Abstract
Indoleamine 2,3-dioxygenase (IDO), a tryptophan catabolizing enzyme, has been implicated in the pathogenesis of various neurological disorders. IDO expression is induced by IFN-γ and leads to neurotoxicity by generating quinolinic acid. Additionally, it inhibits the immune response through both tryptophan depletion and generating other tryptophan catabolites. IL-4 and IL-13 have been shown to control IDO expression by antagonizing the effects of IFN-γ in different cell types. Here, we investigated the effects of these cytokines on IDO expression in microglia. Interestingly, we observed that both IL-4 and IL-13 greatly enhanced IFN-γ induced IDO expression. However, tryptophanyl-tRNA synthetase (WRS), which is coinduced with IDO by IFN-γ, is downregulated by IL-4 and IL-13. The effect of IL-4 and IL-13 was independent of STAT-6. Modulation of IDO but not WRS was eliminated by inhibition of protein phosphatase 2A (PP2A) activity. The phosphatidylinositol 3-kinase (PI3K) pathway further differentiated the regulation of these two enzymes, as inhibiting the PI3K pathway eliminated IFN-γ induction of IDO, whereas such inhibition greatly enhanced WRS expression. These findings show discordance between modulations of expression of two distinct enzymes utilizing tryptophan as a common substrate, and raise the possibility of their involvement in regulating immune responses in various neurological disorders.
Keywords: Indoleamine 2,3-dioxygenase; Tryptophanyl-tRNA synthetase; Cytokine
INTRODUCTION
Indoleamine 2,3-dioxygenase (IDO) is the first and rate-limiting enzyme in the generation of quinolinic acid from tryptophan via the kynurenine pathway outside of the liver. IDO serves immunoregulatory as well as tolerogenic functions (Frumento et al. 2002; Munn et al. 1999; Munn et al. 2004; Terness et al. 2002). Activation of IDO leads to defects in various immunoregulatory mechanisms, including the failure of immune surveillance of tumor cells (Friberg et al. 2002; Munn et al. 2004); development of autoimmune conditions such as multiple sclerosis and autoimmune diabetes (Grohmann et al. 2003; Sakurai et al. 2002); and allogenic fetal rejection (Munn et al. 1998). By depleting the essential amino acid tryptophan and generating tryptophan catabolites, IDO regulates immune responses by inhibiting T-cell functions (Frumento et al. 2002; Meisel et al. 2004; Munn et al. 2004; Terness et al. 2002).
In addition to its effects on the immune system, tryptophan catabolism also affects the central nervous system (CNS). In the brain, IDO is primarily produced by activated macrophages and microglia. As tryptophan is the precursor for the neurotransmitter serotonin, its IDO-induced depletion can cause untoward psychological/psychiatric consequences leading to depression (Widner et al. 2000). During HIV infection, which also has CNS consequences due to the presence of the virus-host interaction in the CNS, the IDO-mediated increased accumulation of tryptophan catabolites such as quinolinic acid (QUIN), has been proposed to have a pathogenic role in inducing neuronal death (Bara et al. 2000; Burudi et al. 2002; Grant et al. 2000; Heyes et al. 1989; Heyes et al. 1992; Stone 2001). In addition to IDO’s potential role in HIV-associated dementia, in Alzheimer’s disease (AD) it has been shown that the inflammatory cytokines and the amyloid β protein Aβ 1-42 induce IDO expression and production of QUIN in neurotoxic concentrations by human macrophages and microglia, leading to neuronal death (Guillemin and Brew 2002; Guillemin et al. 2003).
Tryptophan catabolism and depletion can be quite detrimental to the host cells. Although there are no alternative mechanisms which can rescue cells from QUIN induced cell death, tryptophan starvation can be prevented by another enzyme, tryptophanyl-tRNA synthetase (WRS), which is coinduced with IDO (Boasso et al. 2005) by host cells as a self-protection mechanism. WRS catalyzes the attachment of tryptophan to its cognate transfer RNA molecule, with the resulting Trp-tRNA complex used for protein synthesis (Fleckner et al. 1995;Rubin et al. 1991). Thus, it provides a reservoir of tryptophan directly available for protein synthesis and protects the cells from IDO induced tryptophan depletion by allowing them to use limiting amounts of tryptophan during protein synthesis (Boasso et al. 2005;Murray 2003). Parallel induction of IDO and WRS by IFN-γ has been found in a number of cell types (Boasso et al. 2005); Fleckner et al. 1995; Rubin et al. 1991,Yoshida et al. 1981). On the other hand, anti-inflammatory cytokines can antagonize many cellular responses induced by IFN-γ, including IDO induction (Chaves et al. 2001; Doherty et al. 1993; Musso et al. 1994; Oswald et al. 1992; Yuan et al. 1998), but the effects on WRS are not as well studied.
The understanding of the dynamic interplay between factors controlling IDO and WRS in the CNS is critical. However, the data are scarce as to the mechanism behind the regulation of IDO and WRS in the brain. The present study addresses this issue by evaluating the ability of a stimulatory cytokine (IFN-γ) and anti-inflammatory cytokines (IL-4 and IL-13, related cytokines that share a common receptor component (Kelly-Welch et al. 2003)) to regulate IDO and WRS expression in mouse primary microglia. The regulation of expression as well as signaling mechanisms diverged for IDO and WRS, which can affect the host response to disease.
MATERIALS AND METHODS
Animals
STAT-1 knock out (S1KO) mice were developed by Dr. Robert Schreiber and provided with his permission by Dr. Iain Campbell. The S1KO mice are on a mixed C57BL/6 × 129 background, and F2 wild-type mice between these strains were used for controls to ensure lack of background effects. The STAT-6 knock out (S6KO) mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The S6KO mice are on the BALB/c background, and wild-type BALB/c mice were used for their controls, as well as for all other experiments. Mice were bred and maintained in The Scripps Research Institute’s animal facility. All experiments were conducted under NIH and the Institutional Animal Care and Use Committee guidelines for animal care and use.
Cell Culture and Reagents
Primary mouse microglia cultures were developed from the brains of newborn to 2-day old pups. Brains were carefully freed from meninges followed by trituration in DMEM containing trypsin and EDTA. Following a 20 min incubation at room temperature, trypsinization was stopped by dilution with 5 times the volume of DMEM containing 10% FBS. The cells were centrifuged, resuspended in HEPES buffered DMEM containing 10% FBS, penicillin-streptomycin-fungizone (complete or cDMEM) and plated in 75 cm2 flask. The following day, media was replaced with cDMEM containing 25 ng/ml recombinant mouse GM-CSF (Peprotech, Rocky Hill, NJ) and incubated for 10 days. Cells were then detached by mild scraping and microglial cells were isolated using CD11b MAC column separation as per the manufacturer’s protocol (Miltenyi Biotec, Auburn, CA).
