Abstract
X-chromosome inactivation (XCI) depends on the noncoding Xist gene. Xist transcription is negatively regulated by its antisense partner Tsix, whose disruption results in nonrandom XCI in females. However, males can maintain Xist in a repressed state without Tsix, indicating participation of additional factor(s) in the protection of the single male X from inactivation. Here, we provide evidence that the histone methyltransferase Eed is also involved in the process. Male embryonic stem cells with Eed-null and Tsix mutations (XΔY Eed−/−) showed Xist hyperactivation upon differentiation, whereas cells with either mutation alone did not. Impaired X-linked gene expression was observed in the XΔY Eed−/− ES cells at the onset of differentiation. The Xist promoter in the XΔY Eed−/− cells showed elevated histone H3-dimethyl lysine 4 modifications and lowered CpG methylation, which are characteristics of open chromatin. Hence, we identified Eed as an additional major player in the regulation of Xist expression. The synergy of Polycomb group proteins and antisense Tsix transcription in Xist gene regulation explains why males can repress Xist without Tsix.
Keywords: antisense, DNA methylation, Eed, histone modification, Xist
Introduction
X-chromosome inactivation (XCI) is a sex chromosome dosage compensation mechanism employed by female mammals. During the process, one of two active X-chromosomes (Xa) in female embryonic cells is randomly chosen and inactivated during development (Lyon, 1961; reviewed by Heard and Disteche, 2006). The noncoding gene Xist (Brockdorff et al, 1992; Brown et al, 1992) has been shown to be critical for XCI (Penny et al, 1996). It is encoded on the X-chromosome and is transcribed at a very low level in the undifferentiated condition in both females and males (Panning and Jaenisch, 1996; Lee et al, 1999). Upon differentiation, it is exclusively expressed from the inactive X-chromosome (Xi) and coats Xi in females (Clemson et al, 1996), whereas Xist transcription is soon terminated on the future Xa and in males. The choice of Xi is achieved by Xist upregulation in cis (Wutz and Jaenisch, 2000). Xist is believed to function as an RNA entity, because of its characteristic repeat sequence (Wutz et al, 2002) and distribution pattern in the nucleus. Xist is negatively regulated by its antisense partner Tsix, which overlaps the Xist gene (Lee et al, 1999; Sado et al, 2001; Shibata and Lee, 2003). Xist is always upregulated at the mutant Tsix allele in heterozygous female embryonic stem (ES) cells, resulting in nonrandom inactivation of the Tsix-mutated X-chromosome (Lee and Lu, 1999; Luikenhuis et al, 2001; Sado et al, 2001; Shibata and Lee, 2004). In contrast, Tsix mutation does not lead to Xist expression in male ES cells upon differentiation. Previous reports described ectopic Xist accumulation in a minor portion of Tsix-mutant male ES cells (0–13%) (Lee and Lu, 1999; Luikenhuis et al, 2001; Sado et al, 2002), whereas another study observed ectopic Xist accumulation more frequently (39%) (Vigneau et al, 2006). Importantly, male embryos carrying a Tsix mutation on the single X-chromosome develop to term when the extraembryonic tissues are complemented by wild-type tetraploid cells (Ohhata et al, 2006), indicating that most embryonic cells in males can maintain Xist gene repression without Tsix. These observations suggest the presence of additional or alternative factor(s) that inhibit the activation of Xist gene in male embryos.
Recent studies have shed light on the role of Tsix in regulating chromatin structure in the Xist locus. Sado et al (2005) indicated that disruption of Tsix caused impaired establishment of repressive chromatin structure at the Xist promoter and exon 1 in developing embryos. Navarro et al showed that the Xist promoter region, flanked by CTCF-binding sites, was maintained in a heterochromatic state by Tsix. Tsix truncation resulted in altered modification at lysine 4 of histone H3 (H3K4) and lysine 9 to resemble a pseudoeuchromatic state (Navarro et al, 2006). Sun et al (2006) reported that Tsix downregulation induced a transient heterochromatic state, characterized by histone H3 trimethyl-lysine 27 (H3K27m3) modification in undifferentiated female ES cells. These reports suggest that Tsix transcription influences the chromatin structure at the Xist promoter in different ways depending on the differentiation stage and position within the locus. We focused on H3K27m3, because this modification is clearly elevated when Tsix transcription is absent in both female and male undifferentiated ES cells (Navarro et al, 2006; Sun et al, 2006; Shibata and Yokota, 2008). In addition, the biological significance of the regulation by Tsix of the H3K27m3 modification is still unclear. The H3K27m3 modification is generally considered to be a repressive chromatin mark; however, the loss of Tsix transcription paradoxically results in Xist gene activation in females.
Methylation of the histone H3 lysine 27 (H3K27) is conferred by the Polycomb repressive complex 2 (PRC2), which is composed of the Eed, Ezh2 and Suz12 proteins (Cao and Zhang, 2004). Eed is essential for the histone methyltransferase (HMTase) activity, because Eed−/− ES cells lack the H3K27m3 modification (Montgomery et al, 2005). Eed is necessary for development (Faust et al, 1995) and regulates developmental control genes as well as a subset of imprinted genes (Mager et al, 2003). In ES cells, PRC2 occupies genes encoding transcription factors crucial for development, and Eed mutations result in their premature expression (Azuara et al, 2006; Boyer et al, 2006). These loci were termed bivalent domains due to the special modification pattern consisting of trimethylated H3K27 and H3K4, a repressive and active chromatin mark, respectively (Bernstein et al, 2006). Hence, Eed has an important function in gene regulation in undifferentiated and differentiating cells in conjunction with other chromatin factors. Eed and H3K27 methylation are also involved in the establishment and maintenance of XCI. Eed localizes on the Xi in female trophoblast stem cells, and reactivation of Xi is found in Eed−/− trophoblast stem cells when they are differentiated (Kalantry et al, 2006). Recruitment of Eed and H3K27 methylation are also observed on Xi in female embryos and ES cells at early stages of XCI (Plath et al, 2003; Silva et al, 2003). However, recent findings indicate that Eed is dispensable for the initiation of random XCI (Kalantry and Magnuson, 2006; Schoeftner et al, 2006). XCI without Eed is explained by a contribution of Polycomb repressive complex 1 (PRC1) that ubiquitinates histone H2A (Schoeftner et al, 2006). These reports focused mainly on the role of Eed in inducing global heterochromatin formation on Xi, but as was shown by Sun et al (2006), Eed is likely to have additional roles in the regulation of local Xist chromatin structure in concert with Tsix. Therefore, we disrupted Tsix in an Eed−/− male ES cell line to investigate the role of Eed in regulating Xist chromatin structure and to examine the biological significance of the H3K27m3 modification that is observed when Tsix transcription is absent. The role and relationship of Eed and Tsix in the regulation of Xist are discussed.
