Abstract
The function of the Ets-1 transcription factor is regulated by two regions that flank its DNA-binding domain. A previously established mechanism for auto-inhibition of monomeric Ets-1 on DNA response elements with a single ETS-binding site, however, has not been observed for the stromelysin-1 promoter containing two palindromic ETS-binding sites. We present the structure of Ets-1 on this promoter element, revealing a ternary complex in which protein homo-dimerization is mediated by the specific arrangement of the two ETS-binding sites. In this complex, the N-terminal-flanking region is required for ternary protein–DNA assembly. Ets-1 variants, in which residues from this region are mutated, loose the ability for DNA-mediated dimerization and stromelysin-1 promoter transactivation. Thus, our data unravel the molecular basis for relief of auto-inhibition and the ability of Ets-1 to function as a facultative dimeric transcription factor on this site. Our findings may also explain previous data of Ets-1 function in the context of heterologous transcription factors, thus providing a molecular model that could also be valid for Ets-1 regulation by hetero-oligomeric assembly.
Keywords: Ets-1, stromelysin-1 promoter, X-ray crystallography
Introduction
Controlled transcription in eukaryotes involves the combinatorial action of multiple transcription factors on specific target promoters (Carey, 1998; Sieweke and Graf, 1998; Remenyi et al, 2004). The potential of transcription factors for DNA-mediated oligomerization, either by self-assembly or by complex formation with other transcription factors, presents one of the fundamental mechanisms to control their specificity and activity. Many of these factors require obligatory dimer formation, such as leucine zipper-containing transcription factors (Ferre-D'Amare et al, 1993, 1994; Glover and Harrison, 1995; Nair and Burley, 2003). However, members of the Ets-1 transcription factor family generally bind to single Ets-binding sites (EBS), comprising a 5′-GGAA/T-3′ core motif, by a monomeric Ets-1 DNA-binding (ETS) domain with a winged helix-turn-helix element (Wasylyk et al, 1991; Sharrocks, 2001). The DNA recognition helix H3 of different ETS domains inserts into the DNA major groove, which includes the EBS motif. This helix provides a series of specific base interactions by a set of three highly conserved residues that serve as a marker of this transcription factor family (Garvie and Wolberger, 2001).
The study of Ets factors in general and Ets-1 in particular has also provided a general paradigm showing how such regulation can be achieved by intramolecular auto-inhibition involving two regions flanking the ETS domain (reviewed in Sharrocks, 2001; Pufall and Graves, 2002). Previous structural studies have demonstrated that, in the absence of DNA, these two regions contain two inhibitory α-helices each (HI-1/HI-2 and HI-4/HI-5, respectively). NMR and CD studies have revealed that, upon DNA binding, these regions undergo conformational changes involving the collapse of the first inhibitory helix HI-1 from the N-terminal ETS-flanking region (Petersen et al, 1995; Garvie et al, 2002; Lee et al, 2005). Whereas phosphorylation has been shown to accentuate the inhibitory activity of these domains (Cowley and Graves, 2000; Pufall et al, 2005), it has been postulated that auto-inhibition may be relieved by interaction with heterologous transcription factors (Kim et al, 1999; Goetz et al, 2000; Sharrocks, 2001; Garvie et al, 2002; Pufall and Graves, 2002). Indeed, Ets-1 cooperatively binds to composite DNA-binding motifs with Pax-5 (Fitzsimmons et al, 1996; Garvie et al, 2001), USF-1 (Sheridan et al, 1995; Sieweke et al, 1998), NF-κB (Dickinson et al, 1999), c-Jun (Wasylyk et al, 1991; Bassuk and Leiden, 1995) and Runx-1 (Kim et al, 1999; Goetz et al, 2000; Gu et al, 2000; Garvie et al, 2002). Some of these interactions have been shown to be directly mediated by contact surfaces in the auto-inhibitory domains (Kim et al, 1999; Goetz et al, 2000; Gu et al, 2000).
To date, structural insight into the molecular interactions of these assemblies, however, is still largely lacking. The only ternary complex with an available 3D structure is that of the Ets-1–Pax-5–mb-1 promoter complex, which was crystallized using Ets-1(Δ280) with the ETS domain and the N-terminal-flanking region, and Ets-1(Δ331) with the ETS domain only (Garvie et al, 2001, 2002). In this complex, however, only the ETS domain is involved in interactions with Pax-5. Also worthy of mention is the structure of the ternary SAP-1–SRF–SRE complex, involving the distantly related ETS transcription factor SAP-1 (Hassler and Richmond, 2001; Mo et al, 2001). Another special case is provided by the structure of the ternary protein–DNA complex of the ETS transcription factor GABPα, which assembles with the ankyrin repeat containing GAPBβ subunit (Batchelor et al, 1998). Although the ETS domains of SAP-1 and GABPα adopt different mechanisms of auto-regulation (Buchwalter et al, 2004), they have in common that the ETS domain assembly with heterologous transcription factors is mediated or enhanced by a specific EBS element.
In contrast to promoters with single EBS motifs, the stromelysin-1 promoter comprises a palindromic, inverted repeat arrangement of two EBS separated by four base pairs (Baillat et al, 2002). Stromelysin-1 (matrix metalloproteinase-3) is an extracellular proteolytic enzyme that has an important function in the homoeostasis and morphogenesis of connective tissue and basement membrane components (Nicholson et al, 1989). Regulation of stromelysin-1 transcription is essentially associated with Ets-1 and closely related transcription factors such as Ets-2 (Buttice et al, 1996). This finding is supported by the correlated increase of the coexpression level of stromelysin-1 and Ets-1, observed in pathological processes (Wernert et al, 1994; Naito et al, 2000; Sun and Yokota, 2001) and by a decreasing expression level of stromelysin-1 when Ets-1 or Ets-2 expression is downregulated or depleted (Man et al, 2003; Ozaki et al, 2003; Rothhammer et al, 2004; Baillat et al, 2006). In addition, biochemical data demonstrated that activation of this promoter requires two Ets-1 molecules that bind cooperatively to the promoter (Baillat et al, 2002). In contrast to the auto-inhibitory effects on monomeric EBS promoters, comparison of two Ets-1 isoforms, p51 and p42, revealed that the region encoded by exon VII (245–330, lacking in p42 isoform) is required for recruitment of a second Ets-1 molecule, leading to cooperative binding and transactivation of the stromelysin-1 promoter (Baillat et al, 2002). These findings led to a model where auto-inhibition by this flanking region is relieved through homo-dimeric interactions in the presence of the stromelysin-1 promoter palindromic dual EBS site, as opposed to DNA response elements with single EBS motifs, where heterologous transcription factor partners are usually required to activate ETS. However, the underlying molecular mechanism has remained unknown, to date. Similar data were also obtained for the P53 promoter that contains a similar palindromic EBS element (Venanzoni et al, 1996).
Here, we have selected the palindromic EBS element of the stromelysin-1 promoter (S-EBS) as a prototype to unravel the molecular basis for the DNA-mediated self-assembly of Ets-1 and to study the role of the ETS-flanking regions in this process. The X-ray structure of the (Ets-1)2–S-EBS complex demonstrates how this transcription factor dimerizes by forming a central protein–DNA interface that involves several residues from a loop connecting the N-terminal-flanking region and the ETS domain. Our structural findings are further supported by biochemical data and transactivation data. Unlike the previously established Ets-1 interaction with monomeric EBS motifs (Pufall and Graves, 2002), our findings unravel how the N-terminal ETS-flanking sequence segment enables relief of auto-inhibition by ternary complex formation of Ets-1 with itself, which may serve as a model for interactions with other partners on multi-site DNA response motifs.