The mouse microglial cell line EOC13.31 (CRL-2468) was purchased from the ATCC (Manassas, VA) and was cultured, as recommended by the supplier, in cDMEM containing 20% conditioned media from the LADMAC (CRL-2420, also obtained from the ATCC) cell line.
Before cytokine treatments, cells (105 cells/ml) were incubated in DMEM containing 0.1% FBS for 48 hrs. The cytokines were purchased from Peprotech (Rocky Hill, NJ) and added simultaneously to the culture. The optimal doses of cytokines and the optimal time period for maximal IDO mRNA expression were determined by calibration curves for both dose and time period. A stable plateau of expression was found between 16 and 30 hours, and a 24-hour stimulation period was chosen for assessing levels of mRNA expression. Similarly a plateau was found for IFN-γ treatment at concentrations > 6.25 ng/ml; and at concentrations >10 ng/ml for IL-4/IL-13 co-treatment, thus 10 ng/ml was chosen for IFN-γ and 24 ng/ml for IL-4/IL-13 to assess levels of mRNA expression. Neutralizing anti-mouse IL-4 antisera was obtained from R&D Systems (Minneapolis, MN). LY294002, wortmannin (Calbiochem-Novabiochem, La Jolla, CA) and okadaic acid (A. G. Scientific, Inc, San Diego, CA), were dissolved in dimethyl sulfoxide and used as described. All experiments were performed in triplicate.
FACS and Immunofluorescence
For FACS, microglial cells were suspended at 106 cells/ml in FACS staining buffer (PBS containing 2% FBS and 0.2% sodium azide), and stained with FITC-labeled anti CD11b (BD Bioscience, San Jose, CA) for 20 min in dark at 4° C. Suitable isotype controls were used for calibration. Cells were washed and resuspended in staining buffer and processed through a FACScan flow cytometer before analysis. For immunofluorescence, 105cells were plated on coverslips in a 24 well plate for 48 hrs. Cells were fixed for 25 min with ice-cold 4% paraformaldehyde at 4° C, washed with PBS, and permeabilized by adding 0.2% Tween 20 in PBS. Nonspecific binding sites were blocked by incubation for 1 h with 10% goat serum in PBS containing 0.2% Tween 20. To specifically stain microglia, neurons and astrocytes in primary cell cultures, cells were then incubated for 1 h at room temperature with dilutions of anti-Iba-1 (Wako, Osaka, Japan) at 1:1000, anti-microtubule associated protein-2 (MAP-2; Sigma-Aldrich, St. Louis, MO) at 1:500 or anti-glial fibrillary acidic protein (GFAP; Sigma-Aldrich) at 1:500 respectively. After three washes, the cells were treated with secondary polyclonal antibodies, AlexaFluor 488-conjugated goat anti rabbit (Invitrogen, Carlsbad, CA) at 1:500 or Rhodamine Red-X conjugated goat anti-mouse IgG (Invitrogen) at 1:500 for 1 h at room temperature. For IDO protein expression, EOC13.31 cells were incubated overnight with anti-IDO antibody (Alexis Biochemicals, San Diego, CA) (1:500) at 4° C, washed and incubated with secondary Rhodamine Red-X conjugated goat anti-mouse IgG (Invitrogen) at 1:500 dilution. The cells were then counterstained with fluorescent nuclear stain 4′, 6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). The coverslips containing cells were air-dried, and mounted on glass microscope slides in Vectashield mounting media (Vector Laboratories, Burlingame, CA). Images from coverslips were visualized and acquired using a Zeiss Axiovert 200 inverted microscope at 5× magnification.
Measurement of IDO activity
To monitor IDO enzyme activity, cell supernatants were assayed spectrophotometrically for the presence of kynurenine (Braun et al. 2005), the first stable catabolite downstream of IDO. Briefly, EOC13.31 cells were incubated without cytokines, or with 100 ng/ml of IFN-γ in the presence or absence of 100 ng/ml of IL-4 for 96 h at 37° C. 160 μl of the supernatants were incubated with 10 μl 30% TCA at 50° C for 30 min, vortexed and centrifuged at 10,000 rpm for 5 min. 100 μl of the supernatants were then added to an equal volume of Ehrlich"s reagent (4% p-dimethyl benzaldehyde in glacial acetic acid) in a 96 well microtiter plate. After 10 minutes, the optical density was measured at 492 nm using a Victor3 plate reader (Perkin-Elmer, Waltham, MA). The concentration of kynurenine in the unknowns was calculated by plotting a standard curve of defined kynurenine concentrations (0-1000 μM).
Western Blot Analysis
Following cytokine treatments the cells were washed 3 times in ice-cold PBS, scraped and lysed in ice-cold RIPA lysis buffer (50mM HEPES buffer, sodium orthovandate, 10 mM sodium pyrophosphate, 100 mM NaF, 1% Triton X-100, 30 mM p-nitrophenyl phosphate, 1 mM PMSF, 1X complete protease inhibitor cocktail (Roche Diagnostics, Indianapolis, USA). After incubation for 30min on ice, samples were centrifuged at high speed for 15min and supernatants collected. Total protein content was estimated using the BioRad protein assay. An aliquot corresponding to 25 μg of total protein of each sample was separated by SDS-PAGE and transferred electrophoretically to HY-bond ™ PVDF membranes (Invitrogen). Non-specific antibody binding was blocked by either 5% BSA or non-fat dried milk for 1h at room temperature. Immunoblotting was carried out with antibodies against total STAT-1 (BD Bioscience), pSTAT-1 Tyr701 (BD Bioscience) and pSTAT-1 Ser727 (Biosource International inc., Camarillo, CA) and anti-actin (Sigma) as a loading control followed by secondary antibody (1:10000 HRP conjugated anti rabbit or anti mouse IgG, Invitrogen). Blots were developed with 1:1 solution of Super Signal west Pico Chemiluminescent Substrate and Luminol/Enhancer (Pierce, Rockford, IL).
RNA Isolation and Reverse Transcription (RT)
Total RNA was extracted from the cell pellets. 2 μg of RNA was used for reverse transcription. For a 20 μl reaction, reverse transcription was carried out using the superscript kit (Invitrogen) for 1 h at 42° C followed by 70° C for 5min to inactivate the RT reagents. RNase H (New England Biolabs, Beverly, MA) treatment was given at 37° C for 20 min. The RT product was then diluted with an equal volume of RNAse and DNAse free water.