Results
Generation of male Tsix mutant ES cells with Eed−/− background
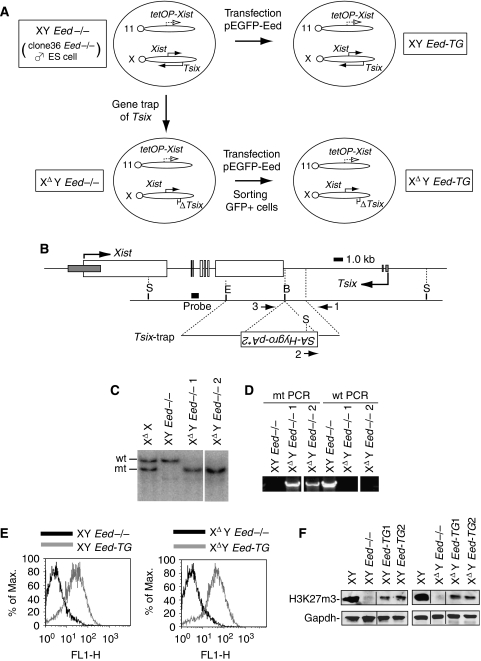
Tsix mutant ES cell lines are summarized in Figure 1A. Firstly, we targeted the clone36 Eed−/− male ES cell line (XY Eed−/−) (Schoeftner et al, 2006) and truncated Tsix transcription to generate male Tsix mutant ES cells with an Eed−/− background (XΔY Eed−/−) (Figure 1B–D). This type of Tsix mutation has been shown to eradicate its function in repressing Xist in female ES cells (XΔX) (Shibata and Lee, 2004). We then rescued Eed in the XY Eed−/− and XΔY Eed−/− cells by transgenic expression of an enhanced green fluorescent protein (EGFP)–Eed fusion protein (XY Eed-TG and XΔY Eed-TG, respectively) (Figure 1E and F). In addition to the western blot for H3K27m3, a quantitative chromatin immunoprecipitation (ChIP) assay was used to examine known H3K27m3-labeled sites in undifferentiated ES cells, the Sox9 and Gata6 promoters (Boyer et al, 2006), and confirmed that the Eed activity was sufficiently rescued in the XΔY Eed-TG cells for these promoters (Supplementary Figure S1). Although the clone36 Eed−/− ES cells have an additional Xist cDNA transgene (Tg) under control of tetracycline-inducible promoter on chromosome 11, the Xist Tg has been shown to be inactive without induction (Wutz and Jaenisch, 2000).
Figure 1.
Generation of Tsix-trap male ES cells in the Eed−/− background (XΔY Eed−/−). (A) Relationship of ES cell lines generated in this study. tetOP-Xist, tetracycline-inducible promoter and Xist cDNA (Tg) with Mus spretus repeat polymorphism; 11, chromosome 11; X, chromosome X. (B) Targeting construct for Tsix. Large open and small gray rectangles show Xist and Tsix exons, respectively. Numbered arrows represent primers for genomic PCR. S, SpeI; E, EcoRI; B, BamHI restriction enzyme sites. (C) SpeI-digested Southern blot showing correct recombination of 5′-homology arm in two independent XΔY Eed−/− clones. All lanes were derived from the same gel. (D) Genomic PCR confirming proper recombination of 3′-homology arm. Primer pair 1–2 was used for mutant (mt) and 1–3 for wild-type (wt) amplification. All lanes were derived from the same gel. (E) Rescuing Eed by transfecting the pEGFP-Eed plasmid (XY Eed-TG and XΔY Eed-TG). Expression of the fusion protein was confirmed by flow cytometry. Results were from the cells at the passage of less than 4 (XΔY Eed-TG) or 8 (XY Eed-TG) after their derivation. (F) Western blot demonstrating reversion of the H3K27m3 modification in the transgenic cell lines. Western blotting was done using cells at the passage of less than 4 (XΔY Eed-TG) or 8 (XY Eed-TG). Four lanes in the right and left panels were derived from the same gel, respectively.
XDY Eed−/− cells display Xist hyperactivation upon differentiation
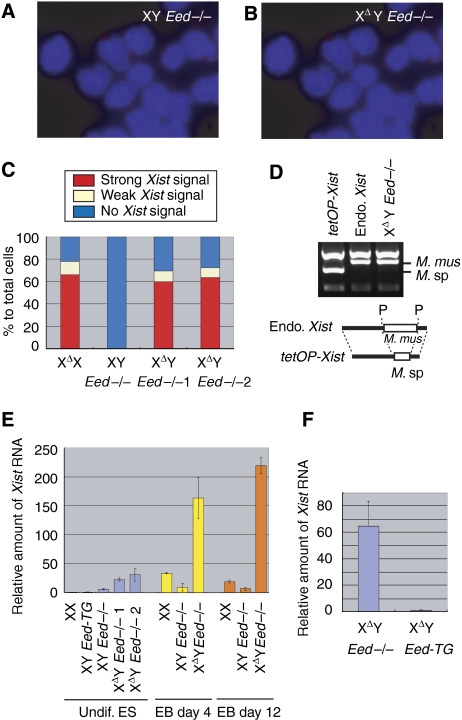
We examined Xist RNA expression in the XΔY Eed−/− cells by fluorescent in situ hybridization (FISH) using a strand-specific riboprobe. We found strong Xist expression in the XΔY Eed−/− cells, but not in the XY Eed−/− embryoid bodies (EB) differentiated for 3 days (Figure 2A and B). The number of Xist-positive nuclei was found significantly elevated in two independent XΔY Eed−/− lines and was similar to that of differentiating XΔX ES cells (Figure 2C and Supplementary Table I). Polymorphism of an Xist RT–PCR product confirmed that the ectopic Xist expression in the XΔY Eed−/− EB was from the endogenous Xist allele, not from the Xist cDNA Tg, which is also present in all clone36-derived ES cells (Figure 2D). The amount of Xist RNA expressed during the course of XCI was further quantified by real-time PCR. The results were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression, and the amount of Xist RNA expression, relative to undifferentiated wild-type female (XX) ES cells, is shown (Figure 2E and Supplementary Table II). Interestingly, the XΔY Eed−/− ES cells showed elevated Xist RNA level even in the undifferentiated condition, and upon differentiation, they expressed five to ten times more Xist RNA than XX cells. The XY Eed−/− EB also displayed elevated Xist level when compared with undifferentiated XY Eed-TG cells, but far less than XX and XΔY Eed−/− EB. We then investigated whether the loss of Eed was the cause of this result, because Xist activation does not generally occur in male Tsix mutant ES cells. We looked for suppression of ectopic Xist hyperactivation in the XΔY Eed-TG cells and confirmed that the rescued Eed successfully inhibited Xist hyperactivation (Figure 2F). Therefore, both Tsix and Eed contribute to the repression of Xist gene, but either of the two is sufficient for preventing ectopic Xist activation in males.
