Abstract
The eukaryotic cytosolic chaperonin containing TCP-1 (CCT) has an important function in maintaining cellular homoeostasis by assisting the folding of many proteins, including the cytoskeletal components actin and tubulin. Yet the nature of the proteins and cellular pathways dependent on CCT function has not been established globally. Here, we use proteomic and genomic approaches to define CCT interaction networks involving 136 proteins/genes that include links to the nuclear pore complex, chromatin remodelling, and protein degradation. Our study also identifies a third eukaryotic cytoskeletal system connected with CCT: the septin ring complex, which is essential for cytokinesis. CCT interactions with septins are ATP dependent, and disrupting the function of the chaperonin in yeast leads to loss of CCT–septin interaction and aberrant septin ring assembly. Our results therefore provide a rich framework for understanding the function of CCT in several essential cellular processes, including epigenetics and cell division.
Keywords: CCT, chaperone, proteomics, septin, SGA
Introduction
The chaperonin of the eukaryotic cytosol, chaperonin containing TCP-1 (CCT) (also known as TRiC (TCP-1 ring complex)) was originally discovered and characterised because of its essential role in folding the cytoskeletal proteins actin and tubulin (reviewed in Frydman, 2001; Willison and Grantham, 2001; Cowan and Lewis, 2002). Similar to all chaperonins, CCT has a double-toroid shape with a central cavity and can, upon binding and hydrolysis of ATP, assist in the folding of non-native proteins or in the assembly of substrates with their respective partners.
CCT has a critical function in cellular protein folding. On the basis of immunoprecipitation experiments, a maximum estimate of 9–15% of all newly synthesised cytosolic proteins transit through CCT and thus may require its assistance for folding and/or assembly (Thulasiraman et al, 1999), although our own previous in vivo pulse-chase experiments identified fewer transiting polypeptides (∼1% of total; Sternlicht et al, 1993; Grantham et al, 2006). Only a relatively small number of proteins (∼30) have been shown to depend on CCT for their biogenesis; these include the cytoskeletal proteins α- and β-actin, an actin-related protein (centractin), α-, β-, and γ-tubulin, the von Hippel–Lindau tumour suppressor, histone deacetylases (HDAC3, Set3p, and Hos2p), and cell cycle regulators (Cdc20p and Cdc55p) (Spiess et al, 2004). Although some CCT substrates possess seven-bladed β-propeller (WD40 repeat-containing) folds and have a high affinity for CCT (Valpuesta et al, 2002; Camasses et al, 2003; Passmore et al, 2003; Spiess et al, 2004; Kubota et al, 2006), most CCT substrates are highly diverse with respect to their functions and structures, and cannot be inferred as such based on homology alone (Valpuesta et al, 2005). Also of note is that some CCT-interacting proteins may not represent substrates, but instead, are cofactors or cochaperones of the chaperonin. For example, several phosducin-like proteins interact with CCT and modulate its abilities to fold actin, tubulin, Gβ and perhaps other substrates (McLaughlin et al, 2002; Lukov et al, 2006; Stirling et al, 2006, 2007). Another important CCT cofactor is prefoldin, a hetero-hexameric chaperone that stabilises newly made forms of actin and tubulin and transiently interacts with the chaperonin to assist in their effective delivery to the folding cavity (Geissler et al, 1998; Vainberg et al, 1998).
Previously, many putative CCT-interacting proteins were uncovered in high-throughput protein interaction screens in yeast (Gavin et al, 2002, 2006; Ho et al, 2002; Graumann et al, 2004; Krogan et al, 2006). However, for the great majority of these proteins, it remains unverified whether they are bona fide CCT substrates, cofactors or cochaperones. In addition, a significant number of proteins that interact with CCT may not have been identified in co-precipitation studies because of the poor accessibility of N- or C-terminal epitope tags, both of which are buried in the central cavity of the chaperonin complex (Pappenberger et al, 2006). Another limitation has been that synthetic-sick or -lethal genetic interaction screens have not been performed on any of the eight CCT subunit genes because they are essential for viability.
In an attempt to identify a comprehensive CCT ‘interactome', we have taken a two-pronged approach using Saccharomyces cerevisiae. We used strains containing CCT complexes with an exposed CBP affinity tag to isolate functional CCT complexes and identify bound proteins and we examined the genetic interaction spectrum of CCT by performing a synthetic genetic array (SGA) screen for synthetic lethal interactions with a temperature-sensitive cct allele. Using these integrative approaches, we identified a large number of novel CCT physical and genetic interactors that define several functional interaction networks. We have validated in vivo the interactions between CCT and septins, a family of GTP-binding filament-forming proteins, and shown that CCT is required for proper septin ring formation. Our data are the first to link globally CCT to specific protein complexes and cellular processes, defining its position in diverse biological networks.
Results
Purification of yeast CCT and identification of bound proteins
Previously, we showed that incorporation of a CBP tag in an exposed region of the CCT complex allows for its effective purification using calmodulin resin chromatography (Pappenberger et al, 2006). To discover novel CCT-interacting proteins from S. cerevisiae, we developed a stringent purification protocol utilising two different versions of tagged CCT, one bearing the CBP tag in the Cct3p subunit (CCT–3CBP: Pappenberger et al, 2006) and another in the Cct6p subunit (CCT–6CBP; see Materials and methods).
The presence of the tag in the Cct3p subunit does not interfere with CCT function in vivo or in vitro (Pappenberger et al, 2006) and CCT–6CBP in haploid yeast is also viable and folding-competent in vitro (data not shown). A conventional 1D SDS gel protein separation and band-excision protocol, as shown for a CCT–6CBP sample in Figure 1A, identified CCT subunits, Hsc70 family members, known substrates such as Act1p, and many other proteins (Supplementary Table S1). Indeed, several proteins were recovered by both Cct3p- and Cct6p-tagged complexes, including actin, as expected, and three of five core septin subunits, proteins not previously known to interact with CCT.
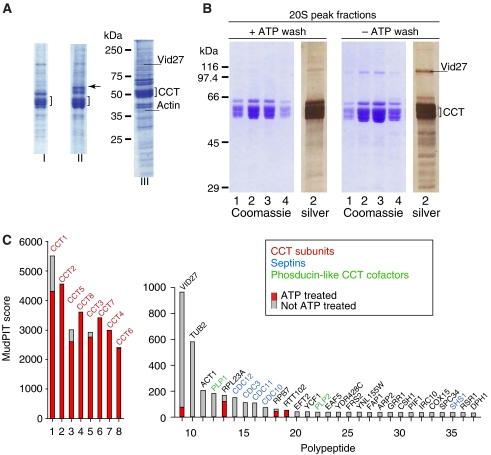
Figure 1.
Isolation of CCT substrate complexes. (A) Affinity purification of CCT–3CBP and CCT–6CBP on calmodulin resin. CaM-resin-purified CCT was analysed by 10% SDS–PAGE. (Lane I) CCT–3CBP complex: the eight CCT subunits migrating around the 60 kDa marker are indicated by the square bracket. (Lane II) CCT–6CBP complex: subunit Cct6p migrating at 70 kDa as a consequence of the inserted tag is indicated by an arrow. (Lane III) CCT–6CBP complex as used for in-house MALDI-MS and PMF-MS analysis. Molecular weight markers in kilodaltons are indicated on the left, Vid27p and actin are indicated as a reference (see also Supplementary Table S1). (B) Purification of CCT–3CBP in the presence and absence of ATP. CCT was incubated ±5 mM ATP at room temperature during the CaM affinity purification procedure. EDTA eluates of CCT were applied to 10–40% sucrose gradients. The panels +ATP wash and −ATP wash show four adjacent sucrose gradient fractions from each 20S CCT peak electrophoresed on 10% SDS gels followed by Coomassie (fractions 1–4) or silver staining (fraction 2 from each peak). (C) Histograms of the CCT–3CBP-bound polypeptides plotted against their MudPIT score (Tables I). Comparisons between the ATP-treated (red bars) and untreated CCT samples (gray bars) show the proteins that are removed by 5 mM ATP treatment during the CaM chromatography step.