Quantitative Real Time PCR
Specific RNA transcripts (mRNA) were quantified by real time PCR using dual-labeled hydrolysis probes (FAM-TAMRA). The primers and probe sequences were designed for mouse sequences using the Genescript online tool (https://www.genescript.com/ssi-bin/app/primer) and obtained from Eurogentec (San Diego, CA). The following primers and probes were used: IDO-F (gagaaagccaaggaaatttttaagag, 0.6 μM), R (gatatatgcggagaacgtggaaa, 0.6 μM), Probe (tgcgtgactttgtggacccagacac, 0.4 μM); WRS-F (tccgagacaggacagatatcca, 0.6 μM), R (cacgtcccttgtcattctgaag, 0.6 μM), Probe (cctcatcccgtgtgccattgagcc, 0.6 μM) and for 18S- F (cggctaccacatccaaggaa, 0.6 μM), R (gctggaattaccgcggct, 0.6 μM), probe (tgctggcaccagacttgccctc, 0.2 μM). To carry out quantitative real time PCR, 2 μl of the (1:10) diluted cDNA was used for assaying the amount of 18S endogenous ribosomal RNA, and 5 μl was used for quantification of IDO as well as WRS mRNA in duplicates. The reaction utilized 12.5 μl of platinum qPCR UDG Supermix (Invitrogen) yielding 0.75 U Taq DNA polymerase, 20 mM Tris-HCl, 50 mM KCl, 3 mM MgCl2, and 200 μM of deoxynucleoside triphosphate. The reaction mixture was brought to a final concentration of 5mM MgCl2.Real time reaction was performed in 96 well plate on a Stratagene MX3000p real time machine (Stratagene, La Jolla, CA). Each assay was optimized by titrating a range of primer and probe concentrations and determining their cycle threshold (Ct) values. The primer and probe combinations that gave the lowest Ct and best amplification plots were used for the final analysis. The reaction was run at an initial temperature of 95° C for 10 min and then at 95° C for 30 sec, 55° C for 1 min followed by 72° C for 30 sec for 45 cycles. The optical signal was recorded at the end of every 72° C extension step. Ct values were determined by the software according to the optimization of the baseline. For computing the relative amounts of IDO and WRS in the samples, the average Ct of the primary signal for 18S was subtracted from that of IDO and WRS to give changes in Cts (dCt, a log2 scale). A baseline dCt of 25 was subtracted and the results multiplied by -1. In this manner, the degree of change in gene expression was determined. Relative units (2dCt) were calculated and used here as a measure of IDO and WRS mRNA expression. The normalization of IDO and WRS with 18S controls for variation in the efficiency of RNA isolation, possible differences in amounts of starting RNA and RT efficiency.
Statistical Analysis
Sgnificance testing was done for treatments with at least 5 biological replicates. Otherwise descriptive statistics were generated. We tested this result across treatments (IFN-γ vs. IFN-γ plus IL-4/IL-13) using one tailed paired t tests, and compared each treatment by marker (IDO vs. WRS) using one tailed unpaired t tests assuming equal variance where the difference between means was interpreted as significant when p < 0.05. Equality of variance was tested using an F test. All parametric tests were done on the log10 transformed values.
RESULTS
IL-4 increases IFN-γ induced IDO expression in microglia
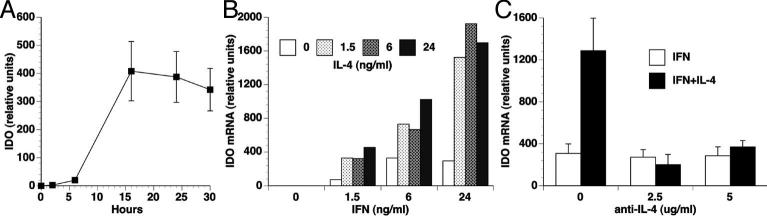
Using the EOC13.31 mouse microglia cell line, we first explored IFN-γ induction of IDO expression in microglia. As expected from other studies on myeloid cells, IFN-γ induced the expression of IDO mRNA (Figure 1A). In the presence of IFN-γ, TGF-β decreased and TNF-α increased IDO expression (data not shown), consistent with others' results (Kwidzinski et al. 2005; MacKenzie et al. 1999; Robinson et al. 2003; Yuan et al. 1998). However IL-4, which has been reported to inhibit IDO expression in a number of cell types, increased IFN-γ induced IDO expression (Figure 1B). This effect could be completely abrogated by the addition of an antiserum against IL-4 (Figure 1C).
Figure 1. IL-4 increases IFN-γ -induced IDO mRNA expression.
(A) EOC13.31 microglia cells were treated with 10 ng/ml IFN-γ for the indicated time periods, and total RNA was analyzed for IDO expression by real time RT-PCR. (B) EOC13.31 microglia cells were treated with the indicated concentrations of IFN-γ and IL-4 for 24 hours, and total RNA was analyzed for IDO expression by real time RT-PCR. (C) EOC13.31 microglia cells were treated with 10 ng/ml of IFN-γ with and without 25 ng/ml of IL-4 for 24 h, in the presence of the indicated concentration of anti-IL-4 antisera. Total RNA was analyzed for IDO expression by real time RT-PCR.