Figure 2.
XΔY Eed−/− cells display Xist hyperactivation upon differentiation. (A) Xist RNA-FISH using strand-specific riboprobe (red) in the XY Eed−/− and (B) XΔY Eed−/− ES cells differentiated for 3 days. (C) The count of Xist-positive nuclei in FISH. More than 80 nuclei for the XY Eed−/− cell line and more than 180 nuclei for other lines were counted. The XΔY Eed−/−1 and XΔY Eed−/−2 are independent clones. (D) Xist cDNA Tg is inactive in the XΔY Eed−/− ES cells, shown by the polymorphism of Xist RT–PCR product digested with PstI restriction enzyme. tetOP-Xist, Xist from the Tg with Mus spretus (M. sp) sequence. RT–PCR was performed using RNA obtained from the XY Eed-TG cells cultured in the presence of doxycyclin for Tg induction. Endo. Xist, endogenous Xist. RT–PCR in the XΔX EB cells in which Xist is predominantly expressed from the Mus musculus (M. mus) allele. XΔY Eed−/−, RT–PCR in the XΔY Eed−/− EB cells differentiated for 12 days. Shown below is a schematic representation of the PCR products with M. mus or M. sp repeat polymorphism (open boxes). P, PstI sites. (E) Quantitative RT–PCR for Xist. Relative amount (mean) of Xist RNA to undifferentiated (undif.) wild-type female (XX) ES cells normalized to Gapdh is shown. Error bars represent s.d. (F) Rescuing Eed inhibited ectopic Xist expression. Relative amount of Xist RNA in the XΔY Eed−/− day 4 EB to that in the XΔY Eed-TG is shown.
Xist hyperactivation in the XDY Eed−/− cells leads to partial XCI upon differentiation
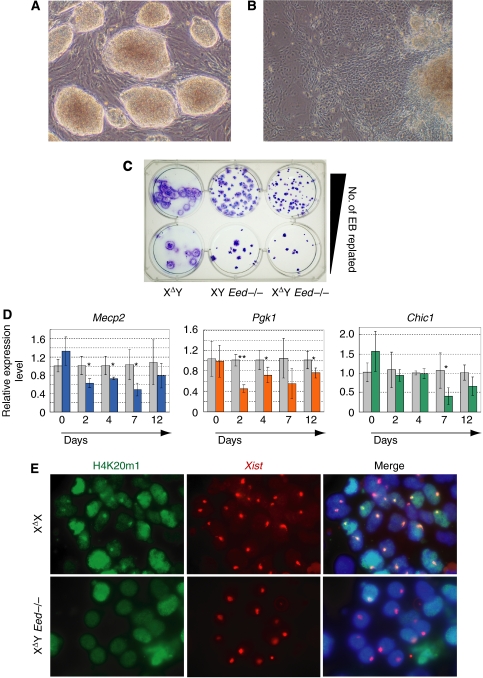
We next investigated the consequence of Xist hyperactivation in the XΔY Eed−/− cells by differentiating the mutant ES cells in vitro. The XΔY Eed−/− ES cells in the undifferentiated condition displayed round, well-packed colony morphology typical of mouse ES cells (Figure 3A). The XΔY Eed−/− EB cells, after long adherent culture, contained flattened cells suggesting differentiation (Figure 3B), but it was not clear whether their X-chromosomes were inactivated or not. The growth of XY Eed−/− EB cells were retarded as compared with the EB cells with intact Eed (Figure 3C). Expression of transgenic Eed rescued their poor growth (Supplementary Figure S2). The growth of XΔY Eed−/− EB cells was further retarded compared with the XY Eed−/− EB cells: they spread less and their EB size was smaller than that of the XY Eed−/− EB cells (Figure 3C and Supplementary Figure S2). This observation suggested that a substantial amount of differentiating XΔY Eed−/− cells were lost from culture due to the inactivation of their single X-chromosome. To examine if XCI occurs in the XΔY Eed−/− cells during differentiation, we studied the expression of X-linked Mecp2, Pgk1 and Chic1 genes by quantitative RT–PCR (qRT–PCR) (Figure 3D). The expression of these genes decreased immediately upon differentiation, and interestingly, the reduction became less obvious at the late stage of EB day 12. We also examined colocalization of the Xi chromatin marker, histone H4 monomethyl-lysine 20 (H4K20m1) (Kohlmaier et al, 2004), with Xist RNA in immuno-FISH. Although Xist RNA deposition was frequently found in the XΔY Eed−/− EB nuclei, co-localization of Xist and condensed H4K20m1 was never detected in the late stages of differentiation in contrast to the XΔX nuclei (Figure 3E and Table I). Careful examination of EB cells at day 2 or 4 revealed weak H4K20m1 staining with Xist paint in a maximum of 10% of the XΔY Eed−/− nuclei (Supplementary Figure S3 and Supplementary Table III). Taken together, these findings indicate that the Xist hyperactivation in the XΔY Eed−/− cells induced partial XCI at the onset of differentiation. However, it was incomplete, presumably due to the absence of Eed, and a substantial number of cells survived and restored their X-linked gene expression after the critical time window for silencing by Xist RNA (Wutz and Jaenisch, 2000) (Supplementary Figure S4).
