Abstract
B-Myb is one member of the vertebrate Myb family of transcription factors and is ubiquitously expressed. B-Myb activates transcription of a group of genes required for the G2/M cell cycle transition by forming the dREAM/Myb–MuvB-like complex, which was originally identified in Drosophila. Mutants of zebrafish B-myb and Drosophila myb exhibit defects in cell cycle progression and genome instability. Although the genome instability caused by a loss of B-Myb has been speculated to be due to abnormal cell cycle progression, the precise mechanism remains unknown. Here, we have purified a B-Myb complex containing clathrin and filamin (Myb–Clafi complex). This complex is required for normal localization of clathrin at the mitotic spindle, which was previously reported to stabilize kinetochore fibres. The Myb–Clafi complex is not tightly associated with the mitotic spindles, suggesting that this complex ferries clathrin to the mitotic spindles. Thus, identification of the Myb–Clafi complex reveals a previously unrecognized function of B-Myb that may contribute to its role in chromosome stability, possibly, tumour suppression.
Keywords: B-Myb, clathrin, filamin, genome instability, mitotic spindle
Introduction
The Myb family of transcription factors has important functions in proliferation and differentiation (Oh and Reddy, 1999). The vertebrate myb gene family contains three members, A-myb and B-myb, in addition to c-myb, the cellular progenitor of the v-myb oncogene (Nomura et al, 1988). The level of c-myb expression is high in immature haematopoietic cells (Gonda and Metcalf, 1984), and analysis of c-myb KO mice revealed that c-myb is required for definitive haematopoiesis and T-cell development at several stages (Mucenski et al, 1991; Bender et al, 2004). A-myb is also highly expressed in the limited types of cells such as testis and breast (Trauth et al, 1994), and is required for spermatogenesis and development of breast tissue following pregnancy (Toscani et al, 1997). In contrast to c-myb and A-myb, B-myb is ubiquitously expressed (Nomura et al, 1988) and the B-myb mutant mice die early in development (Tanaka et al, 1999), suggesting the universal role of B-myb gene product (B-Myb) in many types of cells.
All the three members of vertebrate Myb family proteins have the conserved DNA-binding domain (DBD), which recognizes the specific DNA sequence 5′-AACNG-3′ (Ogata et al, 1994). The transcriptional activation domain containing the acidic amino acid-rich region is also localized adjacent to the DBD in all the three members (Sakura et al, 1989; Nakagoshi et al, 1993), which bind to the transcriptional coactivator CBP (Dai et al, 1996; Bessa et al, 2001). B-Myb is induced at G1/S in the cell cycle and is activated by cyclinA–cdk2-dependent phosphorylation (Robinson et al, 1996), suggesting an important role in cell cycle control.
Of the vertebrate Myb proteins, B-Myb is most closely related to Drosophila Myb (dMyb) (Davidson et al, 2005). Mutants of dmyb and zebrafish B-myb exhibit defects in cell cycle progression (Katzen et al, 1998; Okada et al, 2002) and genome instability (Fung et al, 2002; Manak et al, 2002, 2007; Shepard et al, 2005). A downregulation of B-Myb in mouse megakaryocytes by small interfering RNA (siRNA) also causes genome instability (García and Frampton, 2006). dMyb forms a multisubunit protein complex that is essential for silencing of developmentally regulated genes (Korenjak et al, 2004; Lewis et al, 2004). This complex, referred to as dREAM/Myb–MuvB complex, contained dMyb, RBF, E2F, DP, in addition to the previously identified dMyb-interacting proteins Mip120, Mip130, and Mip40 (Beall et al, 2002, 2004). Interestingly, Caenorhabditis elegans homologues for each component of this are products of the synthetic multivulva class B (synMuvB) genes that function to antagonize Ras signalling in development of the vulva (Fay and Han, 2000; Ceol et al, 2006). The C. elegans synMuvB gene products form a complex termed Dpl–Rb–MuvB similar in composition to the dREAM/Myb–MuvB complex (Harrison et al, 2006). Similar to dMyb, B-Myb forms the dREAM/Myb–MuvB-like complex containing p130, a member of RB family, E2F4, and LIN-9, a homologue of Mip130, and activates transcription of a group of genes such as cyclin B1, which are required for the G2/M cell cycle transition (Zhu et al, 2004; Osterloh et al, 2006; Litovchick et al, 2007; Pilkinton et al, 2007). Recently, dMyb was also shown to activate a cluster of the genes required for the G2/M transition (Wen et al, 2008).
The genome instability caused by the mutations of dmyb and B-myb is speculated to be due to abnormal cell cycle progression, especially at the G2/M transition, but the precise mechanism remains unknown. Here, we have purified a B-Myb complex containing clathrin and filamin, which is required for localization of clathrin at the mitotic spindles. Disruption of this complex has led to genome instability, suggesting that B-Myb directly functions for the mitotic spindle.
Results
Purification of Myb–Clafi complex that is abundant in the M-phase cells
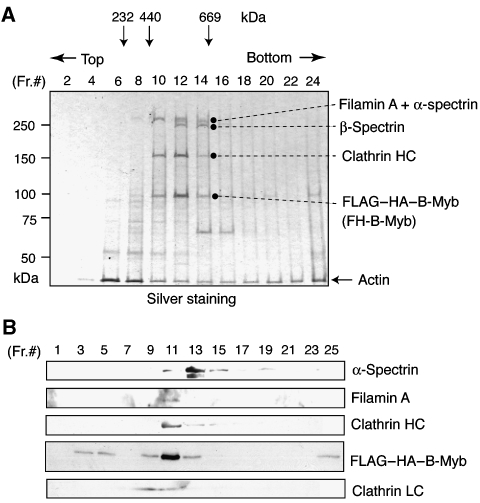
The HeLa S3 cell line, in which FLAG–HA–B-Myb is stably expressed, was generated by retroviral transduction. Western blotting analysis indicated that two clones (clones 7 and 9) among several isolated lines expressed FLAG–HA–B-Myb at levels similar to that of endogenous B-Myb (Supplementary Figure S1A). Clone 7 was cultured to purify the B-Myb complex. Cells were disrupted in hypotonic buffer, and the pellets that contained the nuclei of inter-phase cells and also the condensed chromosomes associated with mitotic spindles of M-phase cells were isolated. The B-Myb complex was purified from the extracts of the pellets by sequential immunoprecipitation with anti-FLAG and anti-HA antibodies, as described by Nakatani and Ogryzko (2003). Silver staining of SDS−PAGE gels of the purified proteins indicated that, in addition to FLAG–HA–B-Myb, the complex contained two high molecular weight bands of 280 and 160 kDa, which were not detected in the purified sample from control HeLa cells not expressing epitope-tagged proteins (Supplementary Figure S1B). Analysis of the FLAG-purified B-Myb complex using glycerol gradient centrifugation indicated that only these two large proteins were detected in the B-Myb-containing fractions (Figure 1A). Mass spectrometric analysis indicated that the 280-kDa band contained filamin A and α-spectrin, whereas the 160-kDa band was the clathrin heavy chain (CHC). Western blotting using specific antibodies indicated that filamin A was detected in the B-Myb-concentrating fractions, whereas α-spectrin was concentrated in the slightly higher molecular weight fraction (Figure 1B). Thus, the B-Myb complex containing clathrin and filamin A was concentrated in fractions 11 and 12, whereas α- and β-spectrin, which may form a heterodimer, were concentrated in the fractions 13 and 14. We designated this complex Myb–Clafi for the Myb complex containing clathrin and filamin. The molecular weight of the Myb–Clafi complex is approximately 500–600 kDa, suggesting that this complex consists of one molecule each of filamin A, CHC, CLC, and B-Myb. In addition to B-Myb complexed with clathrin and filamin A, some amounts of free B-Myb and aggregated B-Myb were also detected in the fractions 3–5 and 25 of the glycerol gradient, respectively (Figure 1B). We have used the extraction buffer containing high salt (0.42 M NaCl) without detergent, under which about 50% of B-Myb was recovered in the lysates (data not shown). B-Myb remained in the chromatin pellet could form the dREAM/Myb–MuvB-like complex.