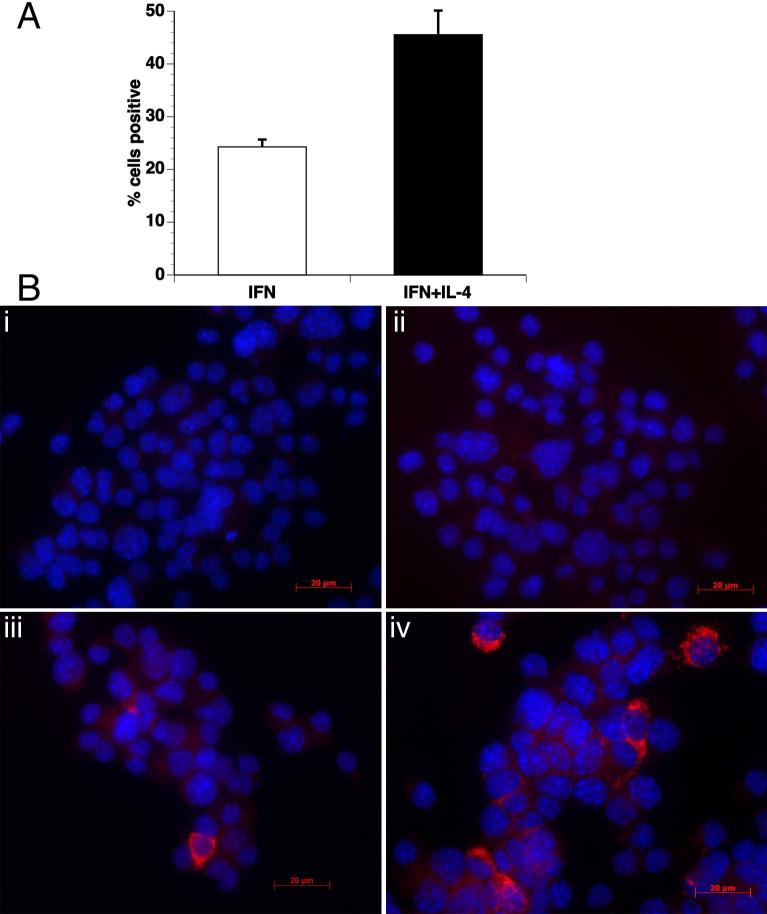
The effect of IL-4 in potentiation of IFN-γ induced IDO expression was also observed at protein levels by immunofluorescence studies. Compared to cells stimulated with IFN-γ alone, a 1.9-fold increase was observed in the number of cells with detectable IDO protein expression when IL-4 was added in the culture along with IFN-γ (Figure 2). Furthermore, IL-4 also enhanced the functional activity of IDO to convert tryptophan into kynurenine. When IL-4 was added to the culture along with IFN-γ an average of 3.2-fold (p<0.001) increase in kynurenine concentration was observed as compared to when the cells were treated with IFN-γ alone. This shows that IL-4 is an enhancer of IFN-γ induced expression of IDO at mRNA, protein as well as at the functional activity level.
Figure 2. IL-4 increases IFN-γ-induced IDO protein expression.
Cellular expression of the IDO protein was examined by immunofluorescence using anti-IDO antibody in EOC13.31 cells. Cells were either untreated, or treated with IFN-γ, IL-4 or IFN-γ+IL-4. (A) Increased numbers of cells had detectable IDO when treated with IFN-γ + IL-4 as compared to IFN-γ alone. Mean and SEM of 5 different fields, data are representative of two independent experiments. (B) Untreated (i) and cells treated with IL-4 alone (ii) did not show any positive staining for IDO. IDO was present in cells treated with IFN-γ alone (iii), and increased in cells treated with IFN-γ+IL-4 (iv).
Primary microglia
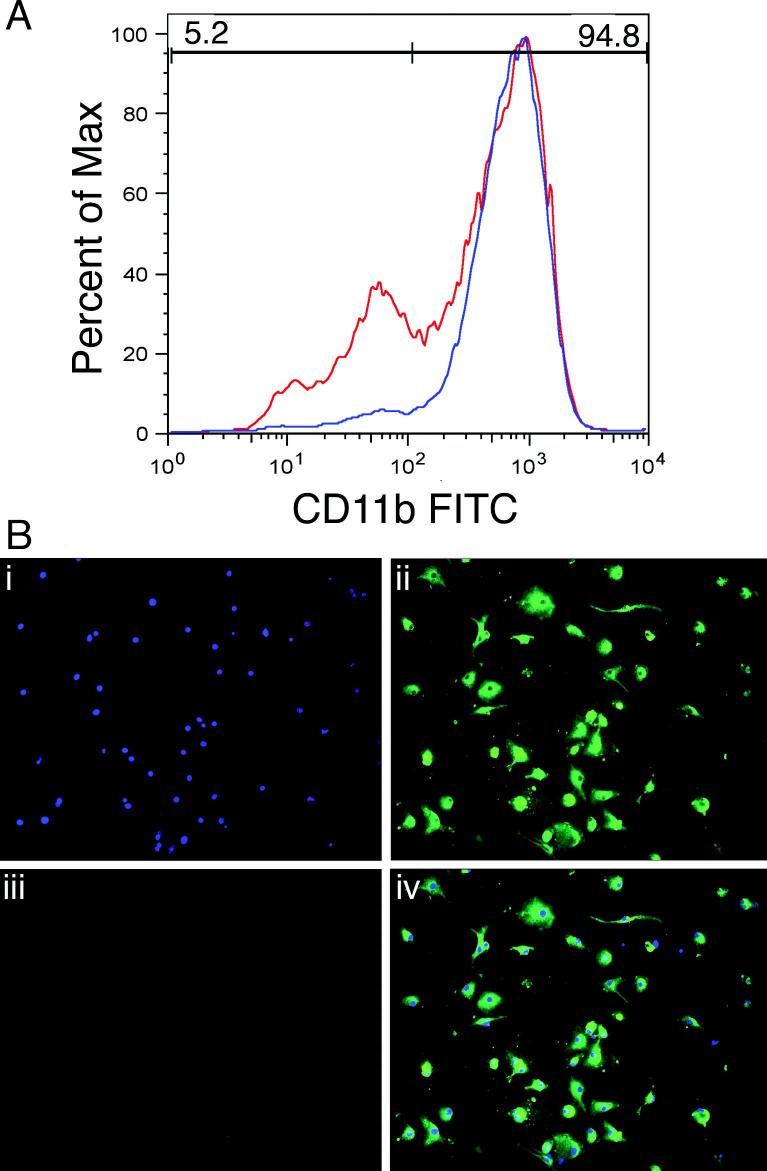
To further assess the mechanisms by which IDO is regulated in microglia, we performed studies on primary mouse microglia. Our method of isolation of microglia from neonatal mouse brains, culture, and subsequent immunomagnetic purification led to higher purity (as indicated by FACS staining) than other methods of isolation that were tried (Sedgwick et al. 1991). The purity of microglial cells using this method was estimated to be approximately 95% based on detection with FACS staining using anti CD11b antibody (Figure 3A). Immunostaining of cultures (Figure 3B) using antibodies against microglia (anti-Iba-1), astrocytes (anti-GFAP), and neurons (anti-MAP-2, not shown) revealed that contaminating cells were primarily astrocytes.
Figure 3. Isolation and purity of primary microglia.
Microglia were isolated from a day-10 mixed culture (obtained from brains of 1-2 days old pups) by magnetic column separation using CD11b beads. The proportion of microglial cells was routinely ∼95% based on detection with FACS staining using anti CD11b antibody. Percentage of cells stained with CD11b before column separation shown in red grey and after column separation shown in blue black (A). Purity was also checked by immunocytochemistry (B) using DAPI nuclear counter stain (i), anti-Iba-1 antibody for microglia (ii) and anti-GFAP antibody for astrocytes (iii). Juxtaposed image of cells stained with DAPI, Iba-1 and GFAP (iv).