Figure 3.
Differentiation and XCI of the XΔY Eed−/− ES cells. (A) Compact colony morphology of the XΔY Eed−/− ES cells in undifferentiated condition. (B) Morphology of the XΔY Eed−/− EB differentiated for 12 days. (C) Gross appearance of day 12 EB in the XΔY, XY Eed−/− and XΔY Eed−/− background. (D) Relative amount of X-linked Mecp2, Pgk1 and Chic1 mRNA in the XΔY Eed−/− cells (colored columns) to those in the XY Eed−/− cells (gray columns) in undifferentiated or differentiating conditions. Error bars show s.d. Asterisks demonstrate statistically significant reduction of the gene expression in the XΔY Eed−/− cells (*P<0.05; **P<0.0005). (E) Immuno-FISH for H4K20m1 (green) and Xist RNA (red) in the XΔX and XΔY Eed−/− EB (days10–12).
Table 1.
The number of nuclei showing colocalization of condensed H4K20m1 and Xist RNA in Xist-positive nuclei of the mutant EB
| Genotype | EB differentiation | H4K20m1 colocalization | No. of Xist-positive nuclei counted |
|---|---|---|---|
| XΔX | 7 days | 44 (40.4%) | 109 |
| XΔY Eed−/− 1a | 7 days | 0 (0%) | 142 |
| XΔY Eed−/− 1a | 18 days | 0 (0%) | 143 |
| XΔY Eed−/− 2b | 18 days | 0 (0%) | 114 |
| XΔY Eed−/− cell clone 1. | |||
| XΔY Eed−/− cell clone 2. | |||
Deregulated antisense transcription in the Xist gene body of the XDY Eed−/− ES cells
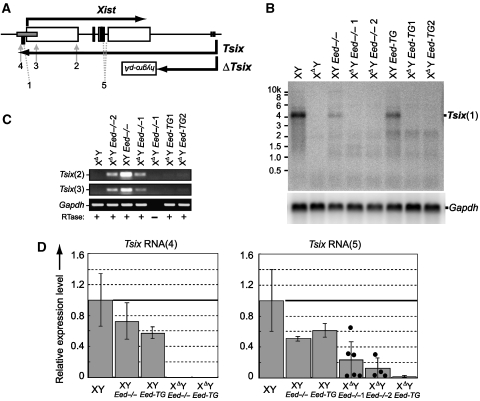
We confirmed that Tsix transcription was successfully truncated in the XΔY Eed−/− ES cells by a northern blot of poly-A purified RNA, using a probe residing in the Xist promoter (Figure 4A and B). However, antisense RNA was detected in the double mutant cells by strand-specific RT–PCR, and it disappeared when Eed was rescued (Figure 4C). To eliminate the possibility that the transcript originated from the Xist cDNA Tg, we performed qRT–PCR for Tsix in amplicons that do not amplify the Tg (Figure 4D). The amplicon at the 3′-end of the Tsix (no. 4) antisense transcript was not detected in the XΔY Eed−/− cells, whereas in the amplicon spanning the Xist introns (no. 5) could be detected. We suggest that the loss of Eed in the Tsix-deficient background resulted in an open chromatin structure that led to deregulated antisense transcription from cryptic promoters to various degrees in the Xist gene body. The absence of an antisense transcript at the 3′-end of Tsix suggested that the transcript was terminated by multiple poly-A signals in the antisense orientation residing near the Xist transcription start site (Shibata and Lee, 2003). Average Tsix expression levels observed by qRT–PCR were lower in the XY Eed−/− and XY Eed-TG lines than the wild-type male ES cells, but the difference was not statistically significant.
Figure 4.
Northern blot and strand-specific or quantitative RT–PCR for Tsix. (A) Positions of northern blot probe (filled rectangle 1), strand-specific RT–PCR amplicons (2 and 3) and qRT–PCR amplicons (4 and 5). (B) Northern blot for Tsix. The XΔY Eed−/−1 and XΔY Eed−/−2, and the XΔY Eed-TG1 and XΔY Eed-TG2 are independent clones. After the initial northern blot for Tsix, the same membrane was stripped and reprobed for Gapdh. (C) Strand-specific RT–PCR for Tsix at the amplicons 2 and 3. (D) qRT–PCR for Tsix at amplicons 4 and 5. Relative Tsix expression levels to the wild-type male (XY) ES cells are shown. Results were from more than three independent samples and error bars indicate s.d. Filled circles in the Tsix RNA (5) graph represent Tsix levels in individual samples of the XΔY Eed−/−1 and XΔY Eed−/−2 lines.
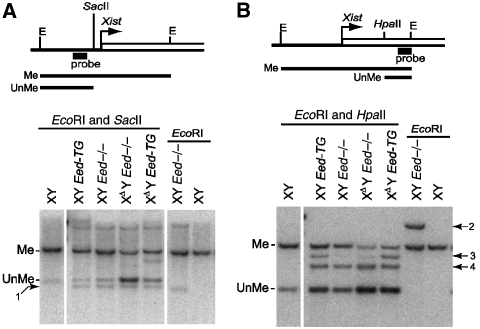
The XDY Eed−/− ES cells display loss of CpG methylation at the Xist promoter
DNA in the Xist locus has been shown to be methylated in undifferentiated male ES cells (Norris et al, 1994). We examined the methylation level of the Xist locus in undifferentiated XΔY Eed−/− ES cells by Southern blot using methylation-sensitive restriction enzymes. The SacII site at the Xist promoter displayed lowered CpG methylation in the XΔY Eed−/− cells (Figure 5A). This was also the case in the HpaII site within Xist exon 1, which was revealed by comparing the intensity of methylated bands (Figure 5B). Note that the unmethylated band in Figure 5B represents both endogenous Xist and Xist cDNA Tg. Extra bands observed in Figure 5B originated from the Xist cDNA Tg, which was obvious to identify due to the absence of an EcoRI site and the presence of multiple HpaII sites in the tetracycline inducible promoter and its flanking sequence. Rescuing Eed in the XΔY Eed−/− cells (XΔY Eed-TG) resulted in partial reversion of CpG methylation, which is in contrast to a previous report showing that a Tsix mutation did not affect the methylation status of Xist locus in males (Sun et al, 2006). Given that the Xist locus in the XΔY Eed−/− ES cells takes an open chromatin configuration, as was shown by reduced CpG methylation and Xist hyperactivation upon differentiation, we suggest that, once opened, the chromatin cannot easily reset to a repressed condition by rescuing Eed activity.