Figure 1.
Identification of the Myb–Clafi complex. (A) Analysis of the Myb–Clafi complex by glycerol gradient centrifugation. The B-Myb complex was immunopurified from HeLa cells expressing FLAG–HA-tagged B-Myb using anti-FLAG. The immunopurified complex was separated on a 10–40% glycerol gradient by ultracentrifugation, resolved by SDS−PAGE, and visualized by silver staining. The polypeptides identified by mass spectrometric analysis are indicated. (B) Immunoblotting of the Myb–Clafi complex. The glycerol gradient-purified B-Myb complex was analysed by immunoblotting with the indicated antibodies.
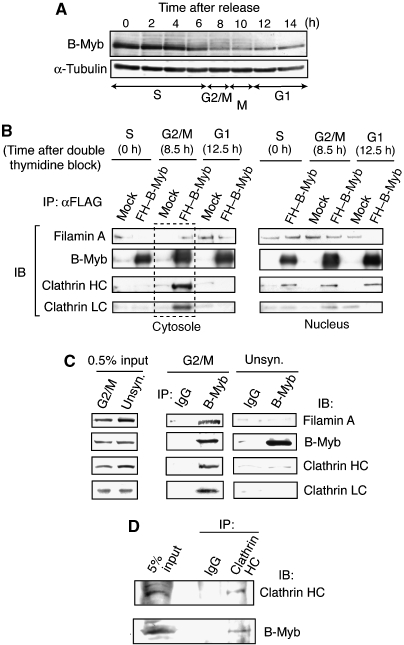
To examine the abundance of the Myb–Clafi complex during cell cycle, we prepared synchronized HeLa cells. HeLa cells expressing FLAG–HA–B-Myb were synchronized at the G1–S boundary by the double thymidine-block protocol, and the cell cycle distribution in G1, S, or G2/M was determined by fluorescence activated cell sorting (FACS) analysis of cells at various times after release of the block (Supplementary Figure S2). The results indicated that a substantial fraction of the cells was synchronized by this protocol. To examine the abundance of B-Myb itself throughout the cell cycle, lysates were prepared at various times after release of the thymidine block. The level of B-Myb was highest in S phase and early G2/M phase (Figure 2A). At M phase, the B-Myb level decreased, but continued to be present at low levels throughout the G1 phase.
Figure 2.
Myb–Clafi complex is abundant in the M-phase cells. (A) Levels of B-Myb during the cell cycle. Whole-cell lysates were prepared from HeLa cells at various times after release from the G1/S block, and used for western blotting with the anti-B-Myb antibody. (B) Co-immunoprecipitation of FLAG–HA–B-Myb with clathrin and filamin A. HeLa cells expressing FLAG–HA–B-Myb (FH–B-Myb) or no epitope-tagged protein (mock) were synchronized by double thymidine block, and subsequently released from the G1–S boundary. At the indicated times after release, cells were lysed and separated into nuclear and cytosolic fractions. Cytosolic (left panels) and nuclear (right panels) fractions were immunoprecipitated with anti-FLAG, and the precipitates were subjected to immunoblot analysis with the antibodies indicated on the left. (C) Co-immunoprecipitation of endogenous B-Myb with clathrin and filamin A. The G2/M-phase-enriched (G2/M) or unsynchronized (unsyn.) parental HeLa cells were lysed and cytosolic fractions were immunoprecipitated with anti-B-Myb or control IgG, and the precipitates were subjected to immunoblot analysis to detect the proteins with the antibodies indicated on the right. (D) Co-immunoprecipitation using anti-CHC. Co-immunoprecipitation experiments were performed as described above except for the use of anti-CHC instead of anti-B-Myb.
Under the same salt concentration as the Myb–Clafi complex purification, CHC, CLC, and filamin A co-precipitated with FLAG–HA–B-Myb only in cytosolic lysates of G2/M-phase-enriched cells, but not in the nuclear fraction (Figure 2B). When nuclear fractions from different stages of the cell cycle were used, small amounts of CHC co-precipitated with FLAG–HA–B-Myb, but neither filamin A nor CLC was present (Figure 2B). These results suggest that CHC may regulate B-Myb-dependent transcription. A similar association between CHC and p53 was recently reported (Enari et al, 2006). To examine whether endogenous B-Myb forms the Myb–Clafi complex, we performed co-immunoprecipitation using HeLa cells. Anti-B-Myb antibody co-precipitated endogenous clathrin and filamin from cytosolic extracts of G2/M-phase-enriched cells, whereas control IgG did not (Figure 2C). When we used lysates of unsynchronized cells, we could not observe this type of specific co-immunoprecipitation, possibly due to small number of M-phase cells. Anti-CHC antibody also co-precipitated endogenous B-Myb from cytosolic extracts of G2/M-phase-enriched cells, although control IgG did not (Figure 2D). Thus, the Myb–Clafi complex is abundant in the cytosolic fraction of M-phase cells. The complex was originally purified from the extracts of pellets of cell lysates in the hypotonic buffer. This may be due to an incomplete separation of the condensed chromosomes associated with the mitotic spindles from the cytosolic fractions of M-phase cells.