Results
Overall structure of the dimeric Ets-1–S-EBS complex
The crystal structure of the dimeric (Ets-1)2–S-EBS complex was determined at 2.58-Å resolution (Table I and Figure 1). For crystallization, we used a truncated Ets-1(Δ280) fragment that includes the DNA-binding ETS domain (331–415) and two auto-regulatory flanking regions (280–330 and 416–441), using previously established definitions (Garvie et al, 2002). For protein–DNA assembly, we selected a 22-bp DNA fragment of the stromelysin-1 promoter with two EBS motifs in a palindromic arrangement, referred to as S-EBS in this article. The refined structure comprises residues 308–436 of Ets-1, the 22 base pairs of the S-EBS oligonucleotide, and 66 ordered solvent molecules. The visible part of the Ets-1 sequence that precedes helix HI-2 of the N-terminal ETS-flanking region is in an extended conformation (308–322), in contrast to a previous structure of the Ets-1–Pax-5–DNA complex where the same Ets-1(Δ280) construct was used for crystallization (Garvie et al, 2002). However, as in the (Ets-1)2–S-EBS complex this region is involved in crystal lattice contacts, it is possible that the observed structure of the N-terminal tail may be biased by these contacts, and therefore, this part of the structure will not be further discussed in this article.
Table 1.
X-r ay structure determination
| X-ray data collection | |
| Wavelength (Å) | 0.9756 |
| Resolution range (Å) | 20.0–2.58 (2.62–2.58) |
| No. of reflections | 20 507 (786) |
| Completeness (%) | 95.8 (75.4) |
| Redundancy | 2.7 (2.7) |
| 〈I/σI〉 | 20.45 (2.12) |
| Rmerge | 0.08 (0.66) |
| Space group | P21212 |
| Cell dimensions (Å) | a=93.6, b=100.8, c=69.8 |
| Structure refinement | |
| Resolution limits (Å) | 20.0–2.58 |
| Number of used reflections | 19 446 |
| Rwork/Rfree (%) | 23.53/27.12 |
| No. of atoms | |
| Protein | 2120 |
| DNA | 895 |
| Solvent | 66 |
| B factors (Å2) | |
| Protein | 67.5 |
| DNA | 67.6 |
| Water | 63.6 |
| R.m.s.d. | |
| Bond lengths (Å) | 0.006 |
| Bond angle (deg) | 1.2 |
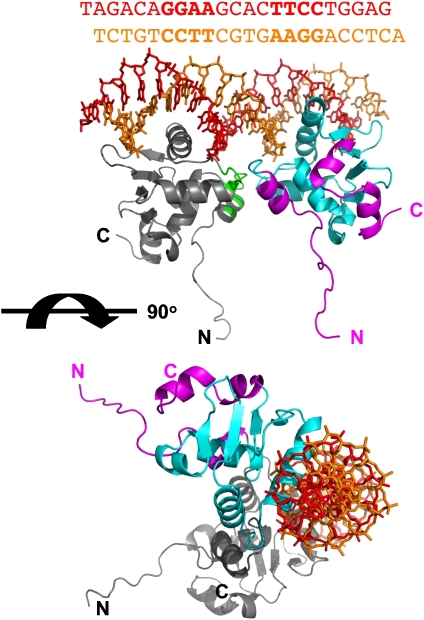
Figure 1.
Overall structure of the (Ets-1)2–S-EBS complex. Ribbon representation, in two different orientations, rotated by about 90°. For one of the two Ets-1 molecules, the ETS domain and ETS-flanking regions are shown in cyan and magenta, respectively. The second Ets-1 molecule is shown in grey. In the upper panel, the protein–protein interaction segment is shown in green for the second Ets-1 molecule. The termini are labelled. The sequence (top) and the structure of the two strands of the S-EBS oligonucleotide are shown in red and orange, respectively. The two EBS motifs in the sequence presentation are shown in bold.
Our structural data on the dimeric (Ets-1)2–S-EBS complex confirm that the sequence segment of the N-terminal ETS-flanking region, which forms α-helix HI-1 in the absence of DNA (Garvie et al, 2002; Lee et al, 2005), is not folded in the presence of the S-EBS element, as indicated by the lack of interpretable electron density of this region. The fold of the visible part of the structure of each Ets-1 molecule from the (Ets-1)2–S-EBS complex is basically identical with those from previous Ets-1–DNA complexes (Garvie et al, 2002) (Figure 3C). We noticed an additional small helix, assigned H1′, covering residues 348–353 (Figures 1, 3A and 4). In previous Ets-1–DNA complexes, the respective residue segment has a helix-like conformation as well.
Each ETS domain of the Ets-1 dimer binds to one of the two EBS (GGAA) core-binding motifs of the S-EBS element. The protein–DNA contacts of each ETS domain are basically identical. Reminiscent of previous binary Ets-1–DNA complexes (Garvie et al, 2002), there are a number of virtually identical base-specific interactions by the residue triplet Arg391, Arg394 and Tyr395 from the DNA-recognition helix H3, flanked by a series of additional interactions with the DNA sugar backbone (Figure 2). Most of the residues involved in protein–DNA contacts are highly conserved within sequences of related ETS transcription factors (Figure 4 and Supplementary Figure 1).
Figure 2.
Protein–DNA interactions in the (Ets-1)2–S-EBS complex. (A) One Ets-1 molecule in ribbon representation (grey), Ets-1 side chains involved in specific interactions with S-EBS are shown by sticks with atom-type specific colours (carbon=cyan, cp. Figure 1; oxygen=red; nitrogen=blue) and are labelled; part of the Ets-1-bound S-EBS oligonucleotide is shown in grey. Those bases and phosphate groups that are involved in specific Ets-1 interactions are shown by sticks with atom-type specific colours (carbon=light red, orange, cp. Figure 1; oxygen=red; nitrogen=blue) and are labelled. Hydrogen bonds are indicated by dashed lines. (B) Schematic presentation of the protein–DNA interactions within the (Ets-1)2–S-EBS complex. Residues involved in base-specific interactions are in bold; the remaining interacting residues are indicated in regular characters. The colour coding and font (bold/regular characters) of the DNA sequence is as in (A). The two EBS motifs are boxed and are grey shaded.
In contrast to previous structures, however, the (Ets-1)2–S-EBS complex allows for the first time to investigate the role of Ets-1 oligomerization in the function of this transcription factor. The two symmetric EBS motifs in the S-EBS element, which are separated by four base pairs, generate an arrangement of the Ets-1 dimer in which the two ETS domains are rotated with respect to each other by about 60° when looking along the DNA axis (Figure 1). When superimposed onto each other, the two Ets-1 molecules differ by a root mean square (r.m.s.) deviation of 0.58 Å (all atoms), thus showing essentially the same conformation (Figure 3B) and generating a symmetric protein–DNA complex (Figures 1 and 2B). The total size of the Ets-1–DNA interface in the presence of the S-EBS element is almost 900 Å2. The overall bending of the bound S-EBS is 4.8o, whereas the bend values for each monomer EBS-binding motif are 14.0° and 16.2°, respectively. Thus, there is a compensating effect from the ternary (Ets-1)2–S-EBS complex formation (Figure 1). This effect emerges from flanking protein interactions of both Ets-1 molecules with several phosphate groups of the DNA backbone, across the palindromic centre of the S-EBS element (Figure 2B), and contrasts with previous observations on binary Ets-1–DNA complexes with large DNA bend values (Garvie and Wolberger, 2001).
