Abstract
CCA-adding enzyme builds the 3′-end CCA of tRNA without a nucleic acid template. The mechanism for the maintenance of fidelity during the CCA-adding reaction remains elusive. Here, we present almost a dozen complex structures of the class I CCA-adding enzyme and tRNA mini-helices (mini-D73N74, mini-D73N74C75 and mini-D73C74N75; D73 is a discriminator nucleotide and N is either A, G, or U). The mini-D73N74 complexes adopt catalytically inactive open forms, and CTP shifts the enzymes to the active closed forms and allows N74 to flip for CMP incorporation. In contrast, unlike the catalytically active closed form of the mini-D73C74C75 complex, the mini-D73N74C75 and mini-D73C74N75 complexes adopt inactive open forms. Only the mini-D73C74U75 accepts AMP to a similar extent as mini-D73C74C75, and ATP shifts the enzyme to a closed, active form and allows U75 to flip for AMP incorporation. These findings suggest that the 3′-region of RNA is proofread, after two nucleotide additions, in the closed, active form of the complex at the AMP incorporation stage. This proofreading is a prerequisite for the maintenance of fidelity for complete CCA synthesis.
Keywords: CCA, fidelity, template-independent polymerase, tRNA
Introduction
Every tRNA has an invariant CCA sequence at positions 74–76. The CCA is required for amino-acid attachment (aminoacylation) to the 3′-terminus of the tRNA by aminoacyl tRNA synthetases (Sprinzl and Cramer, 1979), and is required for peptide bond formation on ribosomes (Green and Noller, 1997; Kim and Green, 1999; Nissen et al, 2000). The CCA is built and/or repaired by the CCA-adding enzyme (ATP (CTP): tRNA nucleotidyltransferase) without the use of any nucleic acid template (Deutscher, 1990; Weiner, 2004). The CCA-adding enzyme is a remarkable, template-independent RNA polymerase, among the large number of nucleotidyltransferases (Holm and Sander, 1995; Martin and Keller, 1996, 2004, 2007; Yue et al, 1996). Unlike other DNA or RNA polymerases, this enzyme does not use any nucleic acid template, but faithfully synthesizes the ordered sequence CCA. Moreover, it recognizes three kinds of tRNAs, tRNA lacking either CCA, CA, or A, and reconstructs the CCA as needed. The CCA-adding enzyme has been identified in all three kingdoms (Yue et al, 1996) and is classified into two classes—archaeal CCA-adding enzyme (class I) and eubacterial and eukaryotic CCA-adding enzymes (class II) (Yue et al, 1996). Although the class I and II CCA-adding enzymes catalyse the same reaction, significant primary amino-acid sequence similarity exists only in the region around the active site signature motif. Moreover, in some ancient and slowly evolving eubacteria, the CCA addition is catalysed by two closely related class II enzymes: one adds CC and another adds A (Tomita and Weiner, 2001, 2002; Martin and Keller, 2004; Bralley et al, 2005).
The crystal structures of both the class I and II CCA-adding enzymes (Li et al, 2002; Augustin et al, 2003; Okabe et al, 2003; Xiong et al, 2003) revealed differences in their overall structures. The class I enzyme adopts a U-shaped structure similar to that of the eukaryotic polyA polymerase (Bard et al, 2000; Martin et al, 2000), whereas the class II enzyme has a seahorse-like structure (Li et al, 2002). However, the structures of the catalytic core domains of both classes are homologous and conserved among the nucleotidyltransferase family, suggesting the divergent evolution of both classes of enzymes (Okabe et al, 2003). The nucleotide selection mechanisms between the class I and II enzymes seem to differ (Li et al, 2002; Xiong et al, 2003). In class I, ATP and CTP bind to the enzyme nonspecifically. In contrast, in class II, CTP and ATP are recognized specifically through Watson–Crick-like base pairing between the bases (cytosine and adenine) and the side chains of aspartic acid and arginine (Asp154 and Arg157 in Bacillus stearothermophilus CCA-adding enzyme). The recent crystallographic analyses of complexes of class I Archaeoglobus fulgidus CCA-adding enzymes with duplex RNA primers revealed that the specificity for CTP and ATP is determined by the arginine side chain (Arg224 in A. fulgidus CCA-adding enzyme (AFCCA)) and the backbone phosphate of tRNA (Xiong and Steitz, 2004; Tomita et al, 2006). The 4-amino group of CTP and the 6-amino group of ATP hydrogen-bond with the phosphate backbone of the discriminator nucleotide, and the O2 and N3 of the CTP base and the N1 of the ATP base hydrogen-bond with the side chain of Arg224. In class II, although the ternary complex of A. aeolicus A-adding enzyme with tRNA was determined, the underlying reaction mechanism for the specificity of CTP and ATP in the conventional CCA-adding enzymes is still obscure (Tomita et al, 2004).