Figure 5.
Methyl-CpG-sensitive Southern blot at the Xist promoter and exon 1. (A) SacII-digested Southern blot at the Xist promoter. Me, methylated; UnMe, unmethylated; E, EcoRI site. Position of the probe is shown in the map. Arrow 1, nonspecific band found in the XY Eed−/− and its derivative lines. (B) HpaII-digested Southern blot in the Xist exon 1. Arrows 2, 3 and 4 indicate bands originated from Xist cDNA Tg. All lanes were derived from the same gel (B) or from twin gels run in parallel (A).
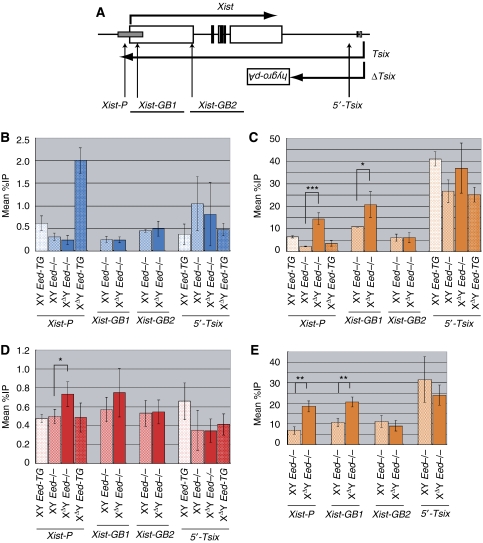
The XDY Eed−/− ES cells display elevated H3K4 methylation at the Xist promoter
To gain further insight into the role of Eed and Tsix in Xist chromatin structure regulation, we examined the methylation of H3K4 and H3K27 and the recruitment of transcription factor IIB (TFIIB) by the ChIP assay. Here, it must be considered again that the XY Eed−/− cells and their derivatives contain an Xist cDNA Tg that includes Xist-GB1 and Xist-GB2 amplicons, but not others (Figure 6A). The H3K27m3 modification was no longer found in the XΔY Eed−/− cells, confirming that Eed is responsible for the modification that appears when Tsix is absent (Figure 6B). Rescuing Eed resulted in a clearly elevated H3K27m3 level at the Xist promoter (Xist-P) in the XΔY Eed-TG cells (P<0.0005). PRC1 and its product monoubiquitinated histone H2A (UbH2A) are linked to Xi (de Napoles et al, 2004; Schoeftner et al, 2006) and they contribute to the control of developmental regulator genes (Stock et al, 2007). We examined if UbH2A modification at the Xist promoter is affected by Tsix mutations in both Eed+/+ and −/− cell lines (Supplementary Figure S5). In all cases, the modification was nearly to the background level, and we did not detect a statistically significant difference between the wild-type and Tsix-deficient lines. The loss of Tsix in the Eed−/− background resulted in a significantly increased dimethyl-H3K4 (H3K4m2) level at the Xist promoter and gene body (Xist-GB1). Both of these amplicons are within the previously reported CTCF-flanked region, and the level of H3K4m2 was comparable with that in the report (Navarro et al, 2006) (Figure 6C). Such augmented H3K4m2 level in the XΔY Eed−/− cells was not clear outside of the CTCF-flanked region (Xist-GB2) or at 5′-portion of Tsix (5′-Tsix). We also found significantly elevated TFIIB recruitment at the Xist promoter in the XΔY Eed−/− cells (Figure 6D), although it might be just reflecting the large transcription difference of Xist gene. Upon differentiation, the H3K4m2 level at the Xist promoter persisted to be higher in the XΔY Eed−/− EB than in XY Eed−/− EB. Enhanced H3K4m2 levels are consistent with the observed Xist hyperactivation (Figure 6E). Because the H3K4m2 modification in the Xist locus disappears upon differentiation in both wild-type and Tsix mutant male ES cells (Shibata and Yokota, 2008), we conclude that the Tsix mutation in the absence of Eed resulted in persistent high H3K4m2 level around the Xist promoter.
Figure 6.
ChIP in male ES cells with mutations in Eed and/or Tsix. (A) Positions of PCR amplicons for ChIP. Those amplicons overlapping with Xist cDNA Tg are underlined. ChIP results (mean %IP to input) for (B) H3K27m3, (C) H3K4m2 and (D) TFIIB in undifferentiated condition. (E) ChIP for H3K4m2 in day 12 EB. Error bars represent s.d. Asterisks in the graphs indicate statistically significant difference between the XY Eed−/− and XΔY Eed−/− lines (***P<0.001; **P<0.005; *P<0.05). Differences between other cell lines are not shown for simplicity.
Tsix and Eed have a synergistic role in repressing Xist
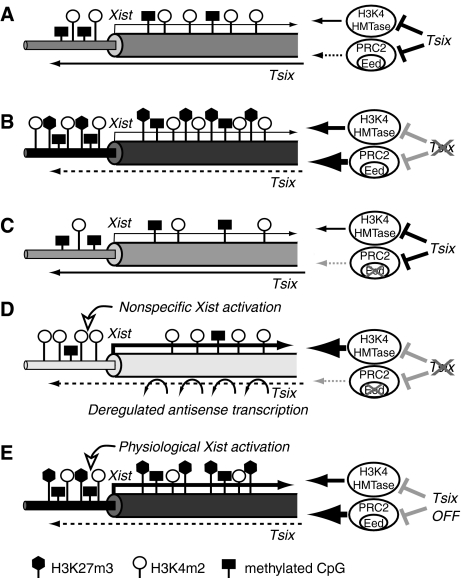
Taken together with data from ChIP, RT–PCR and CpG methylation analyses, we present a summary illustrating the roles of Tsix and Eed in the regulation of Xist chromatin structure (Figure 7). Xist chromatin is most condensed in the XΔY (or XΔY Eed-TG) ES cells, followed by XY (or XY Eed-TG) and XY Eed−/− cells, and becomes highly opened in the XΔY Eed−/− ES cells. The opened Xist chromatin configuration in the XΔY Eed−/− ES cells allows Xist hyperactivation upon differentiation. These results suggest a model that Tsix transcription negatively regulates both PRC2 and H3K4 HMTase at the Xist promoter and exon 1. It has been shown that Tsix transcription prevents Eed/PRC2 recruitment to the Xist promoter in cis (Sun et al, 2006). Tsix transcription has also been reported to inhibit H3K4 methylation (Navarro et al, 2006), whereas the difference in H3K4m2 levels between the XY Eed-TG and XΔY Eed-TG cells was not prominent (Figure 6C). The increased PRC2 recruitment or elevated H3K27m3 modification may inhibit H3K4 HMTase localization or the activity at the Tsix-deficient allele. The loss of Eed alone does not result in highly opened chromatin because Tsix still inhibits H3K4 HMTase. When both Tsix and Eed are absent, augmented H3K4 HMTase activity confers highly elevated H3K4 methylation that induces ectopic Xist activation upon differentiation. The mechanism of Xist hyperactivation in the XΔY Eed−/− cells is in clear contrast to that of the physiological Xist activation in female future Xi. In the latter, Xist RNA yield is limited, and in females, Xist transcription is activated at Tsix-deficient alleles with elevated H3K27m3 modification (Sun et al, 2006). Hence, we suggest that Eed contributes in male cells to inhibit ectopic Xist activation during differentiation when Tsix transcription goes down.