Figure 3.
Superposition of Ets-1 structures. (A) Ribbon representation of one Ets-1 molecule of the ternary (Ets-1)2–S-EBS complex in rainbow colours (N terminus, blue; C terminus, red). Secondary structural elements are labelled. (B) Superposition of the two Ets-1 molecules of the (Ets-1)2–S-EBS complex (magenta and cyan), using the same orientation as for (A). The r.m.s. deviation for 129 aligned residue pairs is 0.58 Å. (C) Superposition of one Ets-1 molecule from the ternary (Ets-1)2–S-EBS complex (magenta), ternary Ets-1–Pax-5–DNA complex (1MDM, green), binary Ets-1–GGAG complex (1K7A, blue). The r.m.s deviations of the superpositioned Ets-1 molecules are: (Ets-1)2–S-EBS—Ets-1–Pax-5–DNA, 0.71 Å for 117 aligned residue pairs; (Ets-1)2–S-EBS—Ets-1–GGAG, 0.86 Å for 104 aligned residue pairs. The superpositions were carried out through the program SSM (Krissinel and Henrick, 2004).
Molecular basis of S-EBS Ets-1 dimer formation
The crystal structure of the (Ets-1)2–S-EBS complex reveals the molecular contributions to DNA binding that require Ets-1 oligomerization. In addition to the two separate ETS–EBS interfaces, a third shared (Ets-1)2–DNA interface of about 370 Å2 is formed within the very centre of the dimeric complex. It involves two identical Ets-1 residue segments and the central EBS-connecting four base pair motif of the S-EBS element (Figure 5). The assembly renders a protein–protein interface with two separate, symmetry-related surface patches. Each interface patch is formed by (a) several residues, located mostly on helix HI-2 of the N-terminal-flanking region and the subsequent loop and by (b) residues from the loop connecting helices 2 and 3 of the ETS domain (Figures 4 and 5B). Apart from one two-fold repeated hydrogen bond, connecting the main chain amino group of Gly333 and the main chain carbonyl group of Asn380, all other protein–protein interface interactions are Van der Waals interactions. Gly333 adopts a conformation (ϕ=99° and φ=161°), which would not be allowed for non-glycine amino acids, and is followed by a proline that, in turn, restricts the conformation of Gly333. Adjacent to this protein–protein interface, another sequence segment from the N-terminal part of each ETS domain (332–338) interacts with the minor groove of the central four base pairs between the two EBS motifs of the S-EBS element (Figure 1). All these interactions are formed with the DNA sugar backbone only and, hence, do not introduce additional specific sequence requirements by the S-EBS element, to allow binding of the Ets-1 dimer.
Figure 4.
Multiple sequence alignment of the Ets-1(Δ280) fragment used for crystallization. The sequences of related Homo sapiens transcription factors Ets-2 and ETV-2, sharing the conserved Gly-Pro motif (black), are shown to the extent they could be unambiguously aligned. The secondary structural elements, as determined from the crystal structure of the (Ets-1)2–S-EBS complex, are shown on top. The colour code is as in Figure 1. The black triangles mark the N-terminal boundaries of the three Ets-1 fragments used in this investigation (cp. Figure 6). Those residues that are involved in the homo-dimeric Ets-1–Ets-1 are marked in green. Residues that are involved in Ets-1–S-EBS interfaces are shown in red (specific DNA contacts) and orange (nonspecific DNA contacts). For comparison, those residues that are involved in the Ets-1–Pax-5 interface of the Ets-1–Pax-5–DNA complex (Garvie et al, 2001) are highlighted in blue. Invariant residues are marked by asterisks. For reference, the sequence alignment of the ETS domain all known human Ets transcription factors is shown in the Supplementary Figure 1. All interface areas were determined with the program AREAIMOL (1994).
Figure 5.
Dimeric Ets-1–Ets-1 interface observed in the (Ets-1)2–S-EBS complex. (A) Surface presentation of one Ets-1 molecule, highlighting and labelling the two protein–protein interfaces in green. For reference, the complete S-EBS oligonucleotide used for crystallization is shown as well. (B) Ribbon representation of one Ets-1 molecule, same orientation and colour code as (A). The secondary structure labels follow the colour codes of Figure 2 to emphasize that the ETS domain-flanking regions (magenta) are involved into the protein–protein interface. (C) Stick presentation of the Ets-1–Ets-1 interface from the (Ets-1)2–S-EBS complex. Atom-type specific colours are used (carbon=green, cyan, cp. Figure 1; oxygen=red; nitrogen=blue). The hydrogen bond connecting Gly333 and Asn380 is shown by a dashed line within each of the two identical protein–protein interfaces. In one of the two molecules (green), the Gly333-Pro334 motif is boxed. Parts of the Cα trace are shown in faint colours, to help orientation.
The presence of the S-EBS element is essential for Ets-1 dimerization
Recent structural data on the apo-form of the Ets-1(Δ301) fragment, comprising part of the N- and the C-terminal ETS-flanking regions, revealed its ability to dimerize in a domain-swapped arrangement under crystallization conditions (Garvie et al, 2002). Moreover, although it had been shown previously that Ets-1(Δ301) does not dimerize in solution (Flannery et al, 1992), a systematic analysis exploring the potential for dimerization of different Ets-1 constructs in the presence or absence of DNA has been lacking, to date.
To confirm whether our structural observations of Ets-1 dimer formation in the presence of the S-EBS element are specific and DNA-mediated, we have analysed the association state of Ets-1 in the absence and presence of this element. To test the specific contribution of the N-terminal ETS-flanking region in dimeric assembly, we used three Ets-1 versions with different N-terminal truncations (Figure 4): Ets-1(Δ280), Ets-1(Δ301) and Ets-1(Δ335). All three variants contain the ETS domain and C-terminal-flanking region. In addition, Ets-1(Δ301) and Ets-1(Δ280) contain partial and complete N-terminal ETS-flanking regions, respectively. The residue segment 280–300, only present in the Ets-1(Δ280) construct, has remained invisible in all published structures of this Ets-1 fragment determined to date.
To assess the association state of the three Ets-1 fragments, we used two independent methods, gel filtration by size exclusion and static light scattering (SLS) (Figure 6A and Table II). The data from both methods consistently demonstrate that the truncated N-terminal ETS-flanking region (301–331) is necessary for Ets-1 dimerization on the S-EBS element, whereas Ets-1 without the N-terminal-flanking region only forms a 1:1 protein–DNA complex, matching previous observations (Baillat et al, 2002). In contrast, all three Ets-1 fragments remained monomeric in the absence of DNA, demonstrating that, under the conditions used for assembly analysis, Ets-1 only dimerizes in the presence of the S-EBS element.
Figure 6.