More recent complete crystallographic analyses of the class I AFCCA in complex with mini-helices containing a TΨC loop revealed that CCA addition proceeds through dynamic changes of the enzyme and the RNA primers during the CCA-adding reaction (Tomita et al, 2006) (Figure 1). In the binary complex with an RNA primer ending in C74, the catalytic carboxylates (Glu59, Asp61, and Asp110) in the head domain are not in the vicinity of the 3′-OH of C74, where the C74 base is stacked down by the β-turn in the head domain and stacks with the base of D73 (discriminator nucleotide). This enzyme state is the catalytically inactive open form. Upon CTP binding, the head domain relocates towards the neck domain, and the catalytic cleft of the enzyme shifts from an open to a closed form. The transition from the open to closed form of the enzyme is accompanied by the conformational change of the β-turn, which allows the 3′-end nucleotide to flip, where the base of C74 stacks with that of the incoming CTP. In this state, the 3′-OH of C74 is in the vicinity of the catalytic carboxylates and the tri-phosphate of CTP, and the complex adopts the active closed conformation for CMP incorporation to proceed. After two CMP incorporations, the enzyme is locked in a closed form and the 3′-end nucleotide is locked in the flipped form. The 4-amino group, O2, and N3 of the C74 base hydrogen-bond with the Oɛ of Glu96, the main chain N of Glu96 and the main chain O of His97, respectively, and N4 and O2 of C75 hydrogen-bond with the phosphate backbone of D73 and Thr130, respectively. The 3′-OH of C75 is in the vicinity of the catalytic carboxylates, and AMP incorporation proceeds in the closed form of the complex. The global motions of the enzyme and the 3′-region of the RNA primer change the conformation of the arginine (Arg224) side chain, which determines the specificity for CTP and ATP.
Figure 1.
Catalytically inactive open form and active closed form of CCA-adding enzyme during the CCA-adding reaction. (A) Transition from the open to closed form of the enzyme–RNA complex for CMP incorporation. For CMP incorporation, the catalytic cleft in the head domain relocates towards the neck domain and the 3′-end nucleotide flips. The complexes with mini-D73C74 and with mini-D73C74 and CTP are coloured magenta and marine blue, respectively. CTP is coloured blue. (B) Closed conformation for AMP incorporation. For AMP incorporation, the enzyme is fixed in the closed form and the 3′-end nucleotide is flipped. The complexes with mini-D73C74C75 and with mini-D73C74C75 and ATP are coloured magenta and green, respectively. ATP is coloured red.
Some template-dependent DNA polymerases remove mis-incorporated nucleotides by using 3′–5′ exo-nuclease domains, which are distinct from the catalytic domains for nucleic acid polymerization (Baker and Bell, 1998). It was previously reported that the reaction catalysed by CCA-adding enzyme is reversible. However, the reverse reaction of CCA addition, pyrohydrolysis, is much slower than the forward polymerization reaction, indicating that the CCA-adding enzyme does not possess an activity to correct a mis-incorporated nucleotide under physiological conditions (Deutscher, 1973; Evans and Deutscher, 1978). Although the molecular basis for the specific selection of CTP and ATP by CCA-adding enzyme has been characterized, the detailed molecular mechanism by which CCA-adding enzyme prevents the synthesis of an incorrect 3′-terminus, other than by selecting the correct nucleotide at the reaction stage, remains obscure.
Here, we analysed nine binary complex structures of the class I AFCCA with nine mini-helix variants, and two ternary complex structures of AFCCA with two mini-helix variants and an incoming nucleotide. Our structural and biochemical studies revealed a unique mechanism for the maintenance of fidelity during the CCA-adding reaction for correct CCA synthesis.
Results and discussion
Molecular basis for CMP incorporation into mini-helices, mini-D73N74
The recent complete crystallographic analysis of CCA sequence addition by the class I AFCCA revealed the detailed dynamics between the enzyme and the tRNA primer during the CCA-adding reaction (Tomita et al, 2006) (Figure 1). In the binary complex structure with mini-D73C74 (mini-D73C74 stage), the enzyme adopts an open form, and C74 is stacked down by the β-turn of the enzyme, through a nonspecific stacking interaction. On the other hand, in the ternary complex structure with mini-D73C74 and CTP (mini-D73C74+CTP stage), the enzyme shifts to a closed form. This conformational transition is accompanied by a drastic conformational change of the β-turn, which in turn allows C74 to flip for CMP incorporation to proceed. The Arg224 side chain hydrogen-bonds with the O2 and N3 of the CTP base. The CTP-induced conformational changes of both the enzyme and 3′-region of the primer are the underlying mechanism for the reaction and the selection of the correct nucleotide at position 75.
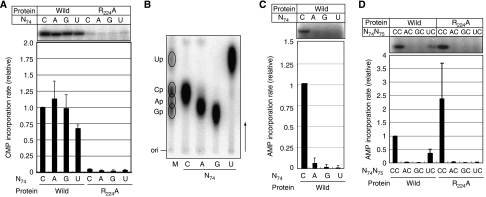
This scheme also occurs in CMP incorporation at position 74 of mini-D73, where D73 (discriminator nucleotide) is stacked down by the β-turn of the enzyme, through a nonspecific stacking interaction. In tRNAs, D73 can be any nucleotide, and it does not affect the CCA-adding reaction. This has raised the possibility that an RNA primer ending with N74 (mini-D73N74, N74 is either A, G, or U) might be able to accept CMP at position 75. We first analysed CMP incorporation into mini-helices ending with N74 as primer substrates by AFCCA. In the presence of 32P-labelled CTP and unlabelled ATP, all of the mini-helices accepted CMP efficiently, to the same extent as mini-D73C74 (Figure 2A). Neighbouring nucleotide analyses of the 32P-labelled product in Figure 2A showed that CMP is incorporated at position 75 in each mini-helix (Figure 2B). Therefore, the observed CMP incorporations into mini-D73N74 were not due to the removal of the nucleotide N74 and the re-incorporation of CMP at positions 74 and 75. The mutation of Arg224 to Ala in AFCCA reportedly reduced the CMP incorporation rate into mini-D73C74 (Tomita et al, 2006) (Figure 2A). The incorporations of CMP into mini-D73N74 were also reduced by the mutation of AFCCA, suggesting that CMP incorporation into the mutant mini-helices might proceed by a similar mechanism as that observed for CMP incorporation into mini-D73C74.