Figure 7.
Summary of the results and a suggested model. Schematic representation of Xist chromatin structure in (A) XY (or XY Eed-TG), (B) XΔY (or XΔY Eed-TG), (C) XY Eed−/−, (D) XΔY Eed−/− ES cells and (E) female future Xi at the onset of XCI. Thick column, Xist exon 1; thin column, Xist promoter; open lollipops, H3K4m2; filled hexagons, H3K27m3; filled rectangles, methylated CpG. The darkness of the columns represents closed chromatin structure.
Discussion
Inability of the XDY Eed−/− ES cells to repress Xist despite intact counting
We demonstrated that the male ES cells with both Eed-null and Tsix mutations underwent ectopic Xist hyperactivation upon differentiation. This result can be attributed to either defective X-chromosome counting or dysfunction in Xist gene regulation. The Tsix mutant allele generated in this study does not lose any DNA elements necessary for X-chromosome counting, because the female ES cells heterozygous for the mutation did not show aberrant counting such as two Xi or no Xi (Shibata and Lee, 2004). The counting function has been ascribed to Xite (Ogawa and Lee, 2003) and an additional region at the 5′-portion of Tsix (Morey et al, 2004; Lee, 2005). Both DNA elements are completely conserved in the XΔY Eed−/− cells, and they are not included in the Xist cDNA Tg. Similarly, the DNA elements required for homologous chromosome pairing at the onset of XCI (Bacher et al, 2006; Xu et al, 2006) are retained in the XΔY Eed−/− cells and are not involved in the Tg. Thus, we conclude that the XΔY Eed−/− cells have a dysfunction in Xist regulation, that is, the mutant male ES cells are unable to repress Xist upon differentiation. It has long been unexplained why and how male ES cells can repress Xist without Tsix, although it is critical in females. Our findings demonstrated a synergistic role of Tsix and Eed in Xist regulation and indicated that Eed alone could effectively block ectopic Xist activation. In this context, our observation that the XΔY Eed−/− cells accumulated much more Xist RNA than wild-type female cells upon differentiation is reasonable, because the mutant cells have lost control of Xist transcription. We have identified Eed as an additional major player in regulating Xist expression and in the protection of future Xa from ectopic inactivation, this being the conceptual advance provided by this report.
Ectopic Xist activation in the XDY Eed−/− cells depends on a different mechanism from physiological Xist activation in females
We suggest that the open chromatin structure at the Xist promoter and exon 1 in the XΔY Eed−/− cells resulted in ectopic Xist hyperactivation, presumably by transcription factors that are not included in the physiological Xist transcription in females. It is also possible that some factors with Xist activator function are derepressed in Eed-null males and caused Xist activation in the XΔY Eed−/− cells, because many developmental regulators are prematurely activated in Eed-null ES cells (Azuara et al, 2006; Boyer et al, 2006). In undifferentiated male ES cells, the loss of Eed resulted in a 4.6-times increment in the Xist RNA level (Supplementary Table II, XY Eed-TG versus XY Eed−/−), whereas the loss of Tsix results in only a 1.6-times increment (Sun et al, 2006). In addition, the XY Eed−/− cells consistently showed an elevated Xist RNA level during differentiation. Therefore, Eed makes a substantial contribution in repressing Xist even in the presence of Tsix. Intriguingly, female ES cells heterozygous for a Tsix mutation can activate Xist transcription at the mutant allele despite the presence of H3K27m3 modification at the promoter (Sun et al, 2006). Hence, unlike nonspecific Xist activation, the physiological transcription factor for Xist can work if the promoter is H3K27 methylated. The transcription factor involved in Xist activation has not been discovered yet, and identification of the factor would be beneficial in understanding the regulation of XCI.
Eed and Tsix have synergistic, but autonomous functions in Xist regulation
Previous reports have shown that, in the absence of Tsix transcription, both active chromatin marker H3K4m2 and repressive marker H3K27m3 increase at the Xist promoter (Navarro et al, 2006; Sun et al, 2006; Shibata and Yokota, 2008), suggesting that Tsix inhibits H3K4 HMTase and PRC2 activity, together. Although we need further study to look into the molecular mechanism, it is likely that antisense transcription has a role in stabilizing local chromatin structure by preventing additional histone modifications. This hypothesis is strengthened by previous observations that the Xist locus in male ES cells is already in a repressed state in terms of CpG methylation (Norris et al, 1994; Sun et al, 2006) that is required for stable Xist repression (Panning and Jaenisch, 1996; Barr et al, 2007). We suggest that, in undifferentiated female cells, prolonged Tsix transcription on the future Xa prevents the reorganization of repressive Xist chromatin structure, whereas immediate Tsix shut down on the future Xi allows conversion to an active state, resulting in transcriptional activation of Xist. Importantly, Tsix transcription can also regulate Xist in an Eed-independent manner because the Xist is not hyperactivated in XY Eed−/− cells. It is still not clear whether the Eed-independent Xist regulation by Tsix is solely based on the inhibition of H3K4 methylation, or whether there is an additional molecular mechanism other than histone modifications. Trithorax group proteins are known to antagonize Polycomb group proteins, and human Trithorax group proteins MLL and ASH1L have been shown to have H3K4 HMTase activity (Milne et al, 2002; Gregory et al, 2007). The di- and trimethyl H3K27 demethylase UTX were reported to associate with MLL complexes and induce H3K4 methylation at the promoters of HOX genes (Lee et al, 2007). Most intriguingly, Drosophila UTX colocalizes with RNA polymerase II (Smith et al, 2008), implying possible association of UTX with Tsix transcription in the regulation of H3K27m3 modification in the Xist locus. We anticipate studies on the involvement of these HMTases and histone demethylases in the control of XCI.