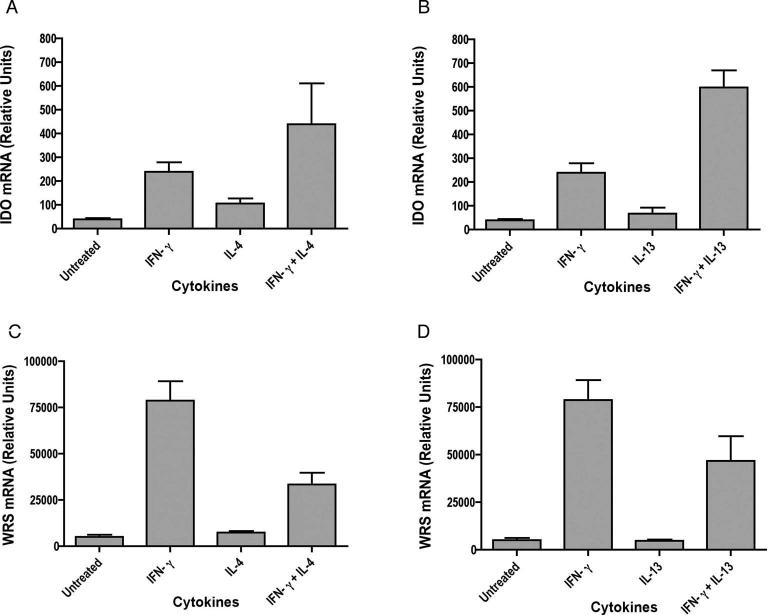
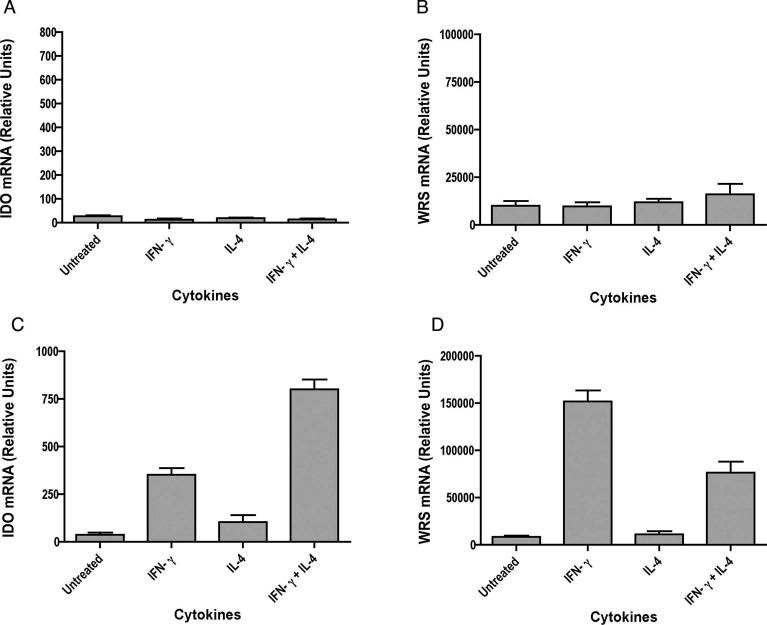
IL-4 and IL-13 enhance IFN-γ induced IDO expression, but reduce WRS expression
Since IL-13 shares many properties with IL-4, we examined the effects of both IL-4 and IL-13 in regulating IFN-γ induced IDO expression in primary microglia. Additionally, since WRS has been reported to be co-induced with IDO by IFN-γ, its expression was studied in parallel. Cultures of primary microglia were treated with IFN-γ, IL-4 or IL-13, and IFN-γ with or without IL-4 or IL-13. Total RNA was isolated and analyzed for expression of IDO and WRS by quantitative real time PCR. Neither IL-4 nor IL-13 alone could induce IDO or WRS, as mRNA levels were similar to those in untreated cells. Treatment with IFN-γ alone greatly enhanced the expression of both IDO and WRS as compared to untreated cells. Treatment of IFN-γ stimulated cells with IL-4 or IL-13 indeed consistently enhanced the expression of IFN-γ induced IDO by an average of 2.0-fold (n =6, p = 0.0068) with IL-4 and 3.2-fold with IL-13 (n =7, p = 0.0148) (Figure 4A, B). In contrast, an average of 3.0-fold decrease was observed in WRS expression when cells were treated with IFN-γ plus IL-4 (n =6, p = 0.0017) and 2.7-fold with IFN-γ plus IL-13 (n =7, p = 0.0004) as compared to treatment with IFN-γ alone (Figure 4 C, D). This result reveals a differential modulation of IFN-γ-induced expression of IDO and WRS genes by IL-4/IL-13 in microglia.
Figure 4. IL-4 and IL-13 enhance IFN-γ -induced IDO expression but reduce WRS expression in mouse microglia.
Microglia were stimulated with cytokines for 24 hrs and total RNA was analyzed for IDO (A, B) and WRS (C, D) expression by real time RT-PCR. Data are representative of six independent experiments with IL-4 and seven independent experiments with IL-13 done in triplicate. IDO expression in IFN-γ stimulated cells was significantly increased by treatment with IL-4 or IL-13, whereas IFN-γ stimulated WRS expression significantly reduced by IL-4 or IL-13.
IFN-γ utilizes STAT-1, but IL-4 and IL-13 act independently of STAT6, in modulating IDO and WRS expression
IFN-γ can exerts its effects on cells by binding to its receptor and activating the JAK / STAT-1 pathway for transcription of IFN-γ responsive genes (Bach et al. 1997). We sought to determine whether IFN-γ mediated induction of IDO and WRS and their modulation by IL-4 or IL-13 is STAT-1 dependent. Microglia from STAT-1 knock out (S1KO) mice were stimulated with IFN-γ with or without IL-4 or IL-13. In the absence of STAT-1, treatment with either IFN-γ alone or in combination with IL-4 or IL-13 could not alter the expression of IDO and WRS (Figure 5A, B). This indicates that in microglia STAT-1 is critical for IFN-γ modulation of IDO and WRS expression.
STAT-6 plays a central role in cellular functions mediated by IL-4 and IL-13. In order to assess whether STAT-6 is involved in IL-4/IL-13 mediated modulation of IDO and WRS expression, we evaluated IDO and WRS expression in the presence of IL-4 or IL-13 in primary microglia isolated from STAT-6 knock out (S6KO) mice. The results observed in S6KO mice were similar to that observed in wild type (WT) mice. In the absence of STAT-6, IL-4 still consistently enhanced IFN-γ induced IDO expression by an average of 2.7-fold (n = 6, p = 0.0004) (Figure 5C) and reduced WRS expression by 3.1-fold (n = 6, p = 0.0073) (Figure 5 D). Similarly, IL-13 enhanced IFN-γ induced IDO expression by an average of 3.7-fold (n = 5, p = 0.0184) and reduced WRS expression by 3.7-fold (n = 5, p = 0.0239). Thus, the IL-4 and IL-13 effects on IDO and WRS expression observed here are independent of STAT-6 activation.