Decrease in B-Myb reduces clathrin at the mitotic spindles and leads to mitotic arrest
Clathrin functions in the generation of vesicles that transfer membrane and proteins between intracellular compartments (Edeling et al, 2006). Independent of this function, clathrin was reported to be localized on and required for the function of the mitotic spindle (Okamoto et al, 2000; Royle et al, 2005). Filamin is an actin crosslinking and molecular scaffold protein (Feng and Walsh, 2004; Popowicz et al, 2006). These results raised the possibility that the Myb–Clafi complex has some role to carry clathrin to the mitotic spindles. To investigate the importance of B-Myb in the localization of clathrin at the kinetochore fibres, we combined cell synchronization with RNAi. HeLa cells were transfected with an siRNA during the interval between the two thymidine blocks and were examined for expression of B-Myb 6–9 h after release into the cell cycle. Transfection of cells with siRNAs specific for B-Myb resulted in a specific reduction in the level of B-Myb (Supplementary Figure S3A). Depletion of B-Myb did not affect the levels of clathrin and filamin (Supplementary Figure S3A), but markedly impaired the localization of endogenous CHC (Figure 3A) and exogenously expressed EGFP-CLC to the kinetochore fibres (Figure 3B). These results raised the possibility that the B-Myb–Clafi complex has a function for the localization of clathrin at the mitotic spindle.
Figure 3.
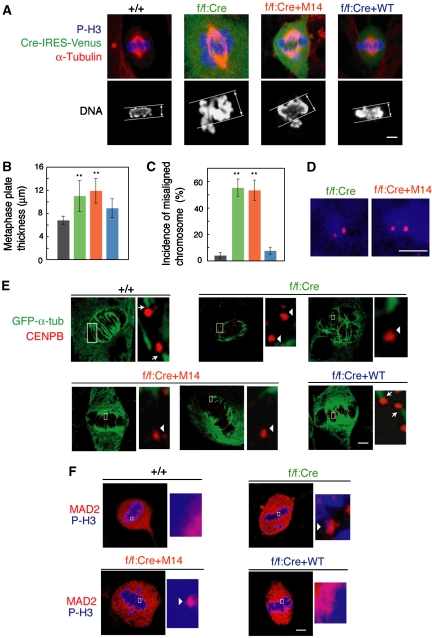
Decrease in B-Myb reduces clathrin on the mitotic spindles and leads to mitotic arrest. (A) Decrease in B-Myb reduces clathrin signals on the mitotic spindles. HeLa cells were treated with a control luciferase siRNA (upper panels) or B-myb siRNA (lower panels), subjected to indirect immunofluorescent staining with anti-clathrin HC (Cy3, red), and anti-α-tubulin (Alexa Fluor 488, green). DNA was stained with TOTO-3 iodide (blue) and cells were visualized using confocal microscopy. Scale bars: 10 μm. On the right, the clathrin signal at the mitotic spindle was quantified and indicated by bar graph with ±s.d. **P<0.01. A value of one represents no specific signal at the mitotic spindle (see Materials and methods). (B) B-Myb depletion decreased clathrin light chain (CLC) signal localization to the mitotic spindle. HeLa cells expressing EGFP-CLC were isolated, and were then transfected with B-myb siRNA or control luciferase siRNA. Cells were extracted with 1% Triton X-100 for 10 min prior to fixation, and immunostained with anti-GFP and visualized using confocal microscopy. Scale bars: 10 μm. (C) B-Myb depletion enhances mitotic arrest. The mitotic incidence of HeLa cells transfected with B-myb siRNA or control luciferase siRNA are indicated in the bar graph. (D, E) B-Myb depletion caused thicker metaphase plates and an increase in misaligned chromosomes. HeLa cells transfected with B-myb siRNA or control luciferase siRNA were fixed, washed with 0.2% Triton X-100 for 10 min, and immunostained with anti-Aurora B or anti-CENPB (Cy3, red), anti-α-tubulin (Alexa Fluor 488, green), and TOTO-3 iodide (blue), and visualized using confocal microscopy. A typical example is shown. The misaligned chromosomes (arrow) and thickness of the metaphase plates are indicated. Scale bars: 10 μm.
Depletion of B-Myb markedly impaired the ability of HeLa cells to progress through mitosis, whereas transfection of cells with a control siRNA had no such effect. The B-Myb-depleted cells took an unusually long time to complete mitosis, displayed marked difficulties in completing cytokinesis, and finally had an apoptosis-like phenomena (Supplementary Figure S3B and Supplementary Videos S1 and S2). The proportion of cells in mitosis increased 4.9-fold in the B-Myb-depleted cells compared with the control siRNA-treated cells (Figure 3C). In B-Myb-depleted cells, the metaphase plate was thicker than in the control cells and there was an increased incidence of misaligned chromosomes (Figures 3D and E). These severe effects of B-myb siRNA on mitosis might be the result of two different mechanisms: disruption of the Myb–Clafi complex and decreased expression of B-Myb target genes mediated by the dREAM/Myb–MuvB-like complex, which were recently shown to be required for G2/M transition (Zhu et al, 2004; Osterloh et al, 2007; Litovchick et al, 2007; Pilkinton et al, 2007).
The Myb–Clafi complex does not tightly associate with the mitotic spindles
To examine the localization of B-Myb–Clafi complex, synchronized HeLa cells were immunostained with anti-B-Myb. At inter-phase, B-Myb was observed in the nucleus in dot-like structures (Supplementary Figure S4A). In M-phase cells, only very weak B-Myb signals were detected at the kinetochore fibres, whereas most of B-Myb was thought to be washed out with Triton X-100 treatment prior to fixation, which is commonly used to detect microtubule-associated proteins (Supplementary Figure S4A). As the endogenous B-Myb signals were weak, we generated a HeLa cell line expressing a fusion of Venus, a GFP derivative (Nagai et al, 2002), with B-Myb (Venus–B-Myb). HeLa cells were infected with a retrovirus vector encoding Venus–B-Myb and then isolated. In the nucleus of inter-phase cells expressing Venus–B-Myb, Venus–B-Myb was observed with some dot-like structures as seen in the case of endogenous B-Myb (Supplementary Figure S4B). In M-phase cells, Venus–B-Myb was again only weakly detected at the kinetochore fibres. As the B-Myb signals at the mitotic spindles were much weaker than that of clathrin, we further investigated whether anti-FLAG and anti-HA antibodies detect FLAG–HA–B-Myb at the mitotic spindles in HeLa cells expressing FLAG–HA–B-Myb. Neither antibody detected FLAG–HA–B-Myb at the mitotic spindles (data not shown). We also could not detect B-Myb in the purified mitotic spindle fractions, although clathrin was detected as reported by Mack and Compton (2001) (data not shown). Further, immunostaining using anti-filamin antibodies indicated the filamin is not specifically concentrated at the mitotic spindles (data not shown). Filamin was uniformly distributed in the cell with enriched signals at the cell periphery. These results suggest that the Myb–Clafi complex does not tightly associate with the mitotic spindles, although it could have a weak trend to gather around the mitotic spindles. On the basis of these results, we speculated that the Myb–Clafi complex may have a function to ferry clathrin to the mitotic spindles.