Association state of Ets-1 in the presence and absence of the S-EBS element. (A) Size exclusion chromatography elution profiles of Ets-1(Δ335) in green, Ets-1(Δ301) in red and Ets-1(Δ280) in blue, in the absence (solid lines) and presence (dashed lines) of the S-EBS element. The elution volume of markers used for the column calibration is indicated (A, MW=670 kDa, V=7.6 ml; B, MW=158 kDa, V=8.4 ml; C, MW=44 kDa, V=10.5 ml; D, MW=17 kDa, V=12.5 ml; E, MW=1.4 kDa, V=17.4 ml). We have interpreted the observed elution volumes of Ets-1 fragments as follows: all Ets-1 constructs in the absence of DNA, monomeric; Ets-1(Δ335) in the presence of the S-EBS element, 1:1 complex; Ets-1(Δ301) and Ets-1(Δ280) in the presence of the S-EBS, 2:1 complex (cp. Table II). (B) Size exclusion chromatography elution profiles of Ets-1(Δ280) WT in blue, G333Q in red, P334Q in grey, G333A in orange and P334A mutant in green, in the absence (solid lines) and presence (dashed lines) of the S-EBS element. The elution profiles indicate that all Ets-1 constructs in the absence of S-EBS are monomeric; Ets-1(Δ335), Ets-1(Δ280, G333Q), Ets-1(Δ280, P334Q), Ets-1(Δ280, G333A) form a 1:1 protein–DNA complex in the presence of the S-EBS element; Ets-1(Δ301, WT) and Ets-1(Δ280, WT) form a 2:1 protein–DNA complex in the presence of the S-EBS; Ets-1(Δ280, P334A) forms a mixture of 1:1 and 2:1 complexes in the presence of the S-EBS.
Table 2.
Determination of Ets-1 association states in the presence/absence of the S-EBS element by static light scattering
| S-EBS | Calculated MW, monomer | Calculated MW, dimer | Experimental MW (SLS) | Interpretation | |
|---|---|---|---|---|---|
| Ets-1(Δ335,WT) | − | 13.9 | 27.8 | 12.8±0.8 | Monomer |
| + | 27.1 | 41.0 | 25.9±2.1 | Monomer | |
| Ets-1(Δ301,WT) | − | 17.6 | 35.2 | 18.3±1.8 | Monomer |
| + | 30.8 | 48.4 | 44.6±5.4 | Dimer | |
| Ets-1(Δ280,WT) | − | 19.9 | 39.8 | 20.8±1.9 | Monomer |
| + | 33.1 | 53.0 | 53.7±3.2 | Dimer | |
| Ets-1(Δ280,G333A) | − | 19.9 | 39.8 | 19.8±2.0 | Monomer |
| + | 33.1 | 53.0 | 33.5±2.7 | Monomer | |
| Ets-1(Δ280,G333Q) | − | 19.9 | 39.8 | 20.6±2.5 | Monomer |
| + | 33.1 | 53.0 | 32.4±2.6 | Monomer | |
| Ets-1(Δ280,P333A) | − | 19.9 | 39.8 | 21.2±3.8 | Monomer/dimer |
| + | 33.1 | 53.0 | 40.5±4.5 | Monomer | |
| Ets-1(Δ280,P334Q) | − | 19.9 | 39.8 | 20.3±3.6 | Monomer |
| + | 33.1 | 53.0 | 33.1±3.0 | Monomer |
Ets-1 homo-dimerization is required for transactivation of the stromelysin-1 promoter
Sequence comparison of the 27 human Ets transcription factor family members revealed that the glycine–proline motif (residues 333–334 in Ets-1), which has a central function in the homo-dimeric assembly of the (Ets-1)2–S-EBS complex, is only present in the sequences of Ets-1, Ets-2 and ETV-2 (Figure 4 and Supplementary Figure 1). To confirm its importance for dimerization of Ets-1, we generated two sets of Ets-1 variants, in which each of the two residues was mutated either into an alanine or glutamine. We assessed the association state of these variants both by size exclusion gel filtration (Figure 6B) and SLS (Table II). As the WT Ets-1(Δ280) protein fragment, all mutants were monomeric in the absence of DNA. In the presence of the S-EBS element, however, only the WT protein was able to dimerize, whereas all mutants either remained monomeric (G333A, G333Q and P334Q) or showed an apparent mixture of different association states (P334A).
To further analyse the ability of these Ets-1 mutants to form dimers on an S-EBS element, we performed an electrophoretic mobility shift assay (EMSA) with WT or mutant Ets-1(Δ280) fragments on the WT or M1 mutant version of the S-EBS element, following a previously published design (Baillat et al, 2002). We used the Ets-1(Δ280) fragment to distinguish dimerization effects from general effects on DNA binding, as in contrast to full-length Ets-1 it can also bind to single EBS (Baillat et al, 2002). Indeed, Ets-1(Δ280) revealed a lower migrating band on the S-EBS(M1) probe with only one intact EBS, indicative of monomeric DNA binding, and a higher migrating band on WT S-EBS, indicative of dimer formation (Figure 7). All Ets-1 mutants showed similar binding as WT protein to the M1 probe, indicating that the selected single-residue mutations did not interfere with DNA binding. By contrast, G333Q, G333A, P334Q and P334A mutants showed a complete loss of Ets-1 dimer formation on S-EBS. Taken together, these results confirm the critical contribution of Gly333 and Pro334 within the DNA-mediated protein–protein interface, as unravelled by the structure of the (Ets-1)2–S-EBS complex, and demonstrate a specific role in homo-dimer formation.
Figure 7.
Electrophoretic mobility shift assay of Ets-1 mutants on the S-EBS element. Variants of purified Ets-1(Δ280) (WT, G333A, G333Q, P334A and P334Q) were incubated with a radiolabelled SEBS element (WT) in the WT configuration or with one EBS of the palindrome mutated (M1). Bands indicative of DNA-mediated Ets-1 monomers and dimers are indicated.
To further test whether homo-dimerization mediated by these specific protein–protein contacts is of functional importance for Ets-1 transactivation activity on the stromelysin-1 promoter, we performed reporter transactivation assays. We transfected HEK293 cells with expression vectors for full-length WT or the G333Q mutant Ets-1 variant and luciferase reporter constructs, driven by the stromelysin-1 promoter in the WT configuration or with a single (M1) or both (M1/M2) sites of the two palindromic EBS motifs mutated (Baillat et al, 2002). Transactivation of the promoter by WT Ets-1 required both EBS sites in the palindrome, whereas the activity was almost completely reduced to background levels on the single EBS-containing M1 reporter. Similarly, the Ets-1 G333Q variant virtually abolishes transactivation activity on the intact stromelysin-1 promoter (Figure 8A). A severe reduction of transactivation activity was also observed for the remaining Ets-1 variants used in this study (Figure 8B), thus confirming the requirement of Ets-1 dimerization for Ets-1 transactivation of the stromelysin-1 promoter. Consistent with the observation that full-length WT Ets-1 cannot bind as a monomer to an M1 site (Baillat et al, 2002), EMSA assays with nuclear cell extracts confirmed that the full-length Ets-1 G333Q variant, which has been shown to prevent dimerization, completely abolished Ets-1 binding to a WT S-EBS (Figure 8C and D). These findings thus indicate that Ets-1 homo-dimerization is essential for binding to a palindromic S-EBS element and transactivation function on the stromelysin-1 promoter.
Figure 8.