Figure 5. IFN-γ utilizes STAT-1 and IL-4 act independent of STAT-6 in modulating IDO and WRS expression.
Microglia from S1KO (A, B) and S6KO (C, D) mice were stimulated with cytokines for 24 hrs and total RNA was analyzed for IDO and WRS expression by real time RT-PCR. Results are representative of three independent experiments for S1KO and six independent experiments for S6KO done in triplicate. In S6KO mice, IL-4 still significantly increased IDO and decreases WRS in IFN-γ stimulated cells.
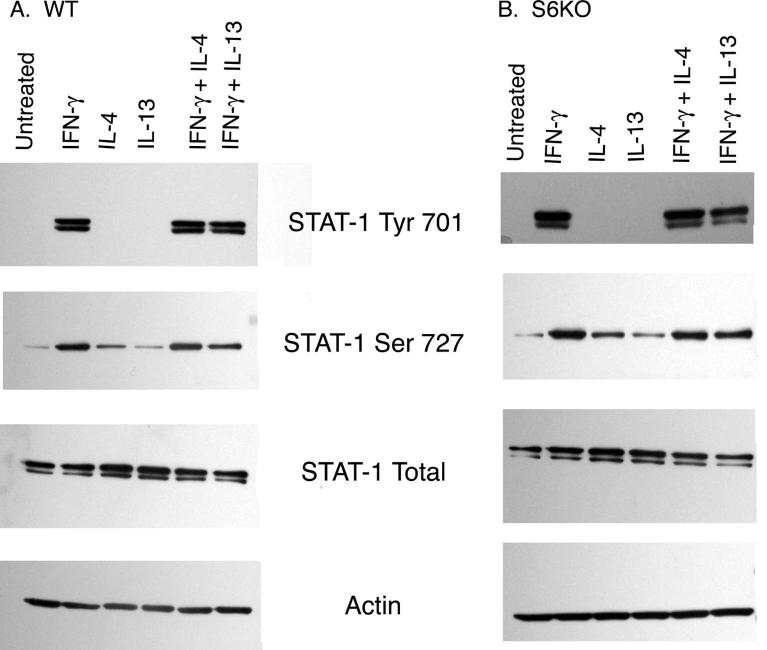
IL-4 and IL-13 do not act by enhancing STAT-1 phosphorylation
Binding of IFN-γ to its receptor activates JAK-1 and JAK-2 tyrosine kinases, which then phosphorylate tyrosine 701 on STAT-1. Consequently, STAT-1 homodimers are formed and bind to IFN-γ -inducible promoters (Darnell et al. 1994). Additionally, STAT-1 phosphorylation on Serine 727 also occurs and leads to a strong enhancement of the STAT-1 induced transcription of IFN-γ responsive genes (Nguyen et al. 2001; Wen et al. 1995). We therefore examined whether IL-4 or IL-13 converge with IFN-γ at the point of enhancing STAT-1 phosphorylation, thus augmenting IDO expression. The degree of phosphorylation of both Tyr 701 and Ser 727 on STAT-1 was assessed by Western blotting in the presence of IFN-γ with or without IL-4 or IL-13 using STAT-1 phosphorylation-site specific antibodies in microglia protein extracts from WT (Figure 6A) as well as S6KO mice (Figure 6B). No difference in either Tyr 701 or Ser 727 phosphorylation of STAT-1 was observed when cells were treated with IFN-γ with IL-4 or IL-13 as compared to IFN-γ alone. Therefore IL-4 and IL-13 aid IFN-γ in upregulating IDO expression by utilizing a mechanism other than enhancing STAT-1 phosphorylation.
Figure 6. IL-4 and IL-13 do not act by altering phosphorylation status of STAT-1.
Microglia from WT (A) and S6KO (B) mice were stimulated with cytokines for 30 min, lysed, and subjected to Western blot analysis using anti total STAT-1, anti pSTAT-1 Tyr701, anti pSTAT-1 Ser727 and anti-actin (as a protein loading control). Blots shown are the representative of one of the three independent experiments.
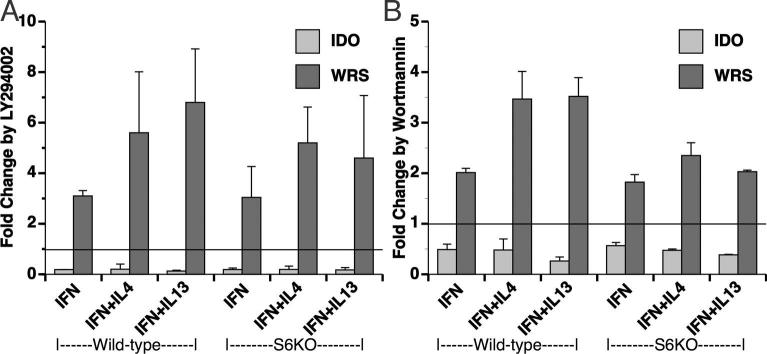
Phosphatidylinositol 3 kinase (PI3K) signaling pathway differentially regulates IDO and WRS
The PI3K system has been implicated in the activating effects of IFN-γ (Hwang et al. 2004) and may also be involved in the phosphorylation of STAT-1 Ser 727 (Nguyen et al. 2001). Interestingly, IL-4 and IL-13 can also activate the insulin receptor substrate family of proteins, leading to activation of the PI3K (Kelly-Welch et al. 2003). In order to assess the role of PI3K in effects of IL-4/IL-13 on IFN-γ induced IDO and WRS expression, microglia were treated with the PI3K inhibitor LY294002 (LY) prior to cytokine treatments. IFN-γ induced IDO expression was decreased by LY an average of 5.6-fold, and the low level of expression detected in the presence of LY in IFN-γ stimulated cells was not altered by the presence or absence of IL-4, IL- 13, or STAT-6 (Figure 7A). In contrast, WRS expression was enhanced by an average of 4.7- fold, and again this enhancement was not significantly altered in the presence or absence of IL-4, IL-13, or STAT-6. To confirm the role of PI3K in these divergent effects, we further used wortmannin, another inhibitor of PI3K. Wortmannin indeed consistently reduced the expression of IFN-γ dependent IDO an average of 2.3-fold, and enhanced WRS expression an average of 2.5-fold (Figure 7B), and similar to the results with LY, no additional effects were found with IL-4 or IL-13, or in STAT-6 knockouts. We note that the magnitude of the effects of wortmannin on IDO and WRS were of lower levels than those found with LY, and believe these are likely due to the relative instability of wortmannin in tissue culture conditions (Vanhaesebroeck and Waterfield 1999).