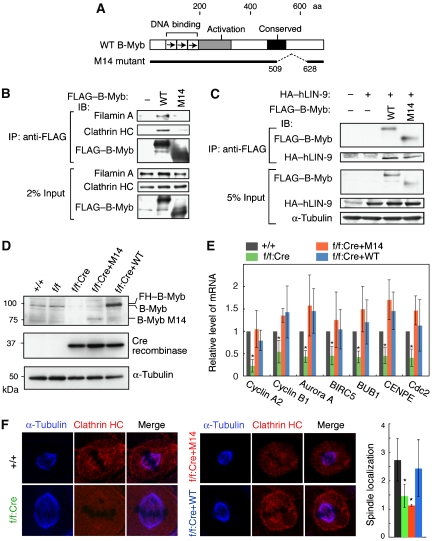
M14 mutant can form the dREAM/Myb–MuvB-like complex, but not the Myb–Clafi complex
To examine the specific role of the Myb–Clafi complex during mitosis, especially by separating it from that of the dREAM/Myb–MuvB-like complex, we have used the B-Myb mutant 14 (M14). The M14 mutant cannot form the Myb–Clafi complex, but still retain the trans-activating capacity mediated by the dREAM/Myb–MuvB-like complex. M14 lacks the 118-amino-acid region (aa 509–628) in the C-proximal region of B-Myb (Figure 4A). Previously we demonstrated that M14 can stimulate transcription from the Myb site-containing promoter in transient transfection assays (Nakagoshi et al, 1993). In co-immunoprecipitations using lysates from HeLa cells transfected with the M14 or wild-type (WT) B-Myb expression vector, clathrin and filamin A co-precipitated with WT B-Myb, but not with M14 (Figure 4B). Human LIN-9 (hLIN-9), which is a component of the human dREAM/Myb–MuvB-like complex (Litovchick et al, 2007), co-precipitated with both WT and M14 B-Myb (Figure 4C), suggesting that M14 retains the trans-activating capacity mediated by the dREAM/Myb–MuvB-like complex.
Figure 4.
The B-Myb mutant M14 retains its trans-activation capacity but cannot form the Myb–Clafi complex. (A) Structures of WT and M14 B-Myb are shown schematically. (B) M14 interacts with neither clathrin nor filamin A. HeLa cells were transfected with the FLAG–HA-tagged WT or M14 B-Myb expression plasmid, and whole-cell lysates were used for coimmunoprecipitation with anti-FLAG, followed by western blotting to detect the proteins indicated on the left. (C) M14 can bind to hLIN-9. HeLa cells were co-transfected with a FLAG-tagged WT or M14 B-Myb expression plasmid and a HA–hLIN-9 expression plasmid, and nuclear extracts were used for coimmunoprecipitation with anti-FLAG, followed by western blotting to detect the proteins indicated on the left. (D) Preparation of MEFs expressing M14 instead of endogenous B-Myb. Whole-cell lysates were prepared from the cells, and subjected to western blotting using anti-B-Myb, anti-Cre, or anti-α-tubulin, as indicated. +/+, WT MEFs; f/f, B-mybf/f MEFs; f/f:CRE, B-mybf/f MEFs infected with the lentivirus encoding Cre recombinase; f/f:Cre+M14 and f/f:Cre+WT, B-mybf/f MEFs infected with the lentivirus encoding Cre recombinase, and then infected with the adenovirus to express FLAG–HA-tagged M14 and WT B-Myb, respectively. The level of reexpressed WT B-Myb is higher than that of endogenous B-Myb, which might be due to some variations in the reexpressed B-Myb among multiple experiments. These variations did not affect the results described below. (E) M14 can trans-activate B-Myb target genes. RNA was prepared from the indicated cells and the mRNA levels of seven B-Myb target genes were measured using real-time RT–PCR (n=6). The mRNA levels of all seven genes in the f/f:Cre cells were significantly lower than levels in the other three cell types (*P<0.05). (F) Reduction of clathrin at the mitotic spindles in M14-reexpressing cells. The indicated MEFs were subjected to indirect immunofluorescent staining with anti-clathrin HC (Cy5, red) and anti-α-tubulin (FITC, green), and visualized using confocal microscopy. DNA was visualized by staining with propidium iodide (blue). Scale bars: 5 μm. On the right, the clathrin signal at the mitotic spindle was quantified and indicated by bar graph with ±s.d. *P<0.05. A value of one represents no specific signal at the mitotic spindle (see Materials and methods).
To generate cells in which M14 was expressed instead of endogenous WT B-Myb, we made mutant mice carrying floxed (flanked by loxP sites) B-myb alleles (B-mybf/f) (Supplementary Figure S5). When the Cre recombinase was expressed in B-mybf/f mouse embryonic fibroblasts (MEFs), endogenous B-Myb was almost completely undetectable (Figure 4D). To exogenously express M14 or WT B-Myb as a control, B-mybf/f MEFs were first infected with the Cre lentivirus, and then re-infected with adenoviruses encoding either M14 or WT B-Myb. The levels of reexpressed B-Myb had some variations among multiple experiments (Figure 4D), but these variations did not affect the results described below. In B-Myb-depleted MEFs, the mRNA levels of seven G2/M genes, which are regulated by B-Myb and hLin-9 (Zhu et al, 2004; Osterloh et al, 2006), were lower than those of control cells (Figure 4E). However, reexpression of either WT or M14 B-Myb efficiently rescued the decrease in these mRNA levels. Thus, M14 can activate the transcription of a cluster of B-Myb target genes. In M14-reexpressing cells (f/f:Cre+M14) and B-Myb-deficient cells (f/f:Cre), the CHC signals at kinetochore fibres were dramatically reduced, while CHC was prominently localized to the mitotic spindle in WT B-Myb-reexpressing cells (f/f:Cre+WT) (Figure 4F). Thus, the ability of B-Myb to form the Myb–Clafi complex is correlated with that of B-Myb to carry clathrin to the kinetochore fibres.