To discover new CCT substrates—those proteins that need CCT for either folding or assembly—we used ATP-dependent release of proteins from the chaperonin as the major determinant. We applied the multidimensional chromatography-mass spectrometry approach, MudPIT, to our CCT preparations (Graumann et al, 2004). Before subjecting CCT to liquid chromatography electrospray ionisation mass spectrometry (LC-ESI-MS), we centrifuged the CaM resin eluates on 10–40% sucrose gradients to further purify the CCT complexes (Figure 1B) and reduce contamination by nonspecific proteins and by bona fide calmodulin-binding proteins. The CaM resin wash step was performed with or without ATP, and in the analysis similar numbers of tryptic peptides were obtained from digestion and LC-ESI-MS analysis of both treatments; CCT-ATP (Table I) and CCT+ATP (Table II)—9352 and 9221 peptides, respectively. Tables I and II show the top matches obtained for the two samples. As expected, the eight CCT subunits dominate both histograms (Figure 1C) and, gratifyingly, the next most abundant ATP-elutable matches include some proteins known to interact with CCT, namely the folding substrates actin and β-tubulin, and folding regulators Plp1p and Plp2p (Stirling et al, 2006, 2007). Immediately following these hits are a previously undiscovered set of CCT-interacting proteins, the septin GTPases (Figure 1C, blue text). We also identified the ATP-elutable, actin-related protein Arp2p just above the arbitrary cutoff for these lists (Melki and Cowan, 1994; Gavin et al, 2002). Thus, it is likely that many of the novel CCT-interacting proteins represent bona fide partners of CCT. Of further note is that additional polypeptides with significant scores appear in the ATP-treated sample including the type V myosin motor, the dynein heavy chain, an mRNA decapping protein (EDC3), and SWI3, a subunit of the SWI1/SNF chromatin remodelling complex (Table II). Finally there are a few proteins that are only partially removed by ATP treatment such as the previously described CCT-binding protein Vid27p (Aloy et al, 2004) and Rpl23Ap, Rpb7p, and Rtt102p.
Table 1.
CCT–3CBP interacting proteins in the absence of ATP, identified by LC-ESI-MS
| Gene name | Score | Mass (Da) | Polypeptide | Copies per cell | Synthesis rate |
|---|---|---|---|---|---|
| TCP1/CCT1 | 5511 | 60 899 | Chaperonin subunit | ND | 0.41 |
| CCT2 | 4409 | 57 510 | Chaperonin subunit | ND | 0.38 |
| CCT5 | 3010 | 61 152 | Chaperonin subunit | ND | 0.81 |
| CCT8 | 2976 | 61 965 | Chaperonin subunit | 2200 | 0.34 |
| CCT3 | 2929 | 59 404 | Chaperonin subunit | ND | 0.48 |
| CCT7 | 2924 | 60 211 | Chaperonin subunit | ND | ND |
| CCT4 | 2554 | 57 967 | Chaperonin subunit | 5530 | 0.34 |
| CCT6 | 2413 | 60 171 | Chaperonin subunit | ND | 0.42 |
| VID27 | 955 | 89 190 | WD40 (yeast/plants) | 3500 | 0.16 |
| TUB2 | 574 | 51 233 | β-Tubulin | ND | 0.40 |
| ACT1 | 201 | 41 906 | Actin | 215 000 | 2.30 |
| PLP1 | 180 | 26 834 | Phosducin-like protein 1 | 2250 | 0.15 |
| RPL23A | 165 | 14 578 | 60S ribosomal subunit | ND | 3.39 |
| CDC12 | 147 | 46 753 | Septin | 1170 | 0.23 |
| CDC3 | 108 | 60 188 | Septin | ND | 0.22 |
| CDC11 | 105 | 47 790 | Septin | 9280 | 0.16 |
| CDC10 | 70 | 37 059 | Septin | 14 100 | 0.26 |
| RPB7 | 58 | 24 330 | RNA pol II subunit 16 | 6420 | ND |
| RTT102 | 43 | 21 177 | SWI/SNF and RSC complex | 1550 | 0.16 |
| EFT2 | 39 | 93 660 | Diphthamide/His target | 160 782 | 2.37 |
| YCF1 | 38 | 172 196 | ABC transporter (cadmium) | 396 | 0.05 |
| PLP2 | 36 | 32 887 | Phosducin-like protein 2 | 7700 | 0.22 |
| EAF5 | 36 | 32 887 | NuA4 acetyltransferase | 937 | ND |
| FRS2 | 35 | 57 511 | Phenylalanyl-tRNA synthetase a | ND | 0.30 |
| YDR428C | 35 | 31 565 | Serine hydrolase | ND | ND |
| YNL155W | 34 | 31 953 | AN1-type zinc-finger | 4280 | ND |
| FAP1 | 33 | 113 668 | Transcription/binds FKBP12 | 589 | ND |
| ARP2 | 33 | 44 217 | Arp2/3 actin nucleation | 6650 | 0.35 |
| GRR1 | 33 | 133 848 | F-box SCF E3 ligase | ND | 0.03 |
| CSH1 | 33 | 44 639 | MIPC synthase | 195 | ND |
| PIF1 | 33 | 97 762 | DNA helicase mitochondrial | 259 | 0.06 |
| IRC10 | 32 | 68 217 | DNA repair | ND | ND |
| COX15 | 32 | 54 794 | Hydroxylation haeme O to haeme A | ND | ND |
| SPC34 | 32 | 34 171 | Dam1/DASH complex | ND | 0.10 |
| SHS1 | 31 | 62 706 | Septin | 5620 | 0.23 |
| RSR1 | 31 | 30 486 | GTPase (CDC24 interaction) | ND | ND |
| DPH1 | 30 | 48 679 | Diphthamide synthesis | 2562 | ND |
| NCBInr Saccharomyces cerevisiae database of 11 143 sequences was used for Mascot searches. CCT subunits (italics) and other proteins listed according to MudPIT score with a cutoff set at 30. Copy numbers are taken from the Swissprot database (http://expasy.org/sprot/), and estimated synthesis rate (in protein per second) are from Arava et al (2003). ND, not determined. | |||||
Table 2.