Antisense RNA in the Xist gene body cannot prevent Xist activation
There is also an issue as to whether Tsix functions as an RNA entity or if transcription itself is sufficient. At present, there is no evidence showing that Tsix works as an RNA molecule (Shibata and Lee, 2004; Sado et al, 2006). Our finding that Xist could not be repressed despite the deregulated antisense transcription in the XΔY Eed−/− cells sheds some light on this issue. One possible interpretation is that continual Tsix transcription over the entire 40 kb length is necessary to organize the local chromatin structure and/or to retain proper subnuclear localization of the locus. Recent transcriptome analysis indicated that paired sense/antisense expression is not restricted to imprinted loci and that overlapping transcript pairs are more widespread in the mammalian genome than was thought previously (Katayama et al, 2005). Various types of interactions between the pairs are suggested, and at least some of them are likely to work through chromatin. Studies on the molecular mechanism of Tsix's action in repressing Xist would provide useful hints for understanding the function of antisense genes.
Materials and methods
Targeting construct
The pBl/E7EBS plasmid having a 6.7 kb EcoRI–BamHI fragment of Xist exon 7 followed by a 3.0 kb fragment of the Xist 3′-end (Shibata and Lee, 2004) was digested by BamHI, blunted-ended, and ligated with SalI-digested and blunted-ended 3.6 kb SA.IRES.hygro.pA2 fragment from pGT1.8IREShygropA2 (Nichols et al, 1998), yielding the targeting vector pBl/E7EBS/SIHA.
Cell culture and generation of cell lines
Clone 36 Eed−/− male ES cells (XY Eed−/−) were electroporated with SalI-linearized pBl/E7EBS/SIHA vector and selected as described (Schoeftner et al, 2006) to generate XΔY Eed−/− lines. Colonies were screened as reported (Shibata and Lee, 2004; Shibata and Yokota, 2008). The EL16.7 female ES cell (XX), EL16.7-derived Tsix-trap cell (XΔX) (Shibata and Lee, 2004), E14.1 male ES cell (XY) and E14.1-derived Tsix-trap cell (XΔY) (Shibata and Yokota, 2008) were used as controls. The J1 male ES cell line was used for EB formation in Supplementary Figure S1 and H3K27m3 ChIP assay in Supplementary Figure S2. Generation of the XY Eed-TG lines was as described (Schoeftner et al, 2006). For generation of the XΔY Eed-TG lines, the XΔY Eed−/− cells were electroporated with 50 μg of pCAG-EGFP-Eed-IREShyg-PA plasmid and cultured in the presence of 260 μg/ml of hygromycin. When cells became confluent, they were trypsinized and GFP-positive cells were sorted using a JSAN cell sorter (Bay Bioscience, Kobe, Japan), replated and grown until the next cell sorting. After three rounds of sorting enrichment, cells were split and GPF-positive single colonies were selected under a fluorescent microscope and isolated. For EB formation, trypsinized ES cells were incubated on a gelatinized dish at 37°C for 1 h to remove feeders, and 5 × 105 cells were split to a non-adherent 6 cm dish and cultured in suspension in ES cell media without leukemia inhibitory factor. After 3–4 days, EB were replated on gelatinized adherent culture dishes for prolonged culture. Gross morphology was examined by staining paraformaldehyde-fixed EB with Giemsa's solution.
Western blot
Whole cell lysate was prepared by dissolving 5 × 105 cells in 100 μl of WCL buffer containing 62.5 mM Tris–Cl, 2% SDS, 10% glycerol and 5% β-mercaptoethanol. The lysate was then boiled for 10 min and sonicated. After treating with SDS–PAGE and transferring to a nitrocellulose membrane, a western blot was performed with anti-H3K27m3 (no. 07–449, Upstate Biotechnology, 1:500 dilution) and goat anti-rabbit IgG-HRP (no. 12–348, Upstate, 1:2000). For reprobing, HRP activity was removed by 15% H2O2/PBS. The western blot was repeated with anti-Gapdh (no. 4300, Ambion, 1:1000) and goat anti-mouse IgG-HRP (no. 12–349, Upstate, 1:2000).
FISH and Immuno-FISH
Xist RNA-FISH was done as described (Lee and Lu, 1999). For immuno-FISH, slides after FISH were post-fixed in 2% paraformaldehyde/PBS and blocked in buffer containing 5% normal goat serum, 0.2% Tween20, 0.2% gelatin in 1 × PBS(−). Slides were incubated with anti-H4K20m1 (no. 07–440, Upstate, 1:50) at 4°C overnight, then incubated in goat anti-rabbit IgG-FITC (AP132F, Chemicon, 1:50) at 37°C for 1 h. Fluorescent microscope images were acquired and adjusted by using the Openlab software (Improvision, Coventry, UK).