Defects in mitosis of MEFs expressing M14 instead of endogenous WT B-Myb
To investigate the role of the B-Myb–Clafi complex during mitosis, we examined mitotic progression in four types of MEFs: control wild-type MEFs (+/+), B-Myb-deficient cells that were generated by infecting B-mybf/f MEF cells with the Cre lentivirus (f/f:Cre), cells expressing M14 instead of endogenous B-Myb that were generated by sequentially infecting B-mybf/f MEF cells with the Cre lentivirus and the M14-encoding adenovirus (f/f:Cre+M14), and B-Myb-reexpressing cells that were generated by sequentially infecting B-mybf/f MEF cells with the Cre lentivirus and the WT B-Myb-encoding adenovirus (f/f:Cre+WT). The B-Myb-deficient cells and the M14-reexpressing cells took an unusually long time to complete mitosis, displayed marked difficulties in completing cytokinesis and had frequent defects in chromosome segregation (Supplementary Figure S6 and Supplementary Videos S3, S4, S5 and S6), although mitosis in the parental B-mybf/f cells appeared normal. The M14-reexpressing cell underwent anaphase, attempted to generate a cleavage furrow but failed to complete a cytokinesis. As a result, the cell appeared to re-enter a G1-like state with doubled DNA content. The M14-reexpressing cell remained at anaphase for a prolonged period of time, resulting in a binucleate cell. These binucleate cells entered the next cell division, but again failed to cleave for a prolonged period, resulting in multinucleation. Thus, the failure of cytokinesis caused the accumulation of centrosomes by duplication in a repeated S phase of the cell cycle. In control experiments, reexpression of WT B-Myb suppressed the abnormalities seen in the B-Myb-deficient cells.
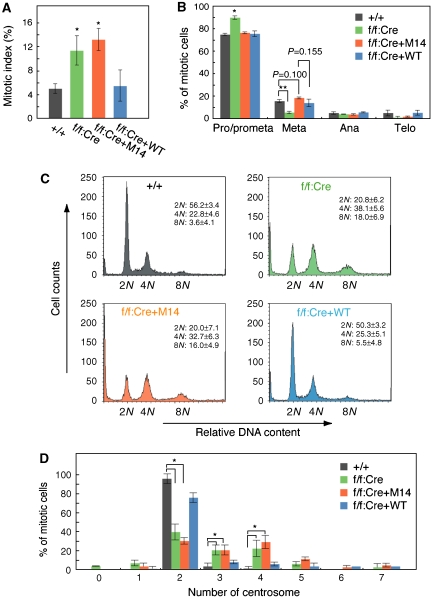
Consistent with these observations, the proportion of cells in mitosis (the mitotic index) increased 2.6- and 2.3-fold in the B-Myb-deficient cells and M14-reexpressing cells, respectively (WT MEFs, 5.0±0.8%; f/f:Cre cells, 11.4±2.4%; f/f:Cre+M14, 13.2±1.8%) (Figure 5A). We examined the proportion of cells at each stage of mitosis. B-Myb-deficient cell populations had significantly greater number of cells in prometaphase compared with wild-type cells (P<0.05) (Figure 5B). M14-reexpressing cells had slightly increased numbers of cells in metaphase compared with wild-type cells or WT-reexpressing cells, although the trends were not significant (P=0.1 and P=0.115, respectively). Thus, M14-reexpressing cells spent longer periods of time in metaphase compared with WT cells.
Figure 5.
The B-Myb M14 mutant delays mitosis and causes aneuploidy. (A) Increase in mitotic index in the M14-reexpressing cells and the B-Myb-deficient cells. The mitotic index of MEFs from the above experiments is shown. n=235–589. *P<0.05. (B) The histogram shows the percentage of mitotic cells, relative to controls, at each stage in mitosis. Values are mean+s.e.m. *P<0.05; **P<0.001. n=300. (C) Multiploidy in M14-reexpressing cells and the B-Myb-deficient cells. The indicated cell types were analysed by FACS (n=3). (D) Abnormal numbers of centrosomes in M14-reexpressing cells and the B-Myb-deficient cells. The number of centrosomes was examined by immunostaining with anti-γ-tubulin. *P<0.05. n=120.
FACS analysis demonstrated that the M14-reexpressing cells and the B-Myb-deficient cells included readily detectable fractions of cells with chromosomal contents of 8N, which were not evident in the WT MEFs or the WT B-Myb-reexpressing cells (Figure 5C). We also quantified the number of centrosomes per cell in MEFs. About 70% of the M14-reexpressing cells had an abnormal number of centrosomes, approximately 1/3 had four centrosomes and 1/6 had three centrosomes (Figure 5D). In contrast, approximately 90% of parental MEFs and 70% of WT B-Myb-reexpressing cells had two centrosomes. Thus, cells expressing M14 B-Myb instead of the normal B-Myb exhibited defects in mitosis and abnormalities in DNA content and centrosome number.
The M14-reexpressing MEFs and the B-Myb-deficient cells exhibited various mitotic abnormalities, which have also been observed in clathrin-depleted cells (Royle et al, 2005). The metaphase plate in the M14-reexpressing cells and the B-Myb-deficient cells was thicker than in parental MEFs and WT B-Myb-reexpressing cells (Figures 6A and B) and there was an increased incidence of misaligned chromosomes in M14-reexpressing cells and the B-Myb-deficient cells (Figure 6C). Misaligned chromosomes were observed in 53.3±7.6% of the M14-reexpressing cells and in 55.4±6.5% of the B-Myb-deficient cells, but only in 7.6±2.7% of the WT B-Myb-reexpressing cells. In the M14-reexpressing cells and the B-Myb-deficient cells, misaligned chromosomes usually consisted of pairs of sister chromatids (Figure 6D). After depolymerization of microtubules by cold treatment, M14-reexpressing cells and the B-Myb-deficient cells often contained centromeres that did not have spindle fibres attached (Figure 6E, arrowheads), suggesting that the stability of the kinetochore–spindle contacts was reduced. In WT B-Myb-reexpressing cells, Mad2 correctly localized to kinetochores at early prometaphase and then became diffusely distributed at metaphase (Figure 6F), indicating that correct attachment of chromosomes to kinetochore fibres was monitored by the spindle checkpoint (Cleveland et al, 2003). In contrast, in M14-reexpressing cells and the B-Myb-deficient cells, Mad2 was found on the kinetochores of misaligned chromosomes as well as chromosomes at the metaphase plate, indicating that the spindle checkpoint was still activated on properly aligned chromosomes. These results indicate that the primary cause of prolonged mitosis in M14-reexpressing cells was the continued activation of the spindle checkpoint, which resulted from destabilization of kinetochore microtubules.