Figure 2.
CTP and ATP incorporation into mutant mini-helix variants. (A) CMP incorporation into mini-D73N74 in the presence of α-32P CTP and unlabelled ATP by the wild-type AFCCA (left part) or the R224A mutant AFCCA (right part). The upper panel shows the autoradiograph of the gel, and the lower graph shows the quantification of the relative initial velocity of CMP incorporation. The absolute CMP incorporation rate was estimated as 1.95 pmol min−1 μg−1, and it was defined as 1.0. (B) Neighbouring nucleotide analysis of 32P-labelled products in (A) by thin-layer chromatography. M indicates the RNase T2 hydrolysate of a uniformly α-32P UTP-labelled E. coli tRNAGlu transcript, as a marker. C, A, G, and U indicate RNase T2 hydrolysates of 32P-labelled products in the gels in the left part of (A). (C) AMP incorporation into mini-D73N74 in the presence of unlabelled CTP and α-32P ATP by the wild-type AFCCA. The upper panel shows the autoradiograph of the gel, and the lower graph shows the quantification of the relative initial velocity of AMP incorporation. The absolute AMP incorporation rate was estimated as 0.256 pmol min−1 μg−1, and it was defined as 1.0. (D) AMP incorporation into mini-D73N74C75 by the wild-type AFCCA (left part) and the R224A mutant AFCCA (right part). The presentation is the same as in (A). The absolute AMP incorporation rate into mini-D73C74C75 was estimated as 3.34 pmol min−1 μg−1, and it was defined as 1.0. The bars on the graphs indicate the standard deviations of more than three independent experiments.
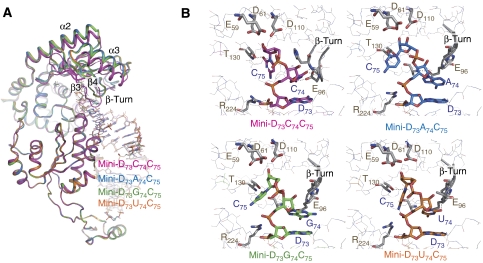
The crystal structures of three complexes of AFCCA and mini-helix variants (mini-D73A74, mini-D73G74, and mini-D73U74) were determined (Supplementary Table 1 and Supplementary Figure 1). As expected from biochemical studies, in all of the complex structures, the enzymes adopt inactive open forms (Figure 3A), and the N74 bases of the mini-helices are stacked down by the β-turn motif, and lack specific hydrogen-bond interactions. The Oɛ of Glu96 in the β-turn hydrogen-bonds with the main chain N of Ala126. The N74 riboses of mini-D73N74 do not superpose on the C74 ribose of mini-D73C74, and the orientations of the N74 riboses of mini-D73N74 are different from that of C74 in mini-D73C74. In the complex with mini-D73C74, the 3′-OH of the C74 ribose hydrogen-bonds with the side chain of Arg224 and the phosphate backbone of D73. On the other hand, in the mini-D73N74 complexes, the 2′-OH of the N74 ribose hydrogen-bonds with the main chain O of Tyr94. As a result, the C74 and N74 bases stack on the D73 base in an inverted manner. Despite the different ribose orientation of N74 in the mini-D73N74 from that of C74 in the mini-D73C74, the bases of N74 of mini-D73N74 superpose with that of C74 of mini-D73C74, and are stacked with D73 and the β-turn. The 3′ CCA end of tRNA reportedly has to have different conformations, depending on the discriminator nucleotide composition (Hou et al, 1998). The different orientations of the riboses of the 3′-end nucleotide of the mini-D73N74 from that of mini-D73C74 might indicate that the interaction between the bases of D73 and N74 is affected by the nucleotide context at positions 73 and 74. A theoretical study on the free energy of base stacking showed that A–C stacking is weaker than those of A–A, A–G and A–U in single-strand A-helices (Friedman and Honig, 1995). Therefore, it is most likely that in the mini-D73C74 complex, the weaker stacking interactions between the D73 (adenosine) base and the C74 base (A73–C74) result in the alternative orientation of the ribose, which is different from those of N74. In agreement with this, the orientation of the D73 ribose in the mini-D73 complex is the same as that of the C74 ribose in the mini-D73C74 complex (Tomita et al, 2006). In the mini-D73 complex, the D73 (adenosine) base stacks with the adjacent C72 base, and this stacking (C72–A73) is also theoretically weaker than others (Friedman and Honig, 1995). It should also be noted that the riboses in the present complex structures can easily flip for CMP incorporation, as described below, and this also explains the efficient CMP incorporation at position 74 (Figure 2A).
Figure 3.
Crystal structures of AFCCA complexes with mini-D73N74. (A) Superposition of the catalytic region of four complexes. The complexes with mini-D73C74, mini-D73A74, mini-D73G74, and mini-D73U74 are coloured magenta, marine blue, green, and orange, respectively. All of the complexes adopted the open form. (B) Superposition of complexes with mini-D73U74, with or without CTP. Complexes without CTP and with CTP are coloured orange and light blue, respectively. The CTP is coloured blue. CTP induces an open to closed conformational transition. (C) Comparison of the catalytic regions of the two complexes. The complex without CTP (upper panel: coloured orange) and that with CTP (lower panel: coloured light blue) are shown.