Transactivation of the stromelysin-1 promoter by Ets-1. (A) Full-length versions of Ets-1 (WT, G333Q) were assayed on luciferase reporters of the WT promoter and two variant versions, where either one (M1) or both EBS motifs (M1/M2) were mutated. (B) All other Ets-1 mutants were assayed on the WT stromelysin reporter. (C) EMSA assays with nuclear extracts from 293 cells transfected with empty vector (EV, lane 1), WT (lane 2) or G333Q (lane 3) full-length Ets-1 on WT S-EBS probe; lane 4, no extract. Arrow, (Ets-1)2–S-EBS complex; asterisk, free probe. (D) Western blot analysis of extracts used in (C) with anti-Ets-1 (top) and anti-β-tubulin (bottom) antibodies. (E) Ets-1 WT and G333Q mutant versions were assayed on a composite E-box/EBS reporter from the HIV-1 LTR. Bars indicate standard error of the mean from normalized duplicates and results are representatives of three independent experiments. For experimental details, see text.
To assess that the observed effects are specific for the palindromic S-EBS element, we also tested the WT Ets-1 and the G333Q variant on a composite E-box/EBS element of the HIV-1 LTR. In contrast to S-EBS, transactivation of this element by Ets-1 requires complex formation of monomeric Ets-1 with the E-box protein USF-1 (Sieweke et al, 1998). We confirmed that mutation of Gly333 had no effect on Ets-1 transactivation in this heterotypic transcription factor complex (Figure 8E). As full-length versions of Ets-1 were used in this assay, we conclude that our structural observations are relevant in the context of the complete transcription factor, demonstrating that the activity of Ets-1 on the stromelysin-1 promoter is regulated by its capacity for DNA-induced dimerization, involving the Gly333–Pro334 interaction motif.
Discussion
Palindromic EBS-mediated Ets-1 dimeric formation is highly specific
A common feature of Ets-1 function, generally in the context of other unrelated transcription factors, is its ability for auto-regulation through two helical sequence segments flanking the ETS DNA-binding domain (Garvie et al, 2002; Pufall and Graves, 2002). The N-terminal ETS-flanking sequence segment is part of the exon VII insert in Ets-1(245–330) that, when differentially spliced, leads to two Ets-1 isoforms, P42 and P51 (Koizumi et al, 1990; Jorcyk et al, 1991). The two isoforms translate into distinct patterns of Ets-1 function (Dittmer, 2003). The previously described auto-inhibitory effect has been attributed to a direct interaction of the two ETS-flanking regions (Jonsen et al, 1996).
In contrast, biochemical data of Ets-1 in the presence of the stromelysin-1 promoter indicated that the presence of the exon VII region leads to cooperative enhancement of DNA binding and transactivation activity (Baillat et al, 2002). On the basis of these findings, it was suggested that this region may counteract auto-inhibition in the presence of the stromelysin-1 promoter (Baillat et al, 2002). A series of truncation experiments revealed that the Ets-1(Δ301) fragment is sufficient to cooperatively bind to this promoter (Baillat et al, 2002). Furthermore, it was shown that competition between cooperative binding to the stromelysin-1 promoter and auto-inhibition of binding to monomeric EBS motifs becomes only fully effective when longer Ets-1 constructs are used, covering the complete exon VII insert. This effect could be due to additional interactions or unfolding transitions from residues beyond the N terminus of the Ets-1(Δ301) fragment (Garvie et al, 2002; Lee et al, 2005).
The crystal structure of the (Ets-1)2–S-EBS complex provides a direct molecular rationale for the previously observed findings on cooperative binding in the presence of the stromelysin-1 promoter. In the structure, a sequence segment N-terminal to the ETS domain directly interacts with residues of the loop connecting helices H2 and H3 from the ETS domain, forming a homo-dimeric Ets-1–Ets-1 assembly. Some of these residues are also in contact with the S-EBS oligonucleotide, thus leading to a ternary (Ets-1)2–DNA interaction site (Figures 2 and 4). Thus, our structural data are in agreement with previous data, demonstrating that the spacing between the two EBS elements is critical for ternary (Ets-1)2–S-EBS complex formation (Baillat et al, 2002).
Conversely, the spacing and orientation of the two Ets-1 molecules imposed by the S-EBS element would not permit a direct protein–protein interface for Ets-1 constructs lacking the N-terminal ETS-flanking region. In contrast to the well-established binary EBS–ETS1 interface, the protein–protein interface within the ternary (Ets-1)2–S-EBS complex is almost void of any specific interactions. One important exception, however, is the interaction of Gly333–Asn380, where sequence specificity is introduced by the peculiar conformation of Gly333, in the context of the next residue, Pro334. Mutation of Gly333 or Pro334 indeed impairs formation of the ternary complex and transactivation activity of Ets-1 on the stromelysin-1 promoter (Figures 6 and 7).
The Gly333-Pro334 motif, however, is only conserved in the most closely related Ets-1 and Ets-2 sequences (Figure 4). This finding may explain why, under in vitro conditions, ternary protein–DNA complexes on palindromic EBS motifs could only be obtained for Ets-1 and closely related family members, such as Ets-2 (Buttice and Kurkinen, 1993; Man et al, 2003). Similar conclusions also hold true for the previously shown activating functions of Ets-1 and Ets-2 on the human p53 promoter with a related palindromic EBS, requiring a spacing of four bases between the two elements as well (Venanzoni et al, 1996).
Moreover, recent in vivo data have demonstrated that activation of the stromelysin-1 promoter is Ets-1 specific in a synovial fibroblast cell line model (HIG-82) (Baillat et al, 2006). On the other hand, it was shown in a mouse model that Ets-2 activity is specifically required for stromelysin-1 expression in macrophages (Man et al, 2003). The different requirements for Ets-1 versus Ets-2 in unrelated cell types suggest that additional levels of regulation, which can discriminate between these closely related transcription factors, may be found outside the Ets-1(Δ280) sequence region investigated in this article.
Conversely, our model predicts that more distantly related ETS factors, in which the Gly333-Pro334 motif is not conserved (Supplementary Figure 1), may not be capable of ternary complex formation with palindromic EBS motifs. Although for Ets-1 and Ets-2 cooperative binding affinity on this element could be shown, the more distantly related factor ERGB/FLI-1 requires a different mechanism, possibly involving additional protein components (Hodge et al, 1996). Further support for Ets-1/Ets-2 specificity on palindromic EBS promoters originates from findings that only these two factors, in contrast to other Ets family members, are capable of recruiting the p300–CBP complex during transcriptional activation of the stromelysin-1 promoter (Jayaraman et al, 1999). Interestingly, a genome-wide analysis of EBS revealed that specificity for Ets-1 binding appears to be achieved by transcription factor partnerships on composite sites (Hollenhorst et al, 2007). Therefore, homo-dimerization on a palindromic S-EBS site could be seen as a special case without a requirement for heterotypic partners. Therefore, it will be interesting to compare an exhaustive panel of Ets proteins and to verify whether sequence variation in the residue positions, equivalent to Gly333-Pro334 in Ets-1, excludes other family members from recognizing an S-EBS element.
Regulation of Ets-1 function via DNA-mediated protein–protein interactions
Our data of the dimeric (Ets-1)2–S-EBS complex and previous structural/functional data on Ets-1 (Baillat et al, 2002; Pufall and Graves, 2002; Lee et al, 2005; Pufall et al, 2005) allow us to draw a number of general conclusions on the involvement of the ETS domain and the N-terminal ETS-flanking domain in auto-regulation of this transcription factor. Although the sequences of ETS domains are highly conserved in many members of this transcription family, the ETS-flanking regions in Ets-1 are unrelated, except for closely related factors, such as Ets-2 (Buttice and Kurkinen, 1993; Man et al, 2003).