CCT–3CBP interacting proteins in the presence of ATP, identified by LC-ESI-MS
| Gene name | Score | Mass (Da) | Polypeptide |
|---|---|---|---|
| CCT2 | 4560 | 57 510 | Chaperonin subunit |
| TCP1/CCT1 | 4308 | 60 899 | Chaperonin subunit |
| CCT8 | 3607 | 61 965 | Chaperonin subunit |
| CCT7 | 3409 | 60 211 | Chaperonin subunit |
| CCT4 | 2992 | 57 967 | Chaperonin subunit |
| CCT3 | 2761 | 59 404 | Chaperonin subunit |
| CCT5 | 2607 | 61 152 | Chaperonin subunit |
| CCT6 | 2371 | 60 171 | Chaperonin subunit |
| RPL23A | 117 | 14 578 | 60S ribosome subunit |
| VID27 | 73 | 89 190 | WD40 (yeast/plants) |
| RTT102 | 49 | 21 177 | SWI/SNF and RSC complex |
| MYO4 | 48 | 170 092 | Type V myosin motor |
| TID3 | 42 | 80 552 | Kinetochore-associated Ndc80 complex |
| RPB7 | 42 | 24 330 | RNA pol II subunit 16 |
| SEY1 | 40 | 89 662 | Membrane organisation—GTP binding |
| YKL088W | 40 | 65 312 | Phosphopantothenoylcysteine decarboxylase |
| EAP1 | 40 | 69 719 | eIF4E-associated protein |
| YOR356W | 39 | 70 103 | Mitochondrion—flavoprotein-type oxidoreductase |
| CCC1 | 36 | 34 229 | Vacuolar Ca2+/Mn2+ transporter |
| HSH155 | 36 | 110 642 | U2-snRNP-associated splicing factor |
| VAR1 | 36 | 47 149 | Mitochondrial ribosomal protein small subunit |
| YRR1 | 36 | 93 321 | Zn2-Cys6 zinc-finger transcription factor |
| RPG1 | 35 | 110 333 | Translation initiation factor 3 complex (eIF3) |
| ENP2 | 35 | 81 983 | Nucleolar protein |
| RPL36A | 34 | 11 117 | N-terminally acetylated protein 60S ribosome |
| MNN1 | 33 | 89 058 | Golgi alpha-1,3 mannosyltransferase |
| DYN1 | 33 | 473 843 | Microtubule motor |
| CPN60 | 33 | 60 999 | Mitochondrial chaperonin |
| DOP1 | 32 | 195 648 | Mitochondria/cell morphogenesis |
| EDC3 | 32 | 195 648 | Enhancer of mRNA decapping protein 3 |
| PSF3 | 32 | 21 952 | DNA replication complex GINS protein PSF3 |
| PEX25 | 31 | 45 111 | Peripheral peroxisomal membrane peroxin |
| SLA2 | 31 | 109 442 | Transmembrane actin-binding protein |
| BCH1 | 30 | 82 567 | Chs5p–Arf1p-binding proteins (ChAPs) |
| PDX1 | 30 | 45 466 | Dihydrolipoamide dehydrogenase (E3BP) |
| SWI3 | 30 | 92 984 | SWI/SNF chromatin remodelling complex |
| NCBInr Saccharomyces cerevisiae database of 11 143 sequences was used for Mascot Searches. CCT subunits (italics) and other proteins listed according to MudPIT score with a cutoff set at 30. | |||
We performed controls for nonspecific binding of yeast proteins to the calmodulin column in the presence and absence of the ATP wash steps by analysing the final washes prior to specific elution of CCT from the CaM resin. We identified 307 hits with MudPIT score >30, our cutoff value (see Supplementary Table S2). Only signals from the most abundant substrates are present in the ATP wash because the spectra are dominated by the most abundant proteins in yeast cytosol (i.e. glycolytic enzymes). The Ssa/Ssb chaperones are highly represented and despite the high affinity that certain Ssa and Ssb family proteins have for calmodulin (Shevchenko et al, 2002) it is likely that some Ssa/Ssb chaperone signal is the result of the specific interactions known to occur with CCT (Lewis et al, 1992). Several mitochondrial protein interactions are also observed; however, all of these hits are encoded by nuclear genes, and are therefore potential clients of CCT, except Var1p, found in the ATP-washed complex (Table II).
Overall, our proteomic analyses of the CCT–3/6CBP fusions revealed a total of 72 protein–protein interactions, many of which are novel (Table I and Supplementary Table S1). Moreover, our functional proteomics experiment (Figure 1C) revealed several ATP-elutable CCT-binding proteins, which may represent substrate proteins or cofactors and regulators of CCT activity. These data significantly expand the known CCT interactome and point to several possible novel cellular roles for CCT.
Genetic interactors of CCT
Genetic interactions can imply that the interacting genes work in the same, or parallel, cellular pathways to execute a common function. This information can be extremely useful for placing a gene in a biochemical pathway or identifying redundancies in vivo. In recent years, SGA technology has allowed the creation of extensive genetic interaction networks based on large-scale synthetic lethal genetic interaction screens (Tong et al, 2001, 2004) and for specific cellular processes (Zhao et al, 2005).
To explore the extent of CCT function in the cell, we used the cct1-2 temperature-sensitive allele to perform an SGA screen with the ∼5000 viable yeast deletion mutants. From all of the possible synthetic interactions identified, a total of 72 gene deletions were validated by random spore analysis as growing more slowly at 30°C (permissive temperature for cct1-2) when in combination with cct1-2 (Supplementary Table S3). The genetic interactions probably result from an overlapping function between the SGA hit and some CCT-interacting protein whose folding is perturbed when CCT is disrupted (Figure 2A). In other words, the SGA hits would not typically interact with CCT physically but instead functionally overlap with a CCT substrate that misfolds in the cct1-2 strain. The exception to this logic would be PLP1, which works in a complex with CCT to promote efficient protein biogenesis (Stirling et al, 2006). Consequently, genetic interactors of CCT point towards those cellular pathways that involve a CCT substrate (Figure 2). We placed SGA interactions into functional groupings along with relevant CCT physical interactions from our studies and the literature (Figure 2; Gavin et al, 2002, 2006; Ho et al, 2002). Our analysis, combined with gene ontology annotations of the interacting genes, reveals strong biases towards the cytoskeleton and chromatin modification (Figure 2B and C, and Supplementary Tables S3 and S4). Additional connections were identified between CCT and the nuclear pore complex (NPC) (Figure 2D), and other functional modules (Figure 2E). Figure 2F shows the remainder of the SGA interactions not grouped in other panels, such that all 72 genetic interactions are shown in Figure 2.
Figure 2.
Genetic and physical interactions of cct1-2 reveal numerous protein complexes functionally connected to the CCT chaperonin. Network diagrams incorporating either physical and genetic interactions (B–D) or only genetic interactions (E, F) are shown. (A) Schematic method used to implicate CCT-binding proteins as putative substrates by integrating SGA and proteomic analyses. The presence of cct1-2 leads to misfolding of a substrate ‘X' that exhibits a known genetic interaction with gene ‘Y'. Therefore, the SGA hit connecting cct1-2 and gene ‘Y' actually reflects loss of function for a CCT substrate. Protein complexes or functional groupings are circled and labelled in each case and sorted into (B) cytoskeletal functions, (C) chromatin remodelling functions, (D) nuclear functions, and (E) other functional groups found within the SGA hits. (F) The remaining 15 SGA interactions with cct1-2 such that all 72 SGA interactions are represented in this figure. Genetic interactions are shown in black, physical interactions are shown in red and interactions within the protein complex groupings are shown in blue. SGA hits are represented by grey nodes and physical interactors are represented by black nodes.
As expected, various components of the actin/tubulin folding machinery were identified, including the prefoldin complex, tubulin folding cofactors C and D (CIN2 and CIN1, respectively), and the phosducin-like protein PLP1. The cytoskeletal connections were expected because of the known participation of CCT on actin and tubulin folding, yet they reveal the depth of physical and genetic connections of CCT to the cytoskeleton and its various effectors (e.g. the endocytic machinery; Figure 2B).
The bias towards chromatin remodelling components may be a partially nonspecific phenomenon, as some of the genes identified have more than 100 synthetic genetic interactions (LGE1, BRE1, CDC73, HTZ1, SWR1; Tong et al, 2004; Collins et al, 2007). Nonetheless, CCT likely assists in the biogenesis of several histone deacetylase complexes (Pijnappel et al, 2001; Guenther et al, 2002). Also, the Set3p, SAGA, and RNAPII–Paf1p complexes that are implicated in chromatin modification contain both genetic and physical connections to the chaperonin, suggesting likely connections to CCT (Figure 2C).