The above-mentioned observations suggest that an incoming CTP can induce the conformational transition of the enzyme from the open to the closed form, which in turn would allow the nucleotide at position 74 to flip for CMP incorporation to proceed. When CTP was soaked into the complex crystal of AFCCA and mini-D73U74, the enzyme adopted the closed form, and U74 flipped (Figure 3B and C; Supplementary Figure 2). The 3′-OH of the U74 ribose is in the vicinity of the tri-phosphate of the incoming CTP. Arg224 hydrogen-bonds with the O2 and N3 of the CTP base, consistent with the result showing that the mutation of Arg224 to Ala in AFCCA reduced CMP incorporation into mini-D73U74, as described (Figure 2A). The mechanism for the selection and reaction of CMP incorporation into mini-D73U74 is exactly the same as that observed in the CMP incorporation into mini-D73C74. In the CMP incorporation stage into mini-D73U74 by AFCCA, the U74 base stacks with the CTP base. The CMP incorporation into mini-D73A74 or mini-D73G74 might proceed in a similar dynamic manner as the CTP selection, as observed in mini-D73C74 and mini-D73U74. These results suggest that a substrate with a mis-incorporated nucleotide (either A, G, or U) at position 74 is not rejected by the enzyme after nucleotide addition, and thus subsequent CMP incorporation at position 75 proceeds.
Molecular basis for the effect of N74 on AMP incorporation at position 76
Although CMP is efficiently incorporated into mini-D73N74, in the presence of unlabelled CTP and 32P-labelled ATP, AMP incorporation into mini-D73N74 was remarkably reduced (Figure 2C). These results imply that, after CMP incorporation into mini-D73N74, subsequent AMP incorporation at position 76 does not proceed. These observations were verified by analysing AMP incorporation into mini-helices, mini-D73N74C75 (mini-D73A74C75, mini-D73G74C75 and mini-D73U74C75). AMP incorporation into mini-D73A74C75 or mini-D73G74C75 was not significantly detectable. AMP incorporation into mini-D73U74C75 was detected, but the AMP incorporation rate was reduced, to less than 30% of that into mini-D73C74C75 (Figure 2D).
To clarify the molecular basis of the sensitivity of subsequent AMP incorporation at position 76 to the nucleotide at position 74, the crystal structures of three complexes of AFCCA and mini-D73N74C75 were determined (Supplementary Table 1). In the complex of AFCCA and mini-D73C74C75, the enzyme adopts the active closed form and the base of C74 hydrogen-bonds with the β-turn of the enzyme (the 4-amino group, O2, and N3 of the C74 base hydrogen-bond with the Oɛ of Glu96, the main chain N of Glu96 and the main chain O of His97, respectively). This interaction between C74 and the β-turn involves Watson–Crick-like base pairing. The O2 of C75 hydrogen-bonds with the Oγ of Thr130, and the 4-amino group of C75 interacts with the phosphate group of D73. As a result, the 3′-nucleotide of the RNA assures the flipped form, and the RNA–enzyme complex adopts the active closed conformation. On the other hand, the enzymes of all complexes with mini-D73N74C75 adopt open forms (Figure 4A). The β-turns of AFCCA in the complexes with mini-D73N74C75 do not hydrogen-bond with the base of the nucleotide at position 74. Instead, the β-turns stack down the nucleotide at position 74 of mini-D73N74C75, and the Oɛ of Glu96 hydrogen-bonds with the main chain N of Ala126. In the complexes with mini-D73A74C75 and mini-D73G74C75, except for the complex with mini-D73U74C75, the 4-amino group of C75 does not interact with the phosphate group of D73, and the O2 of C75 does not hydrogen-bond with the Oγ of Thr130 (Figure 4B and Supplementary Figure 3). The conformations of the Arg224 side chain in the AFCCA complexes with mini-D73N74C75 are similar to that observed in the AFCCA complex with mini-D73C74, rather than mini-D73C74C75 (Figure 4B). These structural features prevent the formation of the closed form of the enzyme.
Figure 4.
Crystal structures of AFCCA complexes with mini-D73N74C75. (A) Superposition of the four complexes. Complexes with mini-D73C74C75, mini-D73A74C75, mini-D73G74C75, and mini-D73U74C75 are coloured magenta, marine blue, green, and orange, respectively. All of the complexes adopted the open form. (B) Structures of the four catalytic regions of the complexes. The complexes with mini-D73C74C75, mini-D73A74C75, mini-D73G74C75, and mini-D73U74C75 are coloured as in (A).
When ATP was soaked into the complex crystals of AFCCA and mini-D73N74C75, no significant electron density corresponding to the ATP was visible, and the enzymes remained in the inactive open forms (data not shown). An ATP model was built into the complex structures of AFCCA and mini-D73N74C75 (Supplementary Figure 4). In the models, the bases of C75 in mini-D73A74C75 and mini-D73G74C75 clash with the ATP. This may explain the lack of detectable AMP incorporation into mini-D73A74C75 and mini-D73G74C75. On the other hand, in the ATP docking model into the complex with mini-D73U74C75, C75 does not clash with the ATP. This reflects the fact that, due to the hydrogen bonds between the 4-amino group of C75 and the phosphate group of D73 and between the O2 of C75 and the Oγ of Thr130, the 3′-end of RNA adopts the flipped conformation. This quasi-open conformation of the complex may explain the significant amount of AMP incorporation into mini-D73U74C75, as described above (Figure 2D). The AMP incorporation is reduced by the Arg224Ala mutation of AFCCA (Figure 2D), suggesting that AMP incorporation might proceed through an open to closed conformational transition of the enzyme, accompanied by the re-orientation of Arg224, as in the CMP incorporation dynamics. This is strikingly different from the mechanism for AMP incorporation into mini-D73C74C75, in which AMP incorporation proceeds in the closed form of the enzyme and the 3′-region of the RNA (Tomita et al, 2006).