Figure 7. Inhibition of PI3 Kinase reduces IDO but enhances WRS expression.
Microglia from WT and S6KO mice were pretreated with the PI3K inhibitors 20 μM LY (A) or 2 μM wortmannin (WM) (B) for 1 h, followed by cytokine treatments as indicated. Total RNA for IDO and WRS expression was analyzed by real time RT-PCR. Data are normalized to that obtained from cells treated with the drug vehicle (DMSO) only, and represent the mean and SEM of three experiments.
As with the STAT-1 results above, we found that PI3K is essential for induction of IDO expression by IFN-γ. However these observations suggest that the PI3K inhibition might activate an alternate pathway, or likely release PI3K-mediated inhibition of different signaling pathway to modulate WRS expression.
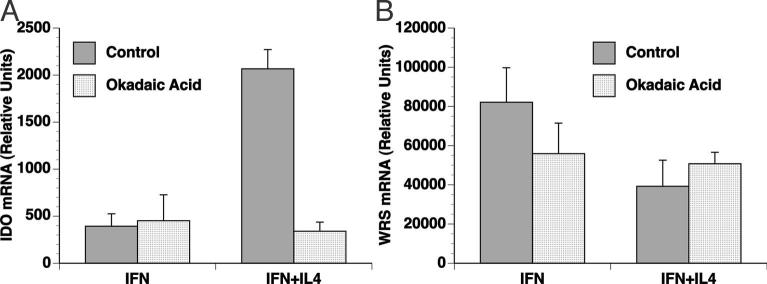
IL-4 acts through a protein phosphatase pathway in potentiating IFN-γ induced IDO expression.
Although we found that both IL-4 and IL-13 act independent of STAT-6, IL-4 has also been found to alternatively act through other mechanisms such as MAP kinase (MAPK), nuclear factor kappa B (NFκB), and protein phosphatase 2A (PP2A). Inhibiting the P38 MAPK and NFκB with pharmacological inhibitors (SB203580 and PDTC, respectively) did not lead to a change in IDO expression in the presence of IFN-γ plus IL-4 (data not shown). However, in the presence of PP2A inhibitor okadaic acid (OA), IFN-γ plus IL-4 induced IDO expression was significantly reduced by an average of 3.5-fold (n=3, p=0.0010), whereas PP2A inhibition did not significantly affect IDO expression in the presence of IFN-γ alone (Figure 8A). Although okadaic acid can also inhibit protein phosphatase 1 (PP1), at the concentration utilized here it is selective for PP2A (Bialojan and Takai 1988; Ludowyke et al. 2000). Thus, these data are consistent with IL-4 acting through PP2A in potentiating IFN-γ induced IDO expression. However no significant change was observed in WRS expression when microglia were treated with OA in the presence of IFN-γ alone or in conjunction with IL-4 (Figure 8B), indicating that the PP2A pathway does not play a role in the IL-4 downregulation of WRS.
Figure 8. IL-4 potentiation of IFN-γinduced IDO expression utilizes a protein phosphatase.
Microglia from WT mice were pretreated with PP2A inhibitor, 50 nM Okadaic acid (OA) for 1 h, followed by cytokine treatment as indicated. Total RNA for IDO (A) and WRS (B) expression was analyzed by real time RT-PCR. Data are mean and SEM of one representative experiment from three independent experiments. OA significantly reduced IDO induction by IFN-γ+IL-4 but not IFN-γ alone, whereas WRS expression did not significantly differ.
DISCUSSION
IDO and WRS are the two enzymes that are involved in tryptophan catabolism and its use in protein synthesis, respectively. Both IDO and WRS are IFN-γ inducible in a wide variety of cell types (Fleckner et al. 1995; Rubin et al. 1991). Thus, IFN-γ exerts a dual effect on tryptophan metabolism by increasing the rate of tryptophan degradation and by enhancing the ability of the cell to use low amounts of tryptophan. The results from the present study in mouse microglia are consistent with previous reports showing that the expression of IDO and WRS is upregulated by IFN-γ. However, many have reported that IL-4 and IL-13 antagonize the effects of IFN-γ (Chaves et al. 2001; Doherty et al. 1993; Musso et al. 1994; O’Keefe et al. 1999; Oswald et al. 1992; Yuan et al. 1998). In distinct contrast, here we show for the first time that in microglia, IL-4 and IL-13 modulate the expression of these enzymes in a distinct fashion (summarized in Figure 9A, B). In the presence of IFN-γ, both IL-4 and IL-13 upregulated the expression of IDO, but downregulated WRS expression. Both cytokines acted in a STAT-6 independent manner, and were also independent of P38 MAPK and NFκB. Inhibition of PP2A abrogated IL-4 mediated potentiation of IDO but not the suppression of WRS, revealing one of the mechanisms of the observed effects. Moreover, in this study, we also demonstrated divergent effects of PI3K inhibition on the IFN-γ induced expression of IDO and WRS, as PI3K inhibition reduced IDO but enhanced WRS expression.
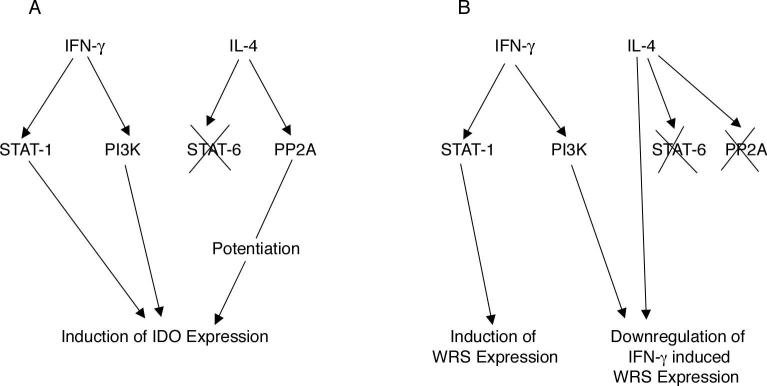
Figure 9. Mechanistic overview of differential modulation of IDO (A) and WRS (B) by IL-4 along with IFN-γ in mouse microglia.