In addition, several physical and genetic interactions suggest that CCT has a functional connection to the NPC and nuclear transport (Figure 2D). This was a totally unexpected result, and, although genetic interactions alone could be considered as secondary cytoskeletal effects, the fact that there are several genetic and physical interactions with CCT (Figure 2D and Supplementary Tables S1 and S3) supports the biological significance of this result.
To expand upon our genomic analysis, we examined the cross-correlation coefficients (CC) of yeast gene expression for the cytoskeletal network (Figure 2B) as calculated by Sceptrans (Kudlicki et al, 2007; shown as a simple heatmap in Supplementary Table S5). The genes in the cytoskeletal network are tabulated as shown in Figure 2, where genes have been grouped in accordance with their functional subgroup, with the addition of the tubulin genes for comparison. The heatmap shows strong correlations between genes within a subgroup, as indicated by hot spots along the diagonal. There are strong correlations between CCT and PLP2, the bud-emergence subgroup, the prefoldin subgroup, and the kinase CLA4 of the septin subgroup. Interestingly, the prefoldin subgroup also correlates strongly with PLP2 consistent with the proposed role of Plp2p in actin folding (Vainberg et al, 1998; Stirling et al, 2007). The correlation between individual CCT subunits and substrates ACT1 and tubulins is variable, with values ranging from 0.51 to −0.36. Positive correlation coefficients are found between CCT subunits and septins, with one septin component, SHS1, having the highest CC. Interestingly, we were able to identify a direct interaction between Cct6p and Shs1p because Shs1p co-elutes specifically with Cct6p in sucrose gradient fractions containing dissociated subunits of the CCT complex (Figure 3E). As exemplified in Tu et al (2005), a positive correlation between CCT and other genes will not necessarily identify substrates per se, but may strongly suggest co-involvement of gene pairs in the same cellular process, akin to a genetic interaction. Hence this information reinforces the idea of CCT involvement in the septin network, possibly mediated through Cla4p. We note the strong correlation between CCT and prefoldin genes, providing another line of support for the transient character of CCT–prefoldin interaction (Hansen et al, 1999), which was not detected in our MS analysis.
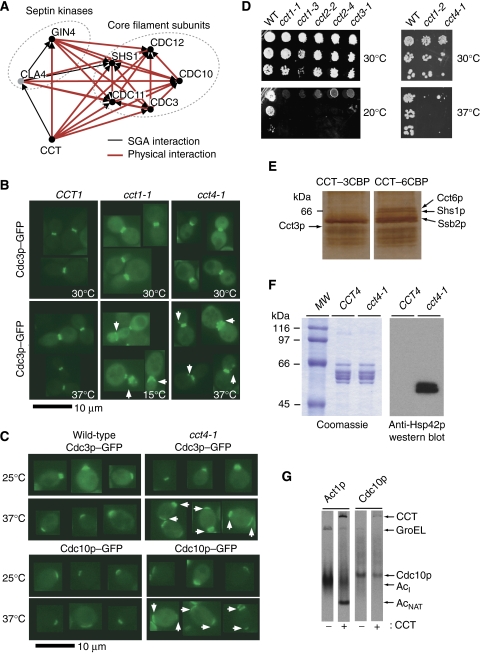
Figure 3.
Genomic and proteomic analyses identify a role for CCT in septin cytoskeleton function. (A) Physical and genetic interactions of yeast CCT with septin core components and septin kinases. (B) Localisation of GFP–Cdc3p in budded wild-type CCT1, cct1-1 cold-sensitive and cct4-1 heat-sensitive yeast cells. White arrows point to mis-shapen septin rings or additional patches of septin signal in cct mutant cells. CCT1 wild-type strain at 15°C which had normal septin localisation is not shown (see Table IIIA). (C) Localisation of GFP–Cdc3p and GFP–Cdc10p in wild-type and cct4-1 cells at permissive (25°C) and non-permissive temperatures (37°C). White arrows point to aberrant sites of septin localisation. (D) Allelic series of CCT cs and ts allele bearing strains to determine relative sensitivities of different alleles. CCT mutant cells were serially diluted, spotted onto rich growth media and grown at the temperatures indicated for 48 or 72 h. (E) Identification of Shs1p–Cct6p interaction. CCT–3CBP and CCT–6CBP were purified by CaM resin and sucrose gradient velocity sedimentation, after which the fraction of 18% sucrose normally containing individual CCT subunits was subjected to another CaM resin purification step. The eluates were run on an SDS gel and silver-stained and key bands were identified by PMF-MS as indicated. (F) Western blot: CaM resin-purified CCT from wild-type cells and mutant cct4-1 cells, blotted with anti-Hsp42p antibody, shows co-migration of Hsp42p with mutant CCT but not with wild-type CCT. (G) In vitro translation of Act1p and Cdc10p in E. coli-derived extract with (+) or without (−) added yeast CCT. AcI and AcNAT indicate the actin folding intermediate and native actin, respectively, according to Pappenberger et al (2006).
Analysis of septin function in yeast bearing mutant CCT subunits
The physical association of CCT with all five septin subunits (Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p) and a septin kinase (Gin4p), as well as the genetic interaction between cct1-2 and the CLA4 septin kinase deletion strain (Figures 1C, 2B and 3A) strongly suggest a role for CCT in modulating septin function. Septins assemble into a defined core complex that polymerises in a GTP-dependent fashion into filamentous structures. During the G1 phase the septins form a patch at the incipient bud site before expanding into a disk during bud emergence and then they re-organise into an hourglass-shaped collar around the bud neck, which then splits during M phase to form a pair of rings before cytokinesis and septin disassembly. This entire process is regulated by the kinases Cla4p, Gin4p, and Swe1p, and the phosphatase Rts1p (Versele and Thorner, 2005).
To explore a role for CCT in septin function, we examined the organisation of two GFP-tagged septin subunits (Cdc3p and Cdc10p) in cells bearing cold- and heat-sensitive alleles of CCT1, CCT2, CCT3, and CCT4. The localisation of Cdc3p and Cdc10p is normally confined to the patch or collar structures described above. We found that the pattern of septin localisation was altered for both Cdc3p and Cdc10p in cold-sensitive cct1-1 and in heat-sensitive cct4-1 cells (Figure 3B and Table III-A). The morphology of the septin collar was disrupted in a significant percentage of cct1-1 and cct4-1 budded cells at the non-permissive temperatures (Figure 3B and Table III-A). Moreover, we observed aberrant cortical GFP-tagged septin patches in the mutant cells (Figure 3B and Table III-A). Interestingly, these phenotypes were observed in cold-sensitive cct1-3 cells for the localisation of Cdc10p but not Cdc3p (Table III). Finally, we observed, in unbudded cct4-1 cells, septin patches that seemed to be retained following cell division while a new septin structure was assembled on the opposite side of the cell (Figure 3C and Table III-B). Importantly, none of the above phenotypes were observed in cct1-2, cct2-4 or cct3-1 alleles or in deletions of the CCT cofactors PLP1 or prefoldin (pac10Δ) (Table III-A). This suggests that the defects specifically arise because of disrupted CCT function, and not simply cytoskeletal protein dysfunction. Even cells lacking both PLP1 and PAC10, which have a severely disrupted actin cytoskeleton (Stirling et al, 2006), had largely wild-type septin localization, although a small but significant percentage of septin collars displayed anomalies in these cells, potentially through an effect on the chaperonin itself, given the functional interactions between CCT and Plp1 and prefoldin (Table III-A).