These results suggest that Watson–Crick-like base pairing between the nucleotide at position 74 of the RNA and the β-turn of AFCCA is a prerequisite for the proper orientation of C75 for subsequent efficient AMP incorporation at position 76 in the closed form of the enzyme. This interaction highlights the importance of the incorporated nucleotide at position 74, after two nucleotides are incorporated at positions 74 and 75. This also explains the absence of significant electron density of ATP in the mini-D73U74C75 complex crystals soaked in the solution containing ATP, although the extent of AMP incorporation into mini-D73U74C75 was approximately 30% of that into mini-D73C74C75 (Figure 2D).
Molecular basis for the effect of N75 on AMP incorporation at position 76
The analyses described above revealed the mechanism for the proofreading of the nucleotide at position 74 by the β-turn, for complete CCA synthesis at the AMP incorporation stage, after the addition of two nucleotides. Now, the question arises as to how the second nucleotide added at position 75 is proofread by AFCCA during the CCA-adding reaction.
When the incorporation of AMP into mini-D73C74N75 (N is either A, G, or U) was analysed, no significant incorporation into either mini-D73C74A75 or D73C74G75 was detected. However, AMP incorporation into D73C74U75 was observed to almost the same extent as that into mini-D73C74C75 (Figure 5A). neighbouring nucleotide analyses of the 32P-labelled product in Figure 5A revealed that AMP is incorporated at position 76 of mini-D73C74U75 (Figure 5B).
Figure 5.
AMP incorporation into mutant mini-helices. (A) AMP incorporation into mini-D73C74N75 by the wild-type AFCCA (left part) and the R224A mutant AFCCA (right part). The upper panel shows the autoradiograph of the gel, and the lower graph shows the quantification of the relative initial velocity of AMP incorporation. The absolute AMP incorporation rate into mini-D73C74C75 was estimated as 3.34 pmol min−1 μg−1, and it was defined as 1.0. (B) Neighbouring nucleotide analysis of 32P-labelled products in (A) by thin-layer chromatography. M indicates the T2 hydrolysate of a uniformly α-32P UTP-labelled E. coli tRNAGlu transcript as a marker. CU indicates the T2 hydrolysate of the 32P-labelled product in the gel shown in the left part of (A). (C) AMP incorporation into mini-D73N74N75. The presentation is the same as in (A). The bars on the graphs indicate the standard deviations of more than three independent experiments.
To clarify the molecular basis of the sensitivity of subsequent AMP incorporation at position 76 to the nucleotide at position 75, the crystal structures of three complexes of AFCCA and mini-helices (mini-D73C74A75, mini-D73C74G75, and mini-D73C74U75) were determined (Supplementary Table 1). Intriguingly, all of the enzymes in the mini-D73C74N75 complexes adopt open forms, with the Oɛ atom of Glu96 hydrogen-bonding with the main chain N of Ala126 (Figure 6A and B; Supplementary Figure 5). The C74 bases in the mini-D73C74A75 and mini-D73C74U75 complexes are not locked by the β-turn of the enzyme through Watson–Crick-like base-pairing, as observed in the mini-D73C74C75 complex (Figure 6B). In the mini-D73C74A75 complex, the 6-amino group of the A75 base hydrogen-bonds with the Oɛ of Glu96, and the O2 of the C74 base interacts with the main chain N of Glu96. In the mini-D73C74U75 complex, the O4 of U75 and the N3 of C74 hydrogen-bond with the 4-amino group of C74 and the main chain N of Glu96, respectively. In the mini-D73C74G75 complex, the conformation of the 3′-region of the RNA is completely different from that in the mini-D73C74C75 complex. The bases of C74 and G75 stack on each other, and the O2 of C74 hydrogen-bonds with the main chain N of Tyr173, the N1 and 2-amino group of G75 hydrogen-bond with the Oδ of Asp61, and the phosphate groups of G75 and D73 hydrogen-bond with Arg224.
Figure 6.
Crystal structures of AFCCA complexes with mini-D73N74C75. (A) Superposition of the four complexes. The complexes with mini-D73C74C75, mini-D73C74A75, mini-D73C74G75, and mini-D73C74U75 are coloured magenta, marine blue, green and orange, respectively. All of the complexes adopt the open form. (B) Structures of the four catalytic regions of the complexes. The complexes with mini-D73C74C75, mini-D73C74A75, mini-D73C74G75 and mini-D73C74U75 are coloured as in (A). (C) Superposition of the mini-D73C74U75 complexes with and without ATP. The complexes without ATP and with ATP are coloured orange and light blue, respectively. The ATP is coloured red. ATP induces an open to closed conformational transition of the enzyme. (D) Comparison of the catalytic regions of the mini-D73C74U75 complexes. The complex without ATP (upper panel: coloured orange) and that with ATP (lower panel: coloured light blue) are shown.