Figure 6.
The Myb–Clafi complex is required for the function of the mitotic spindle. (A) Thicker metaphase plates in M14-reexpressing cells and the B-Myb-deficient cells. The indicated MEFs were immunostained with anti-α-tubulin (red). DNA was stained with TOTO-3 (blue). Cells expressing Cre recombinase together with Venus were immunostained with anti-GFP (green). Signals were visualized with appropriate secondary antibodies using confocal microscopy. Scale bars: 5 μm. (B) Increased thickness of the metaphase plate in M14-reexpressing cells and the B-Myb-deficient cells. The thickness of the metaphase plate ±s.d. (n=30) is shown. **P<0.001. (C) Increase in misaligned chromosomes in M14-reexpressing cells and the B-Myb-deficient cells. The frequency of metaphase-like cells with misaligned chromosomes ±s.d. (n=100–120) is shown. (D) Misaligned chromosomes in M14-reexpressing cells and B-Myb-deficient cells were pairs of sister chromatids. Cells were stained with anti-CENPB (red) and anti-phospho-histone H3 antibody (blue). Misaligned chromosomes were not obviously detected in the wild-type control (+/+) cells. Scale bars: 5 μm. (E) Failure in attachment of kinetochores to microtubules in M14-reexpressing cells and B-Myb-deficient cells. The indicated cells were stained for CENPB (red) and α-tubulin (green), after cold treatment to depolymerize non-kinetochore microtubules. The right panels show higher magnification images of the centromeres delineated by the white box. The arrows indicate the centromeres that are normally connected with α-tubulin, whereas the arrowheads indicate the centromeres that are not connected with α-tubulin. Scale bars: 5 μm. (F) Persistent localization of Mad2 on kinetochores of M14-reexpressing cells and B-Myb-deficient cells. Cells were stained with anti-Mad2 (red) and anti-phospho-histone H3 (blue). The right panels show higher magnification images of the kinetochores delineated by the white box. Mad2-positive kinetochores are indicated by the arrowheads. Scale bars: 5 μm.
Discussion
B-Myb is the member most closely related to dMyb (Davidson et al, 2005). Mutations of dmyb and zebrafish B-myb and also a downregulation of B-Myb in mouse megakaryocytes by siRNA cause genome instability (Fung et al, 2002; Manak et al, 2002; Shepard et al, 2005; García and Frampton, 2006). Furthermore, dMyb was recently shown to contribute to normal separation of centrosomes and also chromosome condensation (Goshima et al, 2007; Manak et al, 2007). However, it remains unknown whether these defects are mediated indirectly through its role in gene transcription or reflect another uncharacterized action of dMyb and B-Myb. In fact, B-Myb and dMyb are required for transcription of a cluster of genes that regulate the G2/M transition and the M-phase events (Zhu et al, 2004; Osterloh et al, 2006; Litovchick et al, 2007; Pilkinton et al, 2007; Wen et al, 2008). The present study strongly suggests that the Myb–Clafi complex has a function to ferry clathrin to the mitotic spindles and regulates the function of mitotic spindles. The B-Myb M14 mutant, which retains the capacity to activate transcription of a cluster of B-Myb target genes but cannot form the Myb–Clafi complex, induced the multiple defects of mitosis, including misaligned chromosomes and unusual number of centrosomes.
B-Myb is known to form the vertebrate dREAM/Myb–MuvB-like complex (Zhu et al, 2004; Osterloh et al, 2006; Pilkinton et al, 2007; Litovchick et al, 2007). One obvious question is why other groups did not detect the Myb–Clafi complex when they analysed the dMyb- or B-Myb-containing complex. First, it is not yet clear at present whether clathrin is localized at the mitotic spindles in Drosophila similar to vertebrate and whether dMyb forms a complex with clathrin. If dMyb would also form the Myb–Clafi complex, the failure to detect the Myb–Clafi complex might be due to its low amounts, because the Myb–Clafi complex exists only at the M phase. In addition, the Myb–Clafi complex may not be extracted by using the condition (0.1 M NaCl), which was used to extract the dREAM/Myb–MuvB complex, because we have used the high-salt condition (0.42 M NaCl) to purify the Myb–Clafi complex. Although the dREAM/Myb–MuvB complex was purified using multiple chromatography (Korenjak et al, 2004; Lewis et al, 2004), the vertebrate dREAM/Myb–MuvB-like complex was characterized only by co-immunoprecipitation (Osterloh et al, 2006; Litovchick et al, 2007). Therefore, it is hard to accurately estimate the ratio of amount of the Myb–Clafi and dREAM/Myb–MuvB-like complexes. An interaction between B-Myb and hLIN-9 (a human homologue of Mip130) or other components of the human dREAM/Myb–MuvB-like complex was detected by co-immunoprecipitation using the low-salt condition (150 mM NaCl) (Osterloh et al, 2006; Litovchick et al, 2007). We have also observed that B-Myb can be co-precipitated with hLIN-9 (Figure 4C) under the low-salt condition. Under the high-salt condition used to purify the Myb–Clafi complex, the dREAM/Myb–MuvB-like complex may be disrupted.
Although we detected the clathrin signals at the kinetochore fibres, neither B-Myb nor filamin signals were clearly detected at the mitotic spindles. Only very faint signals of endogenous B-Myb or exogenously expressed Venus–B-Myb was observed at the mitotic spindles, but FLAG–HA–B-Myb was not detected at the mitotic spindles using anti-FLAG or anti-HA antibody. Further, we could not detect B-Myb in the purified mitotic spindle fraction, and this is consistent with previous work that detected clathrin, but neither B-Myb nor filamin, in the spindle apparatus (Mack and Compton, 2001). In spite of these observations, in the cells expressing M14 B-Myb mutant that could not form the Myb–Clafi complex but still retained the trans-activating capacity, the clathrin signals at the mitotic spindles were strongly reduced. These data suggest that the role of the Myb–Clafi complex is to deliver clathrin to the spindles. The estimated molecular weight of the Myb–Clafi complex (500–600 kDa) indicates that it consists of one molecule each of filamin A, CHC, CLC, and B-Myb. Clathrin can form a triskelion consisting of three heavy chains each with an associated light chain. The recent report using the clathrin mutants suggested that the domains required for the trimer formation is critical for its function in mitotic spindles, suggesting that clathrin functions as a triskelion at the mitotic spindles (Royle and Lagnado, 2006). These results suggest that the Myb–Clafi complex delivers clathrin to the spindles, where clathrin forms triskelion. The domains in clathrin, which are required for trimer formation, could also be required for interaction with B-Myb and filamin to form the Myb–Clafi complex.