Table 3.
In vivo septin defects in cct mutant strains
| Strain | Genotype | Temperature | Normal rings (%) | Abnormal rings (%) | Additional patches (%) | Total no. of cells analysed | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CDC3 | CDC10 | CDC3 | CDC10 | CDC3 | CDC10 | CDC3 | CDC10 | |||
| A. Septin localization in budded cells | ||||||||||
| MLY100 | WT | 15 | 98.2 | 100.0 | 1.8 | 0.0 | 0.0 | 1.0 | 110 | 100 |
| 30 | 100.0 | 97.8 | 0.0 | 1.1 | 0.0 | 1.1 | 98 | 91 | ||
| MLY110 | plp1Δ | 30 | 100.0 | 99.0 | 0.0 | 1.0 | 0.0 | 0.0 | 87 | 100 |
| MLY111 | pac10Δ plp1Δ | 30 | 94.6 | 96.1 | 5.4 | 3.9 | 1.1 | 0.0 | 93 | 102 |
| MLY118 | pac10Δ | 30 | 92.5 | 93.5 | 7.7 | 1.9 | 0.0 | 4.7 | 104 | 107 |
| DUY558 | CCT1 | 15 | 100.0 | 100.0 | 0.0 | 0.0 | 2.9 | 1.9 | 103 | 107 |
| 25 | 94.5 | 99.1 | 5.5 | 0.9 | 1.8 | 0.9 | 109 | 113 | ||
| 30 | 98.0 | 98.1 | 2.0 | 1.9 | 3.0 | 2.8 | 100 | 106 | ||
| 37 | 98.2 | 98.2 | 1.8 | 0.0 | 1.8 | 1.8 | 112 | 111 | ||
| DUY559 | cct1-1 | 30 | 100.0 | 100.0 | 0.0 | 0.0 | 1.0 | 3.6 | 98 | 110 |
| 15 | 55.6 | 62.0 | 44.4 | 38.0 | 18.8 | 12.0 | 117 | 108 | ||
| DUY560 | cct1-2 | 25 | 100.0 | 100.0 | 0.0 | 0.0 | 0.9 | 0.0 | 110 | 104 |
| 37 | 96.2 | 100.0 | 3.8 | 0.0 | 1.9 | 0.0 | 106 | 101 | ||
| DUY561 | cct1-3 | 30 | 100.0 | 100.0 | 0.0 | 1.8 | 2.7 | 5.5 | 111 | 110 |
| 15 | 96.0 | 76.6 | 4.0 | 23.4 | 1.0 | 29.0 | 101 | 107 | ||
| CUY466 | cct2-4 | 30 | 99.1 | 100.0 | 0.9 | 0.0 | 0.9 | 0.0 | 112 | 110 |
| 15 | 94.1 | 98.1 | 5.9 | 1.9 | 4.0 | 3.8 | 101 | 106 | ||
| CUY492 | cct2-2 | 30 | ND | ND | ND | ND | ND | ND | ND | ND |
| 15 | ND | ND | ND | ND | ND | ND | ND | ND | ||
| CUY462 | cct3-1 | 30 | 96.3 | 100.0 | 3.7 | 0.0 | 4.7 | 0.0 | 107 | 104 |
| 15 | 92.3 | 94.6 | 7.7 | 5.4 | 3.3 | 2.2 | 91 | 93 | ||
| DDY299 | cct4-1 | 25 | 98.8 | 99.1 | 1.2 | 0.9 | 3.6 | 5.6 | 83 | 108 |
| 37 | 84.6 | 82.4 | 15.4 | 17.6 | 14.1 | 11.0 | 78 | 91 | ||
| B. Septin localization in unbudded cells | ||||||||||
| Normal single patch (%) | Opposing extra patch (%) | Total no. of cells analysed | ||||||||
| CDC3 | CDC10 | CDC3 | CDC10 | CDC3 | CDC10 | |||||
| MLY100 | WT | 30 | 100.0 | 100.0 | 0.0 | 0.0 | 77 | 57 | ||
| DUY558 | CCT1 | 25 | 91.1 | 93.8 | 8.9 | 6.2 | 79 | 81 | ||
| 37 | 95.4 | 94.9 | 4.6 | 5.1 | 65 | 59 | ||||
| DDY299 | cct4-1 | 25 | 100.0 | 100.0 | 0.0 | 0.0 | 54 | 95 | ||
| 37 | 80.7 | 79.7 | 19.3 | 20.3 | 57 | 59 | ||||
| (A) Aberrant septin ring morphology or additional patches of GFP-septin signal were scored for the strains indicated at the noted temperatures. Scores differing significantly from wild type (P<0.01) are reported in bold. (B) The presence of additional septin patches in unbudded cct4-1 cells was scored and reported as in A. ND, not determined. | ||||||||||
It is possible that septin defects are only seen in some CCT mutant strains because of the relative severity of the alleles. If the cct1-1 and cct4-1 alleles were much stronger than the other alleles tested they may be revealing a general septin defect in response to a dysfunctional CCT holo-complex. In support of the specific nature of the defects, we observed that the ts growth defects in cct1-2 cells was at least as severe as those of cct4-1 cells and that all of the cs alleles tested show strong growth defects when grown at 20°C (Figure 3D). Therefore, at the non-permissive temperatures used in our septin localisation experiments (15 and 37°C), all of the ts and cs alleles should have shown defects if the septin mislocalisation were a nonspecific result of abrogating CCT function. This observation of allele specificity suggests the existence of a physical interaction between the elements under study, a fact we had already observed with purification and mass spectrometry of CCT and septins (Figure 1), as well as with the observation of a direct interaction between Cct6p and the Shs1p septin subunit (Figure 3E). In light of these findings, we note that certain very general downstream effects of septin disruption have been observed previously in CCT mutant yeast, including multi-budded cells and abnormal chitin deposition (Vinh and Drubin, 1994; Stirling et al, 2007).
To assess directly the impact of CCT disruption on the interaction with septins, we analysed the proteins interacting with mutant CCT complexes isolated from cct4-1 cells grown at permissive temperature by LC-ESI-MS in a similar manner as performed for CCT–3CBP (gene lists not shown). Compared to wild-type CCT, two main differences were observed. First, we noted a high level of Hsp42p on the mutant CCT complex, an interaction that was independently confirmed by western blotting with anti-Hsp42p antibodies (Figure 3F and Haslbeck et al, 2004). This interaction is consistent with cct4-1 cells harbouring a functionally abrogated CCT complex, as yeast Hsp42p is a promiscuous chaperone that may be recruited to stabilise or prevent the aggregation of the chaperonin. The second difference observed is that none of the five septins were detected on the mutant CCT complex. Taken together, these data strongly support the idea that loss of CCT interaction underlies the formation of the aberrant septin structures observed in the cct4-1 mutant (Figure 3B and Table III).
We propose that CCT is not involved in the biogenesis and folding of septin polypeptides, but rather performs a regulatory role that is clearly compromised in cct mutant cells. It is unlikely that CCT is required to fold septins de novo because they fold properly and polymerise when co-expressed in Escherichia coli, which lacks CCT (Versele and Thorner, 2005). We translated the septin Cdc10p in an E. coli-derived extract supplemented with yeast CCT, a system we have developed for folding actin polypeptide chains (Pappenberger et al, 2006). Figure 3G shows the analysis for Cdc10p; the presence of CCT during translation and folding has no effect on the yield of the folded septin protein in contrast to Act1p, which is completely dependent upon CCT for folding to the native state. Interestingly, some radiolabelled Cdc10p appears to associate with CCT, consistent with our proteomic and in vivo results (Figures 1 and 3).