These results suggest that the presence of C74 alone is not sufficient for C74 to be locked by the β-turn through Watson–Crick-like base-pairing, and that the locking is affected by the nucleotide composition at position 75. When ATP was soaked into the complex crystals of AFCCA and mini-D73C74N75, the electron density of ATP was visible only in the mini-D73C74U75 complex. In the mini-D73C74U75 complex, ATP shifts the enzyme to the closed form (Figure 6C and D; Supplementary Figure 6). This observation is consistent with the biochemical studies showing that mini-D73C74U75 can accept AMP efficiently (Figure 5A). In the complex of AFCCA, mini-D73C74U75 and ATP, the C74 base hydrogen-bonds with the β-turn, U75 flips, and the O2 of U75 hydrogen-bonds with the Oγ of Thr130. The mutation of Thr130 to Ala reportedly reduces the AMP incorporation into mini-D73C74C75 (Tomita et al, 2006). Arg224 changes its conformation, and it hydrogen-bonds with the N1 of ATP and the phosphate backbone of D73, as observed with the AMP incorporation into mini-D73C74C75. The mutation of Arg224 to Ala in AFCCA reduced the AMP incorporation rate into mini-D73C74U75 by 50%, as compared with that by the wild-type AFCCA (Figure 5A). This is distinct from the AMP incorporation into mini-D73C74C75, where the R224A mutation does not affect the AMP incorporation rate (Figure 5A; Tomita et al, 2006). These observations also suggest that AMP incorporation into mini-D73C74U75 proceeds through the open to closed conformational transition of the enzyme, accompanied by the re-orientation of Arg224, as in the CMP incorporation dynamics. This is distinct from the AMP incorporation reaction into mini-D73C74C75, which proceeds in a static, closed form of the enzyme and the 3′-region of the RNA primer.
A model of ATP was built into the mini-D73C74A75 and mini-D73C74G75 complex structures (Supplementary Figure 7). In the ATP docking model into the mini-D73C74G75 complex structure, ATP clashes with the 3′-terminal region of mini-D73C74G75, thus explaining the inability of mini-D73C74G75 to accept AMP (Figure 5A). On the other hand, in the ATP docking on the mini-D73C74A75 complex structure, ATP can bind to the enzyme without any clashes. When the U75 base, in the AFCCA complex with the mini-D73C74U75 and ATP, is replaced with adenine, the adenine base does not clash with the enzyme. However, in the transition from the open to closed form of the enzyme for AMP incorporation, which is accompanied by the flipping of the nucleotide at position 75 in the complex of mini-D73C74U75 and ATP (Figure 6D), the A75 base is too large to flip for catalysis. The adenine base of A75 would clash with the loop between β5 and α4 (amino-acid residues 125–128) of the closed form of the enzyme (Supplementary Figure 8). Thus, A75 cannot flip for AMP incorporation to proceed.
These results suggest that the enzyme proofreads the nucleotide at position 75 by the size of its base—whether it is a purine or pyrimidine—and by the presence of the 2-oxygen of pyrimidine at position 75 during the AMP incorporation stage for complete CCA synthesis.
Maintenance of fidelity during the CCA-adding reaction for complete CCA synthesis
CCA-adding enzyme synthesizes the CCA sequence without the help of a nucleic acid template. CCA-adding enzyme lacks an activity to remove an incorrectly incorporated nucleotide, and the reverse reaction is much slower than the forward reaction (Deutscher, 1973, 1990). Therefore, the question remained as to how CCA-adding enzyme prevents the synthesis of a tRNA possessing an incorrect sequence at positions 74–76. In this study, we described the detailed molecular basis by which the class I CCA-adding enzyme highlights the mis-incorporated nucleotide during the CCA-adding reaction to prevent the synthesis of a tRNA possessing an incorrect 3′-terminus. The present study has clarified the mechanism for the maintenance of fidelity for the synthesis of the complete CCA, as shown in Figure 7.
Figure 7.
Schematic representation of the mechanism for maintenance of fidelity by the class I CCA-adding enzyme. Proofreading of the nucleotides at positions 74 and 75 occurs after the two nucleotides are incorporated into the closed conformation of the enzyme, during the AMP incorporation stage.
The nucleotide incorporated at position 74 is not proofread by the enzyme at all after it is incorporated. An incorrect nucleotide at position 74 is not removed by CCA-adding enzyme, and the subsequent incorporation at position 75 proceeds (Figure 2A and B). This is consistent with the previous observation that the reverse reaction of CCA synthesis is much slower than the forward reaction (Deutscher, 1990), and that the incorporation of the first two nucleotides proceeds by the same mechanism (Tomita et al, 2006). The first nucleotide addition at position 74 is not affected by the nucleotide composition at position 73 (discriminator nucleotide). This also holds for the nucleotide incorporation at position 75. After the second nucleotide is incorporated at position 75, the first two nucleotides are proofread by the enzyme at the subsequent AMP incorporation stage to complete CCA synthesis. The Watson–Crick-like base-pairing between the β-turn of the enzyme and the base of the nucleotide at position 74 is the primary condition that proofreads the correct nucleotide at position 74. In addition, the size and the presence of hydrogen bonds between Thr130 and the O2 of the pyrimidine of the nucleotide at position 75 are the secondary conditions that proofread the correct nucleotide at position 75. These two conditions in the closed form of the enzyme and RNA complex ensure the synthesis of the CCA end of tRNA. To support this, significant AMP incorporations into mini-helix variants, mini-D73N74N75 (N is either A, G, or U), were not detected (Figure 5C). Mini-D73C74U75 can accept AMP efficiently, and mini-D73C74U75A76 can be synthesized (Figure 5A). This is explained by the aforementioned proofreading mechanism, although the mechanism for AMP incorporation at position 76 proceeds by a dynamic change of the enzyme and the 3′-end of RNA, as in CMP incorporation at positions 74 and 75. This also suggests that the interaction between the 4-amino group of the pyrimidine and the phosphate backbone of D73 does not have a significant impact on the proofreading, although this interaction contributes to the significant AMP incorporation into mini-D73U74C75 (Figure 2D).