Whereas there is no evidence for conformational transitions of the ETS domain upon EBS recognition, comparison of available structures consistently reveals partial unfolding of the N-terminal-flanking region upon DNA binding (Garvie et al, 2002; Lee et al, 2005). Moreover, consistent with previous models of Ets-1 auto-inhibition, the superposition of the previous Ets-1(Δ280) apo structure (Lee et al, 2005) onto the two protein molecules of the (Ets-1)2–S-EBS complex (data not shown) demonstrates that the Ets-1 apo conformation would not generate steric clashes within the ternary protein–DNA arrangement in the presence of the S-EBS element. Thus, unfolding of the N-terminal ETS-flanking region is not a structural requirement for ternary (Ets-1)2–S-EBS assembly, and it rather seems to present an Ets-1 property that is associated with DNA binding independently of the type of the promoter motif containing one or more ETS elements.
Taking the data together, our structure of the (Ets-1)2–S-EBS complex and previous biochemical data (Baillat et al, 2002) are therefore suggestive of enhanced DNA binding of Ets-1 on palindromic EBS elements as opposed to single EBS elements, thus overcoming the previously described auto-inhibitory effect of the N-terminal ETS-flanking region. Therefore, the involvement of the N-terminal ETS domain-flanking region in Ets-1 auto-inhibition is regulated by the oligomerization state of Ets-1 that, in turn, depends on the type of promoter with either a single EBS and dual EBS elements in a specific palindromic arrangement. As the Ets-1 fragments investigated by us remain monomeric in the absence of DNA, this type of Ets-1 activity regulation is DNA dependent (Figure 6). In structural terms, this property, relief of auto-inhibition, may be explained by the specific role of the loop connecting helix HI-2 and the ETS domain as a core motif in mediating ternary complex formation by protein–protein and protein–DNA contacts in the (Ets-1)2–S-EBS complex. In contrast, in the presence of single EBS elements, the same loop is involved in some nonspecific DNA contacts only, flanking the specific core interactions from residues of the ETS domain, and shows a significant degree of conformational variability (Garvie et al, 2002; Pufall and Graves, 2002; Lee et al, 2005). Subtle conformational changes of this loop were previously suspected as the cause of auto-inhibition by the N-terminal-flanking region in Ets-1.
Comparison of the structures of the ternary Ets-1–Pax-5–mb-1 complex (Garvie et al, 2001, 2002) and the homo-dimeric (Ets-1)2–S-EBS complex reveals that the protein–protein interaction sites in both complexes are adjacent to the shared DNA-binding site (Figure 9). In both complexes, there are ETS domain residues that are involved in bifurcated interactions with the DNA EBS motif and with the respective protein binding partner, either Pax-5 or Ets-1. However, although the protein–DNA interactions are limited to the ETS domain in the Ets-1–Pax-5–mb-1 complex, in the dimeric (Ets-1)2–S-EBS complex, these interactions extend into the N-terminal ETS-flanking region. These differences are mirrored in non-overlapping surface patches for protein–protein interactions in the two ternary complexes (Figure 9). The difference also explains why binding of the Ets homo-dimer on the S-EBS element relieves auto-inhibition by the N-terminal ETS-flanking region, whereas no such effect has been reported for Ets-1–Pax-5.
Figure 9.
DNA-mediated homo-dimerization of Ets-1 (A) and heterologous complex formation with Pax-5 (B). For each of the complexes, one Ets-1 molecule is shown in approximately the same orientation, displaying the DNA-binding helix H3 in a horizontal orientation on top. The DNA-binding interfaces of the Ets-1–Pax-5–DNA complex and the (Ets-1)2–S-EBS promoter complex are in red. The homo-dimeric Ets-1–Ets-1 interface of the (Ets-1)2–S-EBS complex is in green. The Pax-5-binding interface of the Ets-1–Pax-5–DNA complex is in blue. The figure illustrates that the protein–protein interfaces in the Ets-1–Pax-5–DNA and (Ets-1)2–S-EBS complexes are distinct but adjacent to the DNA-binding surfaces.
On the other hand, the structures of the two ternary protein–DNA complexes taken together provide a molecular rationale explaining why the presence of DNA is either essential to or will enhance protein–protein assembly. The conserved nature of the ETS domain in many transcription factors suggests that this principle, DNA-mediated recruitment of heterologous transcription factors by ETS domains, may apply for other members of this large family as well. Interestingly, activating effects have also been ascribed to the N-terminal ETS-flanking region of Ets-1, when bound to promoters that bind both Ets-1 and members of the unrelated Runx-1 transcription factor family (Kim et al, 1999; Goetz et al, 2000; Gu et al, 2000; Garvie et al, 2002), thus alleviating auto-inhibition of the same region. Whether relief of auto-inhibition in the presence of heterologous transcription factors such as Runx-1 involves similar contact surfaces and allosteric changes as those observed for Ets-1 dimerization on the stromelysin-1 promoter remains to be explored. Nonetheless, Ets-1 homo-dimerization may be considered as a specific case of general inhibition relief or even activating function of the N-terminal ETS-flanking region that requires the recruitment of additional factors. Although the general role of this region, in the presence of heterologous transcription factors, still awaits more molecular characterization, our findings unravelling the molecular basis of DNA-mediated assembly by Ets-1 homo-dimerization may provide a paradigm that may also be applicable in a broader context of heterologous assemblies involving Ets-1.
Materials and methods
Ets-1 cloning, expression and purification
Full-length Ets (residues 1–441) was cloned by PCR from human cDNA into Rc/CMV (Invitrogen) and verified by sequencing to correspond to the published sequence in NCBI (NM_005238.2). Ets-1 mutant constructs were generated in Rc/CMV by site-directed mutagenesis, as described in QuickChange site-directed mutagenesis protocol by Stratagene, with the exception that KOD hot start polymerase and its buffer (Novagen) were used. For structural and functional in vitro studies, truncated versions of Ets-1 cDNA coding for residues 280–441(Ets-1Δ280), 301–441(Ets-1Δ301) and 335–441(Ets-1Δ335) were cloned into the expression vector pETM10 (Günter Stier, EMBL Heidelberg, Germany). In all bacterial expression constructs, Cys350 and Cys416 were changed into serines by site-directed mutagenesis to decrease the redox sensitivity of expressed protein. The ability to bind to the S-EBS element was fully retained. The same expression and purification procedures were used for all Ets-1 constructs.
The protein fragments were overexpressed in Escherichia coli strain BL21 (DE3) RIL, induced with 1 mM IPTG, at 25°C overnight. Cell pellets were resuspended in lysis buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl and 5 mM imidazole), to which an EDTA-free protease inhibitor mix (Roche), lysozyme and DNase I were added, and sonicated.