Discussion
Efforts to understand the eukaryotic chaperonin CCT have been hampered because its essential and subtle mutant phenotypes are often masked by the pervasive actin and tubulin defects associated with its loss of function. Here, we present the first genome-wide analyses of CCT function using a combination of novel proteomic approaches and a synthetic interaction screen with a ts allele. Our findings provide a rich data set of 136 proteins/genes that interact with CCT, more than doubling the total number of known and putative CCT interactions (now at 227, from 91 previously identified in the literature). Importantly, our study shows for the first time both known and many unexpected links between CCT and specific protein complexes and cellular processes (Figure 2), defining its position in numerous biological networks.
A contentious issue in the CCT field is the number and nature of the substrate proteins actively folded by CCT (Sternlicht et al, 1993; Thulasiraman et al, 1999; Grantham et al, 2006). How does one discriminate experimentally a substrate protein that is completely dependent upon CCT for its folding, such as actin, from one which is partially dependent through kinetic partitioning. This analysis is confounded by the existence of proteins that require CCT for higher order assembly processes, cofactor proteins and other binding activities. Our CCT interactome, combined with functional assays, should assist in the identification and verification of novel substrates and cofactors of the chaperonin.
Cytoskeletal networks
Aside from the expected connections to actin- and tubulin-related processes, our analyses reveal unexpected physical and genetic connections to a third cytoskeletal system—the septin ring. The identification of strong, ATP-elutable interactions between CCT and all of the core septin components in S. cerevisiae suggested a physiologically relevant relationship between CCT and the septin ring (Figure 1). This proposition was also supported by the SGA analysis in which the septin kinase gene CLA4 was identified as an interactor of cct1-2. Indeed, direct examination of GFP–Cdc3p and GFP–Cdc10p in various CCT mutant strains revealed clear septin localisation/assembly defects (Figure 3 and Table III). Septins probably do not need CCT for biogenesis or folding, but CCT may participate in an ATP-dependent septin assembly/disassembly process. Septin organisation is disrupted when CCT function is compromised in mutant cells cct4-1. The ATP-dependent allosteric behaviour of CCT isolated from cct4-1 mutant cells was recently investigated (Shimon et al, 2008), showing that the intra- and inter-ring allosteric cooperativity was abolished in the CCT carrying the mutant subunit. The spectrum of CCT-interacting proteins is also perturbed in the mutant CCT complex from cct4-1 cells (data not shown). The sudden appearance of a small heat-shock protein and the absence of septins in the mutant proteomics data may be a direct consequence of the perturbation of allostery.
Chromatin modification and NPC networks
CCT was previously shown to assist the biogenesis of the SET3 histone deacetylase complex, defining a role for CCT in chromatin modification (Pijnappel et al, 2001; Guenther et al, 2002). We found that a large group of protein complexes seemingly impacted by CCT are involved in modifying or remodelling histones (Figure 2C and Supplementary Table S4). This suggests that the chaperonin impacts chromatin biology and epigenetics to an extent that was not previously appreciated, similar to that recently found for Hsp90 (Zhao et al, 2005). CCT displays interactions with complexes involved in histone methylation (i.e. COMPASS complex; Paf1–RNAPII complex), histone acetylation (i.e. SAGA complex), histone deacetylation (i.e. Rpd3 complex; Set3 complex; type II HDAC complex), chromatin remodelling (i.e. Swr1 complex; SWI/SNF complex), and proteins involved in telomere maintenance (e.g. Yku80p and Pif1p; Supplementary Table S4).
Proteins involved in chromatin modification are usually localised to the nucleus, implying either that CCT performs a protein complex assembly function in the nucleus or that it assists the folding or assembly of nuclear proteins in the cytosol prior to their transit through the nuclear pore. Surprisingly, our data reveal a network of physical and genetic interactions between CCT, core NPC proteins and other factors associated with nuclear transport (Figure 2D). Although the genetic interactions of cct1-2 with NUP170 and NUP188 could be indirect effects of cytoskeletal defects, the association of CCT with Nup1p, Nup2p, and Nsp1p suggests a direct relationship. This raises several possibilities: (1) CCT could be involved in the folding/assembly of the nuclear pore components themselves, (2) CCT could usher nuclear proteins from the ribosome to the NPC, releasing them directly to the pore in a state suitable for nuclear transport, or (3) CCT could transit the NPC to perform protein folding/assembly functions inside the nucleus, consistent with reports of nuclear localisation for CCT subunits (Souès et al, 2003). Further studies should help elucidate the precise role of CCT in nuclear protein folding/transport and/or nuclear pore function.
TOR and cell cycle networks
A role for CCT in cell cycle progression, particularly at the G1/S phase, has been suggested (Yokota et al, 1999; Camasses et al, 2003; Grantham et al, 2006; Stirling et al, 2007). Several CCT substrates are cell cycle proteins (e.g. Cdc20p, Cdc55p, and Cdh1p), and the interaction between CCT and Cdc20p and Cdh1p has been well studied (Camasses et al, 2003). Notably, neither Cdc20p nor Cdh1p were identified in our MS analysis and their absence is most likely due to their low synthesis rate, being respectively 100 × and 366 × lower than Act1p (Table I and Arava et al, 2003). On the basis of Arava et al (2003), we estimate our present detection limit at ∼50 × lower synthesis rate than Act1p. Hence, detecting low abundance CCT substrates such as Cdc20p and Cdh1p could well be outside the current range of state-of-the-art MS technologies.
The connection between CCT function and the G1 phase is not only through CCT-interacting cell cycle proteins. Signalling through the TOR kinase also regulates G1 progression and TOR has been functionally connected to CCT (Kabir et al, 2005). We identified interactions between CCT and proteins related to both TOR and the cell cycle (Supplementary Table S4). Particularly enriched are type IIA protein phosphatase components, including the catalytic subunits Pph3p and Sit4p, the regulatory subunits Cdc55p, Sap155p, and Tap42p, as well as the scaffolding subunit Tpd3p (Ho et al, 2002; Kabir et al, 2005; Gavin et al, 2006; Supplementary Table S4). Cdc55p is a known CCT substrate and the other interactions suggest that CCT may regulate phosphatase trimer assembly or the folding of multiple subunits.
Furthermore, SIT4 has been linked genetically to the CCT cofactor PLP2, as they share numerous suppressors (Stirling et al, 2007). The precise role of CCT function in the G1/S phase of the cell cycle progression remains to be elucidated. Nonetheless, a picture emerges in which CCT is tightly coupled to cell cycle control not only through the biogenesis of actin, tubulin, and the APC activators Cdc20p/Cdh1p but also through the septins and probably several other targets (e.g. type II protein phosphatases).
To investigate a possible evolutionary coupling of CCT function to cell cycle control, we compared our data to the minimal eukaryotic genome of the intracellular parasite Encephalitozoon cuniculi. The tiny, compacted genome of E. cuniculi encodes only 1997 proteins (Katinka et al, 2001). Strikingly, we found that, of the ∼20 conserved CCT-interacting proteins, several cell cycle proteins were present (Supplementary Table S6). This supports an evolutionarily ancient coupling of CCT to cell cycle control, with important implications for human cancer studies. It is difficult unravelling the role of CCT in G1 phase in mammalian cells (Grantham et al, 2006) and yeast (Stirling et al, 2007) because of the ‘nonspecific' actin and tubulin phenotypes caused by disrupting CCT function but with new CCT targets in hand, more directed genetic approaches will be possible.