The extent of nucleotide mis-incorporation by the class I CCA-adding enzyme is quite low. In the presence of all four nucleotides (ATP, GTP, CTP, and UTP), CTP and ATP are exclusively incorporated at positions 74 and 75, and 76, respectively (Yue et al, 1996). In vitro analyses revealed that the extent of UMP incorporation at position 74 of mini-D73 or at position 75 of mini-D73C74 is estimated as ∼2–5 × 10−3 of that of CMP incorporation (Supplementary Figure 9). On the basis of complex crystal structures of AFCCA and the mini-helices, the 3′-region of the RNA does not contribute to its affinity for the enzyme (Tomita et al, 2006). Indeed, the Km values for mini-D73C74C75 and mini-D73C74U75 were estimated as 0.09 and 0.19 μM, respectively, and the affinity of mini-D73C74U75 for the enzyme is within the same order of magnitude as that of mini-D73C74C75 (Supplementary Figure 9). These results suggest that, when tRNA possessing U74C75 or C74U75 end is synthesized by mis-incorporation of UMP, the fraction of synthesized tRNAs possessing U74C75A76 or C74U75A76 would be less than 0.2–0.5% of the total tRNAs in the cells. Moreover, tRNA possessing U74C75A76 or C74U75A76, which is not usable, might be degraded in the cells by a quality control system, such as by polyadenylation (Kadaba et al, 2004; LaCava et al, 2005), as it cannot be functional in the translational system. Therefore, the fraction of tRNA carrying the incorrect 3′-nucleotides remains low in the cell. To synthesize the correct CCA sequence, the CCA-adding enzyme selects the correct nucleotide during each cycle of the polymerization reaction, and the extent of the incorporation of incorrect nucleotides is severely prevented. Nevertheless, the CCA-adding enzyme still possesses an additional proofreading mechanism to identify the mis-incorporated nucleotide and prevent the synthesis of unusable tRNA. The class I CCA-adding enzyme lacks an activity to remove a mis-incorporated nucleotide. Instead, it acquired a unique mechanism to enhance the fidelity of correct CCA synthesis. In addition to the recently revealed mechanism for the dynamic selection of a nucleotide by a protein–RNA complex, the proofreading mechanism for the maintenance of fidelity for CCA synthesis is also based on the function of a protein–RNA complex, confirming the importance of protein–RNA complexes exercising their functions in collaborative manners.
Materials and methods
Preparation of AFCCA and mini-helices
AFCCA was overexpressed in Escherichia coli and purified as described (Tomita et al, 2006). Nine distinct tRNA mini-helices, derived from Thermotoga maritima tRNAPhe, ending in A74, G74, U74, C74A75, C74G75, C74U75, A74C75, G74C75 and U74C75 (referred to as mini-D73A74, mini-D73G74, mini-D73U74, mini-D73C74A75, mini-D73C74G75, mini-D73C74U75, mini-D73A74C75, mini-D73G74C75, and mini-D73U74C75, respectively; thereafter, D is the discriminator nucleoside, A) were synthesized by T7 RNA polymerase using synthetic DNAs as templates, and were purified by polyacrylamide gel electrophoresis under denaturing conditions. The purities of the mini-helices were more than 99%, as judged from electrophoresis.
Crystallization and data collection
We co-crystallized AFCCA complexes with nine distinct tRNA mini-helices, as described (Tomita et al, 2006). All of the binary and ternary complex crystals belong to the space group P43212, and contain one complex molecule in the asymmetric unit. For the preparation of the ternary complex with an incoming CTP or ATP, the crystals were soaked in a reservoir solution containing 2 mM NTP (CTP or ATP) at 20°C for 2 h. The crystals were cryo-protected with 20% (v/v) ethylene glycol and were flash-cooled in a 100-K nitrogen stream, and the data were collected at the beam-lines NW-12A, BL5A, and BL17A of KEK (Tsukuba, Japan). All of the data were processed using the program HKL2000 (Otwinowski and Minor, 1997).
Structure determination of AFCCA complexes with mini-helices
The crystal structures were solved at 2.50- to 3.05-Å resolutions by molecular replacement with the program AMoRe (Navaza, 1994), using the refined complex structures with the mini-D73C74 as a search model (Tomita et al, 2006). The initial model was manually modified using the program O (Jones et al, 1991), and then underwent several cycles of refinement with CNS (Brunger et al, 1998). The refinement statistics are shown in Supplementary Table 1.