Real-time RT–PCR and strand-specific RT–PCR
Total RNA was extracted using Trizol (Invitrogen). For qRT–PCR, total RNA was reverse-transcribed by Superscript III using random primers (Invitrogen). Xist, Tsix (amplicon 4), Mecp2, Pgk1 and Chic1 mRNAs were quantified using TaqMan Universal PCR Master Mix (4324018, Applied Biosystems) and an ABI Prism 7700 instrument. The results were normalized to Gapdh transcript by ΔΔCt method. Predesigned probes and primers for Mecp2, Pgk1, Chic1 and Gapdh were purchased from Applied Biosystems (Assay ID Mm00465017_m1, Mm00435617_m1, Mm01232479_m1 and P/N 4308313, respectively), and other probes and primers are shown in Supplementary Table IV. The qRT–PCR amplicon for Xist spans an intron and it does not detect Tsix. Tsix RNA at amplicon 4 (Figure 4D) was quantified in cDNA primed by a gene-specific primer: AAA GGG AAC TTA GAA CAG. Tsix RNA at amplicon 5 (Figure 4D), spanning from Xist intron 5 to 6 and not detecting transcripts in the Xist cDNA Tg, was quantified by using Brilliant II SYBR Green QPCR Master Mix (600828, Stratagene) as described (Shibata and Lee, 2003). All results were from three independent samples or from three independent cell lines (in XY Eed-TG and XΔY Eed-TG cells). Strand-specific RT–PCR of Tsix was as described previously (Stavropoulos et al, 2001; Shibata and Lee, 2004). Gene-specific primer and PCR primers for Gapdh were TTGGGTGCAGCGAACTTT, GCAGTGGCAAAGTGGAGATTGTTG and CCCTTCCACAATGCCAAAGTTGTC. Gapdh PCR primers for the SYBR green assay were GTAGACAAAATGGTGAAGGTCGGT and CAACAATCTCCACTTTGCCACTGC.
Northern blot
Poly-A tailed mRNA was purified using PolyATract mRNA isolation systems (Promega). A total of 2.5 μg of mRNA per lane was run in denaturing 1.2 % agarose gel in 1 × MOPS and transferred to a nylon membrane, which was firstly hybridized with random-primed MluI–BspMI 0.8 kb fragment (probe 1, Figure 4A) and subsequently reprobed with PCR-amplified Gapdh 0.4 kb fragment. Sequence integrity of PCR-amplified probe was confirmed.
Methyl-CpG-sensitive Southern blot
Genomic DNA was digested with an excess amount of EcoRI, precipitated with ethanol and dissolved in Tris–EDTA. The DNA concentration was determined and 5 μg of DNA was again digested with SacII or HpaII overnight. Double-digested DNA was run in 0.8% agarose gel, transferred and hybridized with MluI–BspMI 0.8 kb fragment at the Xist promoter (Figure 5A) or BamHI–EcoRI 0.6 kb fragment in exon 1 (Figure 5B).
Quantitative ChIP assay
ChIP assay was done as described (Morshead et al, 2003) with little modification. Briefly, fixed cells were aliquoted as 4 × 106 cells in a 1.5 ml tube, sonicated by a BIORUPTOR sonicator (Cosmobio, Tokyo, Japan) and incubated with anti-H3K27m3 (no. 07–449), anti-H3K4m2 (no. 07–030, Upstate), or anti-TFIIB (sc-225, Santa Cruz) at 4°C overnight. A mixture of protein A and G sepharose (GE Healthcare) was then added. Immunoprecipitated chromatin DNA was quantified by real-time PCR. Taqman probes and primers were shown in Supplementary Table IV. All results were from three independent samples. Statistical significance of the difference between the results in the XY Eed−/− and XΔY Eed−/− cells was analyzed by Student's t-test.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figures Legends
Supplementary Tables
Supplementary Methods
Acknowledgments
We thank M Asano (Kanazawa University) for the E14.1 cell line, H Niwa (RIKEN CDB) for the pGT1.8IREShygropA2 plasmid, S Harada (KU) for real-time PCR and C Sun (KU) for flow cytometry. This work was supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (18780250 to SS).
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E (2006) Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8: 293–299 [DOI] [PubMed] [Google Scholar]
- Barr H, Hermann A, Berger J, Tsai HH, Adie K, Prokhortchouk A, Hendrich B, Bird A (2007) Mbd2 contributes to DNA methylation-directed repression of the Xist gene. Mol Cell Biol 27: 3750–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T (1995) The eed mutation disrupts anterior mesoderm production in mice. Development 121: 273–285 [DOI] [PubMed] [Google Scholar]
- Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA (2007) Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 27: 8466–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T (2006) The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T (2006) The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol 8: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A (2004) A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2005) Regulation of X-chromosome counting by Tsix and Xite sequences. Science 309: 768–771 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N (1999) Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R (2001) Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol 21: 8512–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T (2003) Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet 33: 502–507 [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T (2005) The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947 [DOI] [PubMed] [Google Scholar]
- Morey C, Navarro P, Debrand E, Avner P, Rougeulle C, Clerc P (2004) The region 3′ to Xist mediates X chromosome counting and H3 Lys-4 dimethylation within the Xist gene. EMBO J 23: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA (2003) Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci USA 100: 11577–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Page DR, Avner P, Rougeulle C (2006) Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev 20: 2787–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, Sheardown SA, Rastan S (1994) Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 77: 41–51 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT (2003) Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell 11: 731–743 [DOI] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T (2006) Tsix-deficient X chromosome does not undergo inactivation in the embryonic lineage in males: implications for Tsix-independent silencing of Xist. Cytogenet Genome Res 113: 345–349 [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R (1996) DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev 10: 1991–2002 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H (2005) Tsix silences Xist through modification of chromatin structure. Dev Cell 9: 159–165 [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H (2006) Tsix defective in splicing is competent to establish Xist silencing. Development 133: 4925–4931 [DOI] [PubMed] [Google Scholar]
- Sado T, Li E, Sasaki H (2002) Effect of TSIX disruption on XIST expression in male ES cells. Cytogenet Genome Res 99: 115–118 [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E (2001) Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Lee JT (2003) Characterization and quantitation of differential Tsix transcripts: implications for Tsix function. Hum Mol Genet 12: 125–136 [DOI] [PubMed] [Google Scholar]
- Shibata S, Lee JT (2004) Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol 14: 1747–1754 [DOI] [PubMed] [Google Scholar]
- Shibata S, Yokota T (2008) Alteration of histone tail modifications in the Xist locus in wild-type and Tsix-mutant male embryonic stem cells during differentiation. Exp Anim 57: 153–157 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N (2003) Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A (2008) Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT (2001) A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA 98: 10232–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT (2006) A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 21: 617–628 [DOI] [PubMed] [Google Scholar]
- Vigneau S, Augui S, Navarro P, Avner P, Clerc P (2006) An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc Natl Acad Sci USA 103: 7390–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R (2002) Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174 [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT (2006) Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figures Legends
Supplementary Tables
Supplementary Methods