Mining the CCT interaction network reveals putative CCT substrates
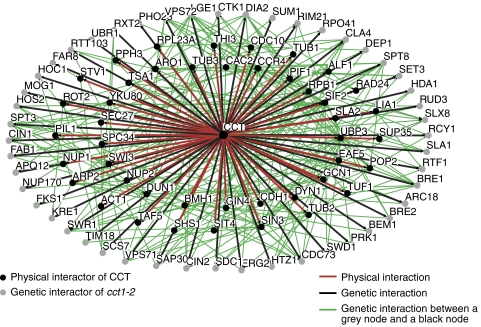
By integrating the CCT physical and genetic interaction networks with one another, we can begin to offer potential explanations for many synthetic sickness interactions we observed in our SGA screen. Figure 2A illustrates how physical interactors of CCT that share a genetic interaction with cct1-2 are more likely to represent substrate proteins than those with no connection to the genetic interaction map. This is based on the assumption that if a CCT-binding protein X is a substrate, it may lose function, by misfolding, when CCT function is compromised. As a result, CCT loss of function phenocopies the loss of individual substrate proteins as evidenced by the actin and tubulin mutant phenotypes in cct-defective cells (Ursic et al, 1994; Grantham et al, 2006). The analysis in Figure 4 (Supplementary Table S7A) expands the functional groupings in Figure 2 by adding another dimension, namely, published genetic interactions between our physical and genetic CCT interaction maps. Assuming that nearly all of the SGA hits arise due to a CCT substrate that misfolds in cct1-2 cells, integrating the SGA data, CCT physical interactions and genetic interactions from the literature identifies CCT-binding proteins that have common genetic interactors with cct1-2 and thus are more likely to be true substrate proteins. A simple example of this type of relationship exists for the known CCT substrate Tub2p—both TUB2 and cct1-2 genetically interact with the tubulin folding cofactor CIN1. Another example of this relationship exists for the CCT-binding protein Nup1p—both NUP1 and cct1-2 genetically interact with NUP170. Figure 4 shows that 44 CCT-binding proteins have reported genetic interactions with at least one of 48 genetic interactors of cct1-2 (see Supplementary Table S7A for a complete list). This group of 44 proteins points to diverse interacting proteins, including known substrates (e.g. actin, tubulins, and Cdh1p), potential substrates, as well as some of the most interesting groups of interactors from our screens and the literature (e.g. septins: Shs1p, Cdc10p, and Gin4p; NPC: Nup1p and Nup2p; TOR signalling: Pph3p, Rot2p, and Sit4p). Also, analogous to Hsp90 (Zhao et al, 2005), the interaction of a protein with CCT and a cochaperone makes it more likely to be a genuine CCT substrate: 36 CCT-interacting proteins interacting with at least one of its putative cochaperones are listed in Supplementary Table S7B. Integrating our data sets in this way is a powerful tool for extracting information about the possible functions of CCT in the cell and will provide a basis for distinguishing CCT-binding proteins from bona fide substrates.
Figure 4.
Integration of the CCT interactome with known interactions reveals putative substrate proteins. Network diagram showing integrated physical and genetic interactors of CCT. Those physical interactors of CCT that exhibit a genetic interaction with at least one genetic interactor of CCT (green lines) are shown. The outer ring (grey nodes) represents SGA hits and the two inner rings (black nodes) represent physical interactors of CCT (see text for description and Supplementary Table S7A).
Conclusion
The chaperonin CCT is essential for the folding of actin and tubulins and has relevance to developmental disease (Bouhouche et al, 2006) and cancer pathways (Camasses et al, 2003; Coghlin et al, 2006). In spite of its importance, most other major molecular chaperone families have been better studied than CCT. Our innovative method for isolating CCT-interacting proteins, together with our integrative, genome-wide analysis of chaperonin function substantially expands the CCT ‘interactome' and points to several previously unknown cellular roles for the chaperonin. In addition to the two classical CCT-interacting cytoskeletal proteins, actin and tubulin, we identified a novel physical connection of CCT to a third cytoskeletal system, the septin ring, which requires CCT activity for its proper assembly and function in vivo. On the whole, our data will help to initiate a multitude of studies aimed at unravelling the role of CCT not only in the cell cycle but also in septin function, nuclear pore function and other cellular processes.
Materials and methods
Purification of CCT
CCT–3/6CBP complexes were purified according to Pappenberger et al (2006) using calmodulin affinity chromatography. CCT was analysed for its binding partner proteins immediately after calcium chelation elution or after further size fractionation on 10–40% sucrose and 15% glycerol gradients. The ATP incubation step was performed on CCT bound to the resin in the presence of nucleotide (5 mM ATP/0.5 mM ADP/15 mM MgCl2) followed by two incubations of five column volumes of wash buffer (20 mM Hepes pH 8.0, 150 mM KCl, 0.2 mM CaCl2, 2 mM DTT, 0.01% LDAO, 20% glycerol) supplemented with 5 mM ATP/0.5 mM ADP/15 mM MgCl2 at 20oC for 20 min each.
Construction of CCT–6CBP strain
The CCT–6CBP strain was constructed using an analogous scheme to the CCT–3CBP tagging method in which tags were inserted into loops predicted to be located on the outside surface of CCT based on crystallographic models (Pappenberger et al, 2006). A 55 amino-acid residue triple tag, containing the Strep-, His8- and calmodulin-binding peptide (CBP) sequences, was inserted into a loop located within residues 368TENTDPKSCTILIK381 at the base of the apical domain of CCT6p. This insertion site in CCT6p at P373∣K374, is exactly equivalent to CCT3-P374∣K375 (Pappenberger et al, 2006).
Mass spectrometry
For peptide mass fingerprinting (PMF)-MS, CCT–6CBP yeast lysate was spun down for 1 h in the presence of protease inhibitors and 1 mM EDTA, then 1.0 ml was added to 100 μl of Calmodulin resin (Stratagene) in the presence of 5 mM CaCl2 and incubated at 4°C overnight. The resin was washed three times with equilibration buffer (2 mM CaCl2) and wash buffer (0.2 mM CaCl2). The protein was eluted in 2 × 100 μl elution buffer containing 10 mM EDTA, and 24 μl was loaded onto a 10% SDS gel, stained with SimplyBlue and bands were cut-out for PMF-MS analysis. In-house analysis was performed on an ABI Voyager STR MALDI-MS as previously described (Passmore et al, 2003). In-house peptide sequencing was performed using a Waters PMF-electrospray MS. Purified CCT samples of wild-type cells and cct4-1 mutant cells were analysed by LC-ESI-MS peformed by Dr Thomas Halder, Toplab, Munich, Germany. Peptide data are available upon request.
Yeast strains and manipulations
Yeast strains used are listed in Supplementary Table S8. Yeast plasmid transformations were performed as described (Amberg et al, 2005). Yeast were grown on rich YPD media or on synthetic complete media lacking uracil to select for plasmids (Adams et al, 1997). For the CCT allelic series, yeast were grown to late log phase before serial dilution and pinning onto YPD plates. The plates were grown at the permissive or non-permissive temperatures for the alleles for 24–72 h.
The cct1-2 strain used for SGA (Supplementary Table S8) was derived from the heat-sensitive tcp1-2 strain described by Ursic et al (1994). The SGA screens were performed in triplicate as described (Tong et al, 2001, 2004). All SGA replica pinning steps were conducted at 22°C with the exception of the final double mutant selection step, which was conducted at 30°C. Genes with the highest statistical probability of representing a true interaction were confirmed by random spore analysis at the cct1-2 permissive temperature of 30°C. Anti-Hsp42p antibodies were a kind gift of Prof J Buchner.