The protein fragments were purified from the soluble cellular fraction by Ni-NTA affinity chromatography and eluted with lysis buffer containing 400 mM imidazole. The eluate was dialysed against a buffer containing 200 mM NaCl, 20 mM Tris–HCl (pH 8.0), and subsequently diluted with the same volume of a solution containing 20 mM Tris–HCl (pH 8.0) and 20% glycerol. For purification of the apo protein, the sample was concentrated and applied onto a Superdex 75 16/60 (Amersham) column, pre-equilibrated with 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 10% glycerol. For protein–DNA complex formation and purification, a 22-bp double-stranded DNA fragment (−219/−198) with 5′ TA overhangs from the palindromic EBS element of the human stromelysin-1 promoter (S-EBS) (Wasylyk et al, 1991) was incubated in a molar 2:1 (protein/DNA) ratio. The sample was concentrated and applied onto a Superdex 75 16/60 (Amersham) column, pre-equilibrated with 20 mM Tris–HCl (pH 8.0), 100 mM NaCl and 10% glycerol. The samples were concentrated up to 10 mg/ml, by using an Amicon concentrator MWCO 5.000 or 10.0000 (Millipore) depending on the Ets-1 construct used. The protein purity was examined by SDS–PAGE electrophoresis.
Analysis of Ets-1 association state
The association state of the Ets-1(Δ335), Ets-1(Δ301) and Ets-1(Δ280) constructs and single residue mutants was analysed by SLS combined with size exclusion chromatography in the absence and presence of the 22-bp S-EBS element used for crystallization. The samples were analysed with a miniDAWN Tristar detector (Wyatt Technology), which was connected with the protein purification system (Äkta Purifier). The protein samples were concentrated to 2 mg/ml and applied onto a Superdex 75 HR 10/30 gel filtration column (Amersham), pre-calibrated with the molecular weight standards (Bio-Rad). The time-averaged scattering intensities from the eluting peaks were measured at λ=690 nm in time intervals of 0.125 s and analysed with the ASTRA software, version 4.90.08 (Wyatt Technology).
EMSA
Double-stranded synthetic oligonucleotides (−223/−194) corresponding to WT or the M1 version of the S-EBS element (Baillat et al, 2002) were incubated with Klenow fragment DNA polymerase in the presence of [α32P]CTP and purified on Qiaquick Spin Columns (Qiagen). Recombinant protein (4 pmol) corresponding to WT or mutant Ets-1(Δ280) was incubated with 0.5 ng of probe in 20 μl binding reaction buffer (20 mM Tris–HCl (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 5% glycerol, 0.1% Triton X-100, 0.02% BSA, 0.5 μg poly d[I-C]) for 20 min. Nuclear extracts were prepared as described (Schreiber et al, 1989) and used at 10 μg with 1.5 μg poly d(I-C), 0.4 μg salmon sperm DNA and 100 fmol of labelled S-EBS probe per reaction. Complexes formed were resolved on a 6% polyacrylamide (acrylamide/bisacrylamide ratio, 29:1) non-denaturating gel (Bio-Rad) in 0.5 × Tris-glycine (purified protein) or 0.5 × TBE buffer (extracts) at 4°C. Gels were dried and autoradiographed at −80°C.
Transient transfection and reporter gene assay and western blotting
HEK293 cells were grown in Dulbecco's modified eagle's medium supplemented with 10% fetal bovine serum in six-well plates to reach 60–80% confluence at the time of transfection. DNA was transfected by calcium phosphate precipitation procedure as described (Sieweke et al, 1996). pGL3 luciferase reporter constructs (200 ng) (Promega) containing a WT stromelysin-1 promoter fragment (−478/+4) from the transcriptional start site or mutant versions (M1 and M1/M2) with one or both EBS mutated, respectively (Baillat et al, 2002) or a reporter containing two composite E-box/EBS elements from the −170 to −138 region of the HIV-1LTR (Sieweke et al, 1998) were co-transfected with 500 ng of Rc/CMV (Invitrogen) constructs driving the expression of WT and mutant full-length Ets-1 or no transgene (vector control). Assays were performed in duplicate. The transfection efficiency was normalized by assaying for β-galactosidase activity from a co-transfected CVM-LacZ construct, and luciferase activity was analysed as described (Sieweke et al, 1996). Immunoblotting was performed with anti-Ets-1 (1/1000; Santa Cruz, , CA; sc-350), anti-tubulin (1/10000; Sigma) and secondary anti-rabbit (Santa Cruz) or anti-mouse antibodies conjugated to horseradish peroxidase (Jackson, MN), using an ECL detection kit (Amersham) as described previously (Kelly et al, 2000).
X-ray structure determination
Crystals of Ets-1(Δ280) in the presence of the 22-bp S-EBS element were grown by the vapour diffusion method in sitting drops. Crystallization drops were prepared using 1 μl solution of concentrated protein–DNA complex and 1 μl of reservoir solution, containing 0.2 M ammonium acetate, 0.1 M sodium citrate–HCl (pH 5.6), and 28% (w/v) PEG-2000. Crystals grew within 5 days at room temperature.
A native X-ray data set was collected at beam line ID-29 at the ESRF (Grenoble, France), at a wavelength of 0.98 Å, according to the strategy using the program BEST (Popov and Bourenkov, 2003) (Table I). The data set was autoindexed, reduced and scaled with the HLK suite (Otwinowski, 1997). On the basis of the found cell parameters and space group, the solvent content amount was estimated to be 58% (Matthews, 1968), assuming one (Ets-1)2–DNA complex per asymmetric unit. The X-ray structure was solved by molecular replacement with the MOLREP program (Vagin and Teplyakov, 1997), using a previously determined binary Ets-1–DNA complex (PDB code 1K79) as template. The structure of the (Ets-1)2–S-EBS complex was refined, using the graphics program O (Jones et al, 1991) and the CNS suite (Brunger et al, 1998). Further statistics of the refined structure are presented in Table I.
Supplementary Material
Supplementary Figure 1
Acknowledgments
MHS and MW greatly acknowledge support from the Volkswagen Foundation (1/79 996). This study was also supported by the European Integrated Project SPINE-2 complexes to MW (031220), as well as the AICR grant 05-0079 and the ARC grant 3857 to MHS. LV received a fellowship from the Association pour la Recherche sur le Cancer (ARC). We thank Gleb Bourenkov and Alexander Popov for assistance during X-ray data acquisition. We thank Francisco C Fernandez for the provision of Ets-1 constructs in expression vectors. We also thank Young-Hwa Song for stimulating discussions.
PDB deposition The coordinates of the (Ets-1)2–S-EBS complex have been deposited at the Protein Data Bank (entry code: 2NNY).