Clathrin is speculated to form a relatively rigid connection between microtubules that strengthens kinetochore fibres (Royle et al, 2005). Filamin functions not only as an actin-crosslinking protein but also as the molecular scaffold protein by interacting with various proteins (Feng and Walsh, 2004; Popowicz et al, 2006). The Myb family proteins are also known to interact with various proteins through the surface of the DBD (Ness, 1999). Therefore, both B-Myb and filamin may exert an effect as scaffold proteins to carry clathrin to the mitotic spindles. We found that disruption of the Myb–Clafi complex causes aneuploidy, a form of genomic instability that leads to cancer (Jallepalli and Lengauer, 2001), suggesting that a small deletion or the point mutations in the C-terminal region of B-Myb, which do not affect its trans-activating capacity, could be sufficient to cause some types of cancers. Not only mutation and decreased expression but also overexpression of B-Myb may cause cancers by disrupting the Myb–Clafi complex due to an imbalance in the components. Identification and characterization of the Myb–Clafi complex may further our understanding of the role of B-Myb in mitotic events, as well as in cancer development.
Materials and methods
Purification and characterization of the Myb–Clafi complex
The complex was purified essentially as described by Nakatani and Ogryzko (2003). The HeLa S3 cell clone expressing FLAG/HA–B-Myb was generated as described in the Supplementary data. Cells from a 4 l culture were disrupted in hypotonic buffer, and the nuclear pellet was collected by centrifugation at 25 000 g for 20 min. The pellet was extracted with buffer C (20 mM HEPES, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, and 0.5 mM DTT) and lysates were collected by centrifugation. The B-Myb complex was purified by using anti-FLAG M2 monoclonal antibody (mAb)-conjugated agarose beads followed by anti-HA 12CA5mAb-conjugated agarose beads in wash buffer. For glycerol gradient sedimentation, FLAG-purified material was loaded on the top of a 4.2-ml 10–40% glycerol gradient in the wash buffer and centrifuged for 5 h at 55 000 r.p.m. After centrifugation, fractions were collected from the top of the gradient. The purified proteins were separated by 4–20% gradient SDS–PAGE and silver stained. The protein bands were excised and analysed by mass spectrometry at the RIKEN Brain Science Institute Mass Spectrometry Facility. Anti-B-Myb (N-19; Santa Cruz Biotechnology), anti-filamin A (MAB1692; Chemicon), anti-CHC (X22; Affinity BioReagents), or anti-CLC (CON.1; Santa Cruz Biotechnology) was used for immunoblotting.
Synchronization of HeLa cells and siRNA treatment
Cells were treated with 2 mM thymidine for 20 h, and released for 8 h. Cells were then retreated with 2 mM thymidine for 14 h. At various times after release, cells were used for FACS analysis, co-immunoprecipitation, or immunocytochemistry. HeLa cells were transfected with siRNAs (final 200 nM) (see Supplementary data for the sequence) using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Co-immunoprecipitation
To examine the abundance of the Myb–Clafi complex, synchronized parental HeLa cells or HeLa cells expressing FLAG–HA–B-Myb were lysed in hypotonic buffer, and the cytosolic fraction and nuclear pellet were separated by centrifugation at 17 400 g for 10 min. Nuclei were extracted in buffer C for 30 min at 4°C and centrifuged at 17 400 g for 10 min. Cytosolic or nuclear fractions were incubated with anti-B-Myb (N-19; Santa Cruz Biotechnology) followed by Protein G-Sepharose or M2 anti-FLAG agarose (Sigma) for 4 h with rotation. After five washes with wash buffer, the bound proteins were separated by SDS−PAGE and were analysed by immunoblot analysis with the antibodies described above. The co-immunoprecipitation conditions using the transfected HeLa cells are described in the Supplementary data.
Immunocytochemistry and imaging
For M-phase cells, cells were washed with 1% Triton X-100, and then fixed with 4% paraformaldehyde as indicated in the figure legends. In the case of inter-phase cells, cells were permeabilized with 0.2% Triton X-100 treatment for 10 min after fixation. The cells were incubated with the following primary antibodies: anti-GFP (598; Medical Biological Laboratories); anti-B-Myb (N-19 and H115; Santa Cruz Biotechnology); anti-CHC (X22; Affinity BioReagents); anti-filamin A (MAB1692; Chemicon), anti-α-tubulin (DM1A; Sigma); anti-CENPB (rabbit polyclonal, a gift from H Masumoto) (Suzuki et al, 2004); phospho-histone H3 (Ser10) (06-570; Upstate); and anti-Aurora B (ab2254; Abcam). The cells were then incubated with Alexa Fluor 488- or Cy3- or rhodamine-conjugated anti-rabbit or anti-mouse secondary antibodies (Molecular Probes or Chemicon). Chromatin was labelled with TOTO-3 iodide (Molecular Probes). Confocal and Nomarski images were obtained using an LSM510 (Zeiss) laser scanning microscope. For quantitative immunostaining experiments, identical laser power and acquisition settings were used. The mean pixel density of images was measured by LSM510 3.2 software at a depth of 8 bit. The signal located specifically at the spindles was assayed by dividing the mean pixel density measured in the region of interest (1 μm × 1 μm) placed over the spindle by that measured in a region outside the spindle. Therefore, a value of one represents no specific localization at the spindles. For image quantification and counting experiments, 4–100 cells were analysed and 100–1000 cells were counted from experiments performed 3–5 times.
Generation of MEFs expressing M14 instead of endogenous B-Myb
B-mybf/f MEFs were prepared as described in the Supplementary data. To express the Cre recombinase, B-mybf/f MEFs were infected with lentivirus at an MOI of 10 for 12 h. Cells were then re-infected with adenovirus encoding WT or M14 B-Myb. Cells were used for RNA preparation or immunocytochemistry 72 h after the lentivirus infection.