Microscopy
N-Terminal GFP fusion plasmids containing CDC3 and CDC10 were a kind gift of Dr Christopher Beh. Cells bearing one of these plasmids were grown to log phase before shifting to the non-permissive temperature, 15°C for cold-sensitive cells or 37°C for heat-sensitive cells, and grown for 16 or 4 h, respectively, before live cell imaging on a Leica DM 6000 epifluorescence microscope with the appropriate fluorescence filter. Images were analysed in Openlab 5.0.2 (Improvision). Cellular defects were scored manually and the χ2 analysis was used to determine the significance of any differences from wild-type cells under the same conditions.
Interaction analyses
Network diagrams in Figures 2, 3 and 4 were generated using Osprey 1.2.0 (Breitkreutz et al, 2003a). Interactions between CCT-interacting nodes were overlayed automatically by Osprey according to the GRID database (Breitkreutz et al, 2003b).
Supplementary Material
Supplementary Tables
Supplementary Data
Acknowledgments
KRW was funded by Cancer Research UK and HFSP. MRL acknowledges funding from the Canadian Institutes of Health Research (CIHR; grant BMA121093), and scholar awards from CIHR and Michael Smith Foundation for Health Research (MSFHR). PCS was supported by scholarships from NSERC and MSFHR. CB was funded by grants from the CIHR, Genome Canada, and Genome Ontario.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T (1997) Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbour, New York, USA: Cold Spring Harbour Laboratory Press [Google Scholar]
- Aloy P, Böttcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB (2004) Structure-based assembly of protein complexes in yeast. Science 303: 2026–2029 [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN (2005) Methods in Yeast Genetics. Cold Spring Harbour, New York, USA: Cold Spring Harbour Laboratory Press [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhouche A, Benomar A, Bouslam N, Chkili T, Yahyaoui M (2006) Mutation in the epsilon subunit of the cytosolic chaperonin-containing t-complex peptide-1 (Cct5) gene causes autosomal recessive mutilating sensory neuropathy with spastic paraplegia. J Med Genet 43: 441–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M (2003a) Osprey: a network visualization system. Genome Biol 4: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M (2003b) The GRID: the general repository for interaction datasets. Genome Biol 4: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W (2003) The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell 12: 87–100 [DOI] [PubMed] [Google Scholar]
- Coghlin C, Carpenter B, Dundas SR, Lawrie LC, Telfer C, Murray GI (2006) Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol 210: 351–357 [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Cowan NJ, Lewis SA (2002) Type II chaperonins, prefoldin and the tubulin-specific chaperones. Adv Prot Chem 59: 73–104 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier M-A, Hoffman V, Hoefert C, Klein K, Hudak M, Michon A-M, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E (1998) A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17: 952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham J, Brackley KI, Willison KR (2006) Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res 312: 2309–2324 [DOI] [PubMed] [Google Scholar]
- Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, Wold BJ, Deshaies RJ (2004) Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol Cell Proteomics 3: 226–237 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA (2002) Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev 16: 3130–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ (1999) Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol 145: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J (2004) Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Kabir MA, Kaminska J, Segel GB, Bethlendy G, Lin P, Della Seta F, Blegen C, Swiderek KM, Zoladek T, Arndt KT, Sherman F (2005) Physiological effects of unassembled chaperonin Cct subunits in the yeast Saccharomyces cerevisiae. Yeast 22: 219–239 [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivarès CP (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kubota S, Kubota H, Nagata K (2006) Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc Natl Acad Sci USA 103: 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicki A, Rowicka M, Otwinowski Z (2007) SCEPTRANS: an online tool for analyzing periodic transcription in yeast. Bioinformatics 23: 1559–1561 [DOI] [PubMed] [Google Scholar]
- Lewis VA, Hynes GM, Zheng D, Saibil H, Willison K (1992) T-Complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature 358: 249–252 [DOI] [PubMed] [Google Scholar]
- Lukov GL, Baker CM, Ludtke PJ, Hu T, Carter MD, Hackett RA, Thulin CD, Willardson BM (2006) Mechanism of assembly of G protein betagamma subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J Biol Chem 281: 22261–22274 [DOI] [PubMed] [Google Scholar]
- McLaughlin JN, Thulin CD, Hart SJ, Resing KA, Ahn NG, Willardson BM (2002) Regulatory interaction of phosducin-like protein with the cytosolic chaperonin complex. Proc Natl Acad Sci USA 99: 7962–7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Cowan NJ (1994) Facilitated folding of actins and tubulins occurs via a nucleotide-dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol Cell Biol 14: 2895–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenberger G, McCormack EA, Willison KR (2006) Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/γ subunit. J Mol Biol 360: 484–496 [DOI] [PubMed] [Google Scholar]
- Passmore L, McCormack EA, Au SWN, Paul A, Willison KR, Harper JW, Barford D (2003) Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J 22: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15: 2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Schaft D, Roguev A, Pijnappel WW, Stewart AF, Shevchenko A (2002) Deciphering protein complexes and protein interaction networks by tandem affinity purification and mass spectrometry: analytical perspective. Mol Cell Proteomics 1: 204–212 [DOI] [PubMed] [Google Scholar]
- Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A (2008) ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J Mol Biol 377: 469–477 [DOI] [PubMed] [Google Scholar]
- Souès S, Kann ML, Fouquet JP, Melki R (2003) The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germline and somatic cells. Exp Cell Res 288: 363–373 [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J (2004) Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol 14: 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison KR, Yaffe MB (1993) The t-complex polypeptide 1 complex is a chaperonin for tubulin and acton in vivo. Proc Natl Acad Sci USA 90: 9422–9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Cuellar J, Alfaro GA, El Khadali F, Beh CT, Valpuesta JM, Melki R, Leroux MR (2006) PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J Biol Chem 281: 7012–7021 [DOI] [PubMed] [Google Scholar]
- Stirling PC, Srayko M, Takhar KS, Pozniakovsky A, Hyman AA, Leroux MR (2007) Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol Biol Cell 18: 2336–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J 18: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Evangelista M, Parsons AB, Xu VH, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L et al. (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL (2005) Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 310: 1152–1158 [DOI] [PubMed] [Google Scholar]
- Ursic D, Sedbrook JC, Himmel KL, Culbertson MR (1994) The essential yeast Tcp1 protein affects actin and microtubules. Mol Biol Cell 5: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93: 863–873 [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Carrascosa JL, Willison KR (2005) Structure and function of the cytosolic chaperonin CCT. In Protein Folding Handbook, Part II, Buchner J, Kiefhaber T (eds), pp 725–755. Weinheim: Wiley-VCH [Google Scholar]
- Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR (2002) Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett 529: 11–16 [DOI] [PubMed] [Google Scholar]
- Versele M, Thorner J (2005) Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol 15: 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh DB, Drubin DG (1994) A yeast TCP-1-like protein is required for actin function in vivo. Proc Natl Acad Sci USA 91: 9116–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willison KR, Grantham J (2001) The roles of the cytosolic chaperonin, CCT, in normal eukaryotic cell growth. In Molecular Chaperones in the Cell, Lund P (ed), pp 90–118. Oxford: Oxford University Press [Google Scholar]
- Yokota S, Yanagi H, Yura T, Kubota H (1999) Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem 274: 37070–37078 [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 120: 715–727 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Data