In vitro CMP and AMP incorporation assays
In vitro CCA adding assays were performed as described (Tomita et al, 2006), with slight modifications. For AMP incorporation into mini-D73N74N75, reaction mixtures containing 50 mM glycine-KOH, pH 8.5, 15 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 100 μM ATP, 100 nM α-32P ATP (3000 Ci/mmol; GE Healthcare), 5 μM mini-helix, and 1 μg/ml AFCCA were incubated at 45°C for 5 min. The assay procedures for CMP incorporation into mini-D73N74 were the same as those described above, except that 100 μM CTP, 100 μM ATP, and 100 nM α-32P CTP (3000 Ci/mmol; GE Healthcare) were used instead of the 100 μM ATP and 100 nM α-32P ATP. The reaction was stopped by adding an equal volume of stop buffer (9 M urea, 0.02% BPB, 0.02% XC). Under these conditions, the reaction proceeds in a linear range. The products were separated by 12% (w/v) polyacrylamide gel electrophoresis under denaturing conditions, and the intensity of 32P-labelled RNAs was quantified by a BAS-2500 imager (Fuji Film, Japan).
Neighbouring nucleotide analysis by thin-layer chromatography
32P-labelled products were separated by 12% (w/v) polyacrylamide gel electrophoresis under denaturing conditions, and were excised from the gels and eluted from the slices. The 32P-labelled products were completely digested in a 10 μl solution containing 50 mM Tris-Cl, pH 7.0, 10 000 cpm 32P-labelled RNA, and 1 unit of RNase T2 (Invitrogen, Japan) at 37°C for 4 h. The hydrolysed products were separated by thin-layer chromatography (Kodak) using a developing solution (2-propanol/HCl/water (70:15:15 v/v/v)) (Kuchino et al, 1987), and the 32P-labelled nucleotides were visualized by a BAS-2500 imager (Fuji Film).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Alan Weiner of the University of Washington (Seattle) for facilities and support during the initial biochemical experiments. We thank the beam-line staffs of BL-5A, AR-NW-12A and BL-17A (KEK, Tsukuba, Japan) for technical assistance during data collection, and Azusa Hamada of AIST for technical assistance. This study was supported by grants from JSPS to young scientists, MEXT for the priority area of science, and the PRESTO program of JST to KT. The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 2ZH1, 2ZH2, 2ZH3, 2ZH4, 2ZH5, 2ZH6, 2ZH7, 2ZH8, 2ZH9, 2ZHA, and 2ZHB).
References
- Augustin MA, Reichert AS, Betat H, Huber R, Mörl M, Steegborn C (2003) Crystal structure of the human CCA-adding enzyme: insights into template-independent polymerization. J Mol Biol 328: 985–994 [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP (1998) Polymerases and the replisome: machines within machines. Cell 92: 295–305 [DOI] [PubMed] [Google Scholar]
- Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A (2000) Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science 289: 1346–1349 [DOI] [PubMed] [Google Scholar]
- Bralley P, Chang SA, Jones GH (2005) A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases. J Bacteriol 187: 5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Deutscher MP (1973) A novel nucleolytic activity associated with rabbit liver tRNA nucleotidyltransferase. Biochem Biophys Res Commun 52: 216–222 [DOI] [PubMed] [Google Scholar]
- Deutscher MP (1990) Transfer RNA nucleotidyltransferase. Methods Enzymol 181: 434–439 [DOI] [PubMed] [Google Scholar]
- Evans JA, Deutscher MP (1978) Kinetic analysis of rabbit liver tRNA nucleotidyltransferase. J Biol Chem 253: 7276–7281 [PubMed] [Google Scholar]
- Friedman RA, Honig B (1995) A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J 69: 1528–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Noller HF (1997) Ribosomes and translation. Annu Rev Biochem 66: 679–716 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci 20: 345–347 [DOI] [PubMed] [Google Scholar]
- Hou YM, Lipman RSA, Zarutskie JA (1998) A tRNA circularization assay: evidence for the variation of the conformation of the CCA end. RNA 4: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou YY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DF, Green R (1999) Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4: 859–864 [DOI] [PubMed] [Google Scholar]
- Kuchino Y, Hanyu N, Nishimura S (1987) Analysis of modified nucleosides and nucleotide sequence of tRNA. Methods Enzymol 155: 379–396 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Li F, Xiong Y, Wang J, Cho HD, Tomita K, Weiner AM, Steitz TA (2002) Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 111: 815–824 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J 15: 2593–2603 [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W (2004) Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases. RNA 10: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W (2007) RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W, Doublié S (2000) Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J 19: 4193–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Okabe M, Tomita K, Ishitani R, Ishii R, Takeuchi N, Arisaka F, Nureki O, Yokoyama S (2003) Divergent evolutions of trinucleotide polymerization revealed by an archaeal CCA-adding enzyme structure. EMBO J 22: 5918–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Cramer F (1979) The -C-C-A end of tRNA and its role in protein biosynthesis. Prog Nucleic Acid Res Mol Biol 22: 1–69 [DOI] [PubMed] [Google Scholar]
- Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O (2004) Structural basis for template-independent RNA polymerization. Nature 430: 700–704 [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishitani R, Fukai S, Nureki O (2006) Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature 443: 956–960 [DOI] [PubMed] [Google Scholar]
- Tomita K, Weiner AM (2001) Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science 294: 1334–1336 [DOI] [PubMed] [Google Scholar]
- Tomita K, Weiner AM (2002) Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J Biol Chem 277: 48192–48198 [DOI] [PubMed] [Google Scholar]
- Weiner AM (2004) tRNA maturation: RNA polymerization without a nucleic acid template. Curr Biol 14: 883–885 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Li F, Wang J, Weiner AM, Steitz TA (2003) Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes. Mol Cell 12: 1165–1172 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA (2004) Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430: 640–645 [DOI] [PubMed] [Google Scholar]
- Yue D, Maizels N, Weiner AM (1996) CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2: 895–908 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information