observed that the secY205 mutation selectively abolished the ATP- and proOmpA-dependent reaction and that this defect was overcome by the SecA36 alteration (19). We have shown here that the SecA36 alteration enhanced the preprotein-dependent mode of SecA insertion into the ΔsecG IMV (Fig. 3). The higher than normal ATPase activity of SecA36 (Fig. 2) is consistent with this idea but not with a lowered-deinsertion reaction being the cause of an apparent enhancement of the steady-state insertion level.
These results, taken together, establish that SecA can be mutated to a form that no longer requires SecG for its function at 20°C. It is in turn suggested that wild-type SecA requires the assistance from SecG to function effectively at this temperature.
In this study, we have shown that low ATP concentration and the absence of SecG resulted in the synergistic inability of translocase. The ATP requirement profiles in translocation were different between the normal IMV and those lacking SecG. The latter IMVs were almost inactive even at 37°C when the ATP concentration was lowered to 30 μM or lower, where the secG+ IMVs were only moderately affected. Although the SecA36 protein (in the presence of sufficient ATP) circumvents the translocation defect of the ΔsecG IMV, it did not alleviate the synthetic defect caused by the low ATP concentration and the lack of SecG. The simplest explanation ascribes this inability to the lack of sufficient ATP for SecA36 to work in the absence of SecG. In contrast, the SecA36-stimulated translocation into the secY205 mutant IMV was “normal” with respect to the ATP-dependence profile. The inability to function at lower ATP concentrations seems to be a specific feature of translocase that lacks SecG. It might be argued that SecG itself has a role in utilizing ATP efficiently, but no SecG–ATP interaction is known. Rather, SecG may compensate for the lowered activity of SecA. Hanada et al. (35) suggested that pmf and SecG have overlapping functions; the impairment of both results in severe defects in translocase function. Although the present assay system precluded the contribution of pmf, increased ATP dependence (Fig. 4) and increased pmf dependence (35) of the ΔsecG IMV might have a common ground. When the preprotein is released from SecA, pmf facilitates its forward movement. Pmf also prevents the backward movement of the translocating chain (8, 9).
We propose that SecG also facilitates effective movement of the preprotein. The consequence of the lack of this function may become apparent either at 20°C where protein movement is restricted intrinsically (40) or under conditions where the ATP supply declines below a critical level. One possible mechanism for the SecG function might be that SecG directly interacts with a preprotein (25), preventing its backward movement. Another possibility is that SecG facilitates protein movement in close conjunction with SecA. This cooperation would not only facilitate SecA insertion but also increase the yield of protein movement per insertion/deinsertion cycle of SecA. The observation of Duong and Wickner (17) that the absence of SecG results in the futile ATPase activity at 37°C is consistent with this role of SecG.
Although SecG is required for translocation at 20°C, this requirement can be alleviated by SecA36 alteration in the presence of sufficient ATP. Thus, the SecA36 protein may have an advantage in catalysis of protein translocation. Why was such a form of SecA not selected during evolution? It is conceivable that a translocation system with such a hyperactive SecA without SecG may actually be disadvantageous because of, for instance, a lack of regulation in channel gating or in controlled delivery of a hydrophobic stretch into the lipid phase during translocation. It is also true that the SecA36 form of SecA does not substitute for SecG completely, although we do not know under what conditions the cellular level of ATP declines to below 30 μM. Our measurements so far showed that the ATP content of KN370 cells was about one-fourth of that of cells of the MC4100 background (G.M., unpublished results).
Further elucidation of the mechanisms underlying the cooperation between SecA and SecG, the two most dynamic components of protein translocase (6, 22), will be important for our understanding of how translocase activities have been optimized in living organisms.
Acknowledgments
We thank Yoshinori Akiyama for bacterial strains and for critical discussion, Hajime Tokuda for the ΔsecG∷kan strain, Takayuki Homma for help in strain constructions, Tohru Yoshihisa for discussion, and Kiyoko Mochizuki, Yusuke Shimizu, and Toshiki Yabe for technical support. This work was supported by grants from The Ministry of Education, Science and Culture, Japan, from Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, and from the Human Frontier Science Program Organization.