References
- Baillat D, Begue A, Stehelin D, Aumercier M (2002) ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J Biol Chem 277: 29386–29398 [DOI] [PubMed] [Google Scholar]
- Baillat D, Leprivier G, Regnier D, Vintonenko N, Begue A, Stehelin D, Aumercier M (2006) Stromelysin-1 expression is activated in vivo by Ets-1 through palindromic head-to-head Ets binding sites present in the promoter. Oncogene 25: 5764–5776 [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Leiden JM (1995) A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity 3: 223–237 [DOI] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C (1998) The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279: 1037–1041 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B (2004) Ets ternary complex transcription factors. Gene 324: 1–14 [DOI] [PubMed] [Google Scholar]
- Buttice G, Duterque-Coquillaud M, Basuyaux JP, Carrere S, Kurkinen M, Stehelin D (1996) Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos–Jun complex. Oncogene 13: 2297–2306 [PubMed] [Google Scholar]
- Buttice G, Kurkinen M (1993) A polyomavirus enhancer A-binding protein-3 site and Ets-2 protein have a major role in the 12-O-tetradecanoylphorbol-13-acetate response of the human stromelysin gene. J Biol Chem 268: 7196–7204 [PubMed] [Google Scholar]
- Carey M (1998) The enhanceosome and transcriptional synergy. Cell 92: 5–8 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cowley DO, Graves BJ (2000) Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev 14: 366–376 [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Trauger JW, Baird EE, Dervan PB, Graves BJ, Gottesfeld JM (1999) Inhibition of Ets-1 DNA binding and ternary complex formation between Ets-1, NF-kappaB, and DNA by a designed DNA-binding ligand. J Biol Chem 274: 12765–12773 [DOI] [PubMed] [Google Scholar]
- Dittmer J (2003) The biology of the Ets1 proto-oncogene. Mol Cancer 2: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J 13: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK (1993) Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363: 38–45 [DOI] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodson W, Wheat W, Maira S-M, Wassylyk B, Hagman J (1996) Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell specific promoter. Genes Dev 10: 2198–2211 [DOI] [PubMed] [Google Scholar]
- Flannery CR, Lark MW, Sandy JD (1992) Identification of a stromelysin cleavage site within the interglobular domain of human aggrecan. Evidence for proteolysis at this site in vivo in human articular cartilage. J Biol Chem 267: 1008–1014 [PubMed] [Google Scholar]
- Garvie CW, Hagman J, Wolberger C (2001) Structural studies of Ets-1–Pax5 complex formation on DNA. Mol Cell 8: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Pufall MA, Graves BJ, Wolberger C (2002) Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem 277: 45529–45536 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C (2001) Recognition of specific DNA sequences. Mol Cell 8: 937–946 [DOI] [PubMed] [Google Scholar]
- Glover JN, Harrison SC (1995) Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373: 257–261 [DOI] [PubMed] [Google Scholar]
- Goetz TL, Gu TL, Speck NA, Graves BJ (2000) Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol Cell Biol 20: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TL, Goetz TL, Graves BJ, Speck NA (2000) Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1). Mol Cell Biol 20: 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler M, Richmond TJ (2001) The B-box dominates SAP-1–SRF interactions in the structure of the ternary complex. EMBO J 20: 3018–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge DR, Robinson L, Watson D, Lautenberger J, Zhang XK, Venanzoni M, Seth A (1996) Interaction of ETS-1 and ERGB/FLI-1 proteins with DNA is modulated by spacing between multiple binding sites as well as phosphorylation. Oncogene 12: 11–18 [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ (2007) Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R, Weitzman S, Buttice G, Thimmapaya B (1999) p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem 274: 17342–17352 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Jonsen MD, Petersen JM, Xu QP, Graves BJ (1996) Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol 16: 2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorcyk CL, Watson DK, Mavrothalassitis GJ, Papas TS (1991) The human ETS1 gene: genomic structure, promoter characterization and alternative splicing. Oncogene 6: 523–532 [PubMed] [Google Scholar]
- Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T (2000) MafB is an inducer of monocytic differentiation. EMBO J 19: 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J 18: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Fisher RJ, Fujiwara S, Jorcyk C, Bhat NK, Seth A, Papas TS (1990) Isoforms of the human ets-1 protein: generation by alternative splicing and differential phosphorylation. Oncogene 5: 675–681 [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP (2005) The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem 280: 7088–7099 [DOI] [PubMed] [Google Scholar]
- Man AK, Young LJ, Tynan JA, Lesperance J, Egeblad M, Werb Z, Hauser CA, Muller WJ, Cardiff RD, Oshima RG (2003) Ets2-dependent stromal regulation of mouse mammary tumors. Mol Cell Biol 23: 8614–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33: 491–497 [DOI] [PubMed] [Google Scholar]
- Mo Y, Ho W, Johnston K, Marmorstein R (2001) Crystal structure of a ternary SAP-1–SRF–c-fos SRE DNA complex. J Mol Biol 314: 495–506 [DOI] [PubMed] [Google Scholar]
- Nair SK, Burley SK (2003) X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112: 193–205 [DOI] [PubMed] [Google Scholar]
- Naito T, Razzaque MS, Nazneen A, Liu D, Nihei H, Koji T, Taguchi T (2000) Renal expression of the Ets-1 proto-oncogene during progression of rat crescentic glomerulonephritis. J Am Soc Nephrol 11: 2243–2255 [DOI] [PubMed] [Google Scholar]
- Nicholson R, Murphy G, Breathnach R (1989) Human and rat malignant-tumor-associated mRNAs encode stromelysin-like metalloproteinases. Biochemistry 28: 5195–5203 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Ozaki I, Mizuta T, Zhao G, Zhang H, Yoshimura T, Kawazoe S, Eguchi Y, Yasutake T, Hisatomi A, Sakai T, Yamamoto K (2003) Induction of multiple matrix metalloproteinase genes in human hepatocellular carcinoma by hepatocyte growth factor via a transcription factor Ets-1. Hepatol Res 27: 289–301 [DOI] [PubMed] [Google Scholar]
- Petersen JM, Skalicky JJ, Donaldson LW, McIntosh LP, Alber T, Graves BJ (1995) Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science 269: 1866–1869 [DOI] [PubMed] [Google Scholar]
- Popov AN, Bourenkov GP (2003) Choice of data-collection parameters based on statistic modelling. Acta Crystallogr D Biol Crystallogr 59: 1145–1153 [DOI] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ (2002) Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol 18: 421–462 [DOI] [PubMed] [Google Scholar]
- Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ (2005) Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science 309: 142–145 [DOI] [PubMed] [Google Scholar]
- Remenyi A, Scholer HR, Wilmanns M (2004) Combinatorial control of gene expression. Nat Struct Mol Biol 11: 812–815 [DOI] [PubMed] [Google Scholar]
- Rothhammer T, Hahne JC, Florin A, Poser I, Soncin F, Wernert N, Bosserhoff AK (2004) The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell Mol Life Sci 61: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Robinson L, Thompson DM, Watson DK, Papas TS (1993) Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene 8: 1783–1790 [PubMed] [Google Scholar]
- Sharrocks AD (2001) The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2: 827–837 [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA (1995) Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev 9: 2090–2104 [DOI] [PubMed] [Google Scholar]
- Sieweke MH, Graf T (1998) A transcription factor party during blood cell differentiation. Curr Opin Genet Dev 8: 545–551 [DOI] [PubMed] [Google Scholar]
- Sieweke MH, Tekotte H, Frampton J, Graf T (1996) MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85: 49–60 [DOI] [PubMed] [Google Scholar]
- Sieweke MH, Tekotte H, Jarosch U, Graf T (1998) Cooperative interaction of Ets-1 with USF-1 required for HIV-1 enhancer activity in T-cells. EMBO J 17: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HB, Yokota H (2001) Messenger-RNA expression of matrix metalloproteinases, tissue inhibitors of metalloproteinases, and transcription factors in rheumatic synovial cells under mechanical stimuli. Bone 28: 303–309 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Venanzoni MC, Robinson LR, Hodge DR, Kola I, Seth A (1996) ETS1 and ETS2 in p53 regulation: spatial separation of ETS binding sites (EBS) modulate protein: DNA interaction. Oncogene 12: 1199–1204 [PubMed] [Google Scholar]
- Wasylyk C, Gutman A, Nicholson R, Wasylyk B (1991) The c-Ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J 10: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernert N, Gilles F, Fafeur V, Bouali F, Raes MB, Pyke C, Dupressoir T, Seitz G, Vandenbunder B, Stehelin D (1994) Stromal expression of c-Ets1 transcription factor correlates with tumor invasion. Cancer Res 54: 5683–5688 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1