Analysis of B-Myb target gene expression
Total RNAs were isolated from MEFs using RNAeasy spin columns according to the manufacturer's instructions (Qiagen). The primers and probes were designed using the Primer Express software (Applied Biosystems). Primers and TaqMan probes are shown in Supplementary Table 1. Real-time RT–PCR was performed using the ABI 7500 Real-Time PCR System and QuantiTect Probe RT–PCR Kit (Qiagen) according to the manufacturer's instructions.
Long-term imaging
Cells were grown in poly-D-lysine-coated glass bottom 35 mm dishes (IWAKI). For observation of living cells, medium was replaced with fresh DMEM containing 20 mM HEPES (pH 7.4). Dishes were imaged at 37°C using a Tempcontrol 37-2 chamber (Zeiss). Images were obtained using an LSM510 (Zeiss) laser scanning microscope.
Additional details of the Materials and methods are described in the Supplementary data.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Acknowledgments
We are grateful to T Takagai for his help, Y Nakatani for the pOZ-FH-N vector, the staff of the Research Resources Center of the RIKEN Brain Science Institute for mass spectrometric analysis, H Masumoto for the anti-CENPB antibody, SJ Royle for the EGFP-CLC expression vector, S Gaubatz for the HA–hLIN-9 expression vector, A Miyawaki for the Venus expression vector, H Miyoshi for the lentivirus encoding Cre, and members of the Experimental Animal Division of the RIKEN Tsukuba Institute for maintenance of the mice. This study was supported by Grants-in-aid for Scientific Research and by grant from the Genome Network Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Beall EL, Bell M, Georlette D, Botchan MR (2004) Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 [DOI] [PubMed] [Google Scholar]
- Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K (2004) Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5: 721–729 [DOI] [PubMed] [Google Scholar]
- Bessa M, Saville MK, Watson RJ (2001) Inhibition of cyclin A/Cdk2 phosphorylation impairs B-Myb transactivation function without affecting interactions with DNA or the CBP coactivator. Oncogene 20: 3376–3386 [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Stegmeier F, Harrison MM, Horvitz HR (2006) Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics 173: 709–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Hou DX, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S (1996) CBP as a transcriptional coactivator of c-Myb. Genes Dev 10: 528–540 [DOI] [PubMed] [Google Scholar]
- Davidson C, Tirouvanziam R, Herzenberg L, Lipsick J (2005) Functional evolution of the vertebrate myb gene family: B-Myb, but neither A-Myb nor c-Myb, complements Drosophila Myb in hemocytes. Genetics 169: 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Smith C, Owen D (2006) Life of a clathrin coat: insights from clathrin and AP structures. Nat Rev Mol Cell Biol 7: 32–44 [DOI] [PubMed] [Google Scholar]
- Enari M, Ohmori K, Kitabayashi I, Taya Y (2006) Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev 20: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Han M (2000) The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26: 279–284 [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA (2004) The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 6: 1034–1038 [DOI] [PubMed] [Google Scholar]
- Fung SM, Ramsay G, Katzen AL (2002) Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development 129: 347–359 [DOI] [PubMed] [Google Scholar]
- García P, Frampton J (2006) The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J Cell Sci 119: 1483–1493 [DOI] [PubMed] [Google Scholar]
- Gonda TJ, Metcalf D (1984) Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 310: 249–251 [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N (2007) Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR (2006) Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA 103: 16782–16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C (2001) Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 1: 109–117 [DOI] [PubMed] [Google Scholar]
- Katzen AL, Jackson J, Harmon BP, Fung SM, Ramsay G, Bishop JM (1998) Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev 12: 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A (2004) Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR (2004) Identification of a Drosophila Myb–E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA (2007) Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Mack GJ, Compton DA (2001) Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc Natl Acad Sci USA 98: 14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Mitiku N, Lipsick JS (2002) Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc Natl Acad Sci USA 99: 7438–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Wen H, Van T, Andrejka L, Lipsick JS (2007) Loss of Drosophila Myb interrupts the progression of chromosome condensation. Nat Cell Biol 9: 581–587 [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schereiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677–689 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nakagoshi H, Takemoto Y, Ishii S (1993) Functional domains of the human B-myb gene product. J Biol Chem 268: 14161–14167 [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V (2003) Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 370: 430–444 [DOI] [PubMed] [Google Scholar]
- Ness SA (1999) Myb binding proteins: regulators and cohorts in transformation. Oncogene 18: 3039–3046 [DOI] [PubMed] [Google Scholar]
- Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R (1988) Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res 16: 11075–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79: 639–648 [DOI] [PubMed] [Google Scholar]
- Oh IH, Reddy EP (1999) The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18: 3017–3033 [DOI] [PubMed] [Google Scholar]
- Okada M, Akimaru H, Hou DX, Takahashi T, Ishii S (2002) Myb controls G2/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J 21: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto CT, McKinney J, Jeng YY (2000) Clathrin in mitotic spindles. Am J Physiol Cell Physiol 279: C369–C374 [DOI] [PubMed] [Google Scholar]
- Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B, Hauser S, Gaubatz S (2006) The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26: 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR (2007) Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J Biol Chem 282: 168–175 [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Schleicher M, Noegel AA, Holak TA (2006) Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci 31: 411–419 [DOI] [PubMed] [Google Scholar]
- Robinson C, Light Y, Groves R, Mann D, Marias R, Watson R (1996) Cell-cycle regulation of B-Myb protein expression: specific phosphorylation during the S phase of the cell cycle. Oncogene 12: 1855–1864 [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L (2005) Clathrin is required for the function of the mitotic spindle. Nature 434: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Lagnado L (2006) Trimerisation is important for the function of clathrin at the mitotic spindle. J Cell Sci 119: 4071–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda TJ, Ishii S (1989) Delineation of the three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA 86: 5758–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J, Finley KR, Pfaff KL, Hersey C, Zhou Y, Barut B, Freedman M, Lee C, Spitsbergen J, Neuberg D, Weber G, Golub TR, Glickman JN, Kutok JL, Aster JC et al. (2005) A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci USA 102: 13194–13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Nakano M, Nozaki N, Egashira S, Okazaki T, Masumoto H (2004) CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J Biol Chem 279: 5934–5946 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S (1999) B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem 274: 28067–28070 [DOI] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Letvin J, Orth J, Hatton KS, Reddy EP (1997) Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 386: 713–717 [DOI] [PubMed] [Google Scholar]
- Trauth K, Mutschler B, Jenkins NA, Gilbert DJ, Copelend NG, Klempnauer KH (1994) Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J 13: 5994–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS (2008) Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev 22: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR (2004) E2Fs link the control of G1/S and G2/M transcription. EMBO J 23: 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information