ABBREVIATIONS
- IMV
inverted membrane vesicle
- pmf
proton-motive force
References
- 1.Randall L L. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]
- 2.Hartl F-U, Lecker S, Schiebel E, Hendrick J P, Wickner W. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 3.Fekkes P, van der Does C, Driessen A J M. EMBO J. 1997;16:6015–6133. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lill R, Cunningham K, Brundage L A, Ito K, Oliver D, Wickner W. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lill R, Dowhan W, Wickner W. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 6.Economou A, Wickner W. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 7.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 8.Schiebel E, Driessen A J M, Hartl F-U, Wickner W. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 9.Driessen A J M. EMBO J. 1992;11:847–853. doi: 10.1002/j.1460-2075.1992.tb05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito K. Genes Cells. 1996;1:337–346. doi: 10.1046/j.1365-2443.1996.34034.x. [DOI] [PubMed] [Google Scholar]
- 11.Wickner W, Leonard M R. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 12.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 13.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Proc Natl Acad Sci USA. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiyama K, Mizushima S, Tokuda H. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brundage L, Fimmel C J, Mizushima S, Wickner W. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 16.Douville K, Leonard M, Brundage L, Nishiyama K, Tokuda H, Mizushima S, Wickner W. J Biol Chem. 1994;269:18705–18707. [PubMed] [Google Scholar]
- 17.Duong F, Wickner W. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douville K, Price A, Eichler J, Economou A, Wickner W. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto G, Yoshihisa T, Ito K. EMBO J. 1997;16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taura T, Yoshihisa T, Ito K. Biochimie (Paris) 1997;79:517–521. doi: 10.1016/s0300-9084(97)82744-7. [DOI] [PubMed] [Google Scholar]
- 21.Taura T, Akiyama Y, Ito K. Mol Gen Genet. 1994;243:261–269. doi: 10.1007/BF00301061. [DOI] [PubMed] [Google Scholar]
- 22.Nishiyama K, Suzuki T, Tokuda H. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 23.Nishiyama K, Hanada M, Tokuda H. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bost S, Belin D. EMBO J. 1995;14:4412–4421. doi: 10.1002/j.1460-2075.1995.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bost S, Belin D. J Biol Chem. 1997;272:4087–4093. doi: 10.1074/jbc.272.7.4087. [DOI] [PubMed] [Google Scholar]
- 26.Yoshihisa T, Ito K. J Biol Chem. 1996;271:9429–9436. doi: 10.1074/jbc.271.16.9429. [DOI] [PubMed] [Google Scholar]
- 27.Crooke E, Brundage L, Rice M, Wickner W. EMBO J. 1988;7:1831–1835. doi: 10.1002/j.1460-2075.1988.tb03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato K, Mori H, Yoshida M, Tagaya M, Mizushima S. J Biol Chem. 1997;272:20082–20087. doi: 10.1074/jbc.272.32.20082. [DOI] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 201–205. [Google Scholar]
- 30.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. p. 14. [Google Scholar]
- 31.Akiyama Y, Ito K. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 32.Klionsky D J, Brusilow W S A, Simoni R D. J Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 34.Hanada M, Nishiyama K, Mizushima S, Tokuda H. J Biol Chem. 1994;269:23625–23631. [PubMed] [Google Scholar]
- 35.Hanada M, Nishiyama K, Tokuda H. FEBS Lett. 1996;381:25–28. doi: 10.1016/0014-5793(96)00066-x. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H, Nishiyama K, Tokuda H. Mol Microbiol. 1998;29:331–342. doi: 10.1046/j.1365-2958.1998.00937.x. [DOI] [PubMed] [Google Scholar]
- 37.Huie J L, Silhavy T J. J Bacteriol. 1995;177:3518–3526. doi: 10.1128/jb.177.12.3518-3526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J M. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 39.Eichler J, Rinard K, Wickner W. J Biol Chem. 1998;273:21675–21681. doi: 10.1074/jbc.273.34.21675. [DOI] [PubMed] [Google Scholar]
- 40.Pogliano J K, Beckwith J. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]