Binding of IFN-γ to its receptor leads to activation of STAT-1, which in turn leads to transcription of IDO and WRS genes. IL-4 potentiates IDO expression but downregulates WRS expression in the presence of IFN-γ in a STAT-6 independent manner. PP2A does not play a role in WRS expression by IFN-γ and/or IL-4, however inhibition of PP2A abrogates IL-4 upregulation of IFN-γ induced IDO expression. IFN-γ also utilizes PI3K pathway in inducing IDO expression as shown by LY and WM (PI3K inhibitors). However, use of LY and WM enhance WRS expression unlike suppression of IDO expression again showing differential regulation of IDO and WRS, two enzymes utilizing tryptophan as a common substrate.
The antagonistic effects of various cytokines to IFN-γ on IDO expression in fibroblasts as well as the myeloid lineage dendritic cells and monocytes have been reported (Chaves et al. 2001; Doherty et al. 1993; Musso et al. 1994; Yuan et al. 1998). However, the effects of anti-inflammatory cytokines such as IL-4 and IL-13 on the expression of IFN-γ induced IDO and WRS expression in primary microglia, the resident myeloid cells of the CNS that are involved in various pathological disorders of the brain, have not been previously evaluated. In contrast to previous reports in other cell types, microglial cells responded uniquely, as both IL-4 and IL-13 enhanced IFN-γ induced expression of IDO.
In several brain disorders, such as HIV/SIV encephalitis, there is an increase of IDO in the brain, which is linked to increased levels of IFN-γ, likely expressed in infiltrating T lymphocytes (Burudi et al. 2002). IL-4 and IL-13 expression has been found to be present in the normal brain (Szczepanik et al. 2001), and IL-4 has been found to have a role in the CNS and other pathologies induced by SHIV infection in macaques (Buch et al. 2004; Dhillon et al. 2005). The cellular sources of IL-4 and IL-13 in the brain, however, are uncertain. In multiple sclerosis, IL-4 expression is most prominent in astrocytes (Hulshof et al. 2002), and in rodents, intracerebral LPS injection induces microglial IL-13 expression (Shin et al. 2004). The sources of IL-4 and IL-13 in the brain may also be type I helper and cytotoxic T cells (Mosmann and Sad 1996). In the present study IL-4 enhanced the expression of IFN-γ induced IDO both at mRNA and protein levels and also enhanced the functional activity of IDO. Although the level of expression of IFN-γ is the limiting factor in the context of IDO expression, it is tempting to speculate that the presence of IL-4 and/or IL-13 could be potentiating factors that prolong and exacerbate the effect of IFN-γ on IDO induction leading to neurotoxicity and immunosuppression.
IL-4 and IL-13 both downregulated IFN-γ induced WRS expression. As WRS is coinduced with IDO, it has been suggested that IDO expressing cells avoid the negative effects of IDO induced tryptophan depletion and protect themselves from tryptophan self-starvation by increasing the expression of WRS as a compensatory mechanism (Boasso et al. 2005; Murray 2003). Our results in microglia indeed showed upregulation of IDO and WRS by IFN-γ̃ However, in the presence of IL-4 and IL-13, WRS expression decreased, while IDO increased. In pathologies where IL-4 or IL-13 would be present, IFN-γ induced IDO would be upregulated but WRS would be downregulated. Expression of IFN-γ and IL-4 are found simultaneously in the brain in human neurocysticercosis and mouse cryptocococcal meningoencephalitis (Maffei et al. 2004; Restrepo et al. 2001). IDO expressing cells are indeed present in granulomatous inflammation (Popov et al. 2006). Since granuloma formation allows for control of the pathogen, IDO-mediated T cell inhibition can actually protect the host from the untoward consequences of harmful immune reactivity (Zganiacz et al. 2004). The discordance between the IL-4 and IL-13 mediated enhancement of IDO but suppression of WRS may represent a mechanism by which the immune system can still inhibit an infected cell. Although the enhanced IDO expression leads to immune inhibition, suppression of WRS leads to the cell’s inability to utilize the diminishing amounts of tryptophan, thereby inhibiting needed protein synthesis.
In order to understand the mechanisms of these divergent effects, we first examined STAT-1, central to mediating the effects of IFN-γ. In the absence of STAT-1 there was no induction of either gene indicating the involvement of STAT-1 in modulating IDO and WRS expression. Since STAT-1 was essential, we further investigated whether IL-4 or IL-13 enhance STAT-1 phosphorylation to potentiate IDO expression. However, both IL-4 and IL-13 were not active in altering IFN-γ induced patterns of STAT-1 phosphorylation. Second, we examined the role of STAT-6, the main signaling pathway triggered by IL-4 and IL-13. The data reported here reveal that the effects of IL-4 and IL-13 on IFN-γ induced IDO and WRS expression were STAT-6 independent.
The inhibition of TNF-α and IL-12 by IL-4 in macrophages has been shown to be in part STAT-6 independent, but the pathway has not been identified (Levings and Schrader 1999). Another report showed that IL-4 utilizes PP2A but not STAT-6 in inhibition of TNF-α induced formyl peptide receptor 2 expression (Iribarren et al. 2005). Here we found that inhibition of PP2A significantly reduced the expression of IDO in the presence of IFN-γ plus IL-4, but did not affect IDO expression in the presence of IFN-γ alone. Although the inhibitor used, OA, is known to show selectivity for PP2A over its other potential target, PP1, further studies are needed to define the exact role of kinases and phosphatases in the regulation of IDO. A large body of evidence suggests that PP2A acts at multiple points in the signal transduction cascade (Gomez and Spitzer 1999). Therefore, it is possible that PP2A might be deactivating an inhibitory factor in the pathway where IL-4 converges with IFN-γ in potentiation of IDO expression. On the other hand, PP2A inhibition did not alter the expression of WRS under the same conditions, indicating that the modulation of WRS expression by IL-4 is independent of PP2A. Thus, this underlines the fact that IL-4 differentially regulates the expression of IDO and WRS.
PI3K is an important factor in cell proliferation and plays a crucial role in mediating survival signals in a wide range of cells (Ahmed et al. 1997; Dudek et al. 1997). IFN-γ can also lead to activation of PI3K (Hwang et al. 2004). Inhibition of PI3K indeed prevented the IFN-γ induced upregulation of IDO, however it accentuated the IFN-γ induced expression of WRS. Few reports show that inhibition of PI3K leads to enhanced gene transcription. For example, PI3K inhibition led to enhanced expression of urokinase like plasminogen activator (Mochizuki et al. 2002) and an increase in inducible nitric oxide synthase production (Pahan et al. 1999). In our study, one possibility of enhanced WRS expression in the presence of PI3K inhibitors may be that the inhibitors may be activating some other, yet undescribed, pathway for WRS induction, or they could more likely be releasing a PI3K mediated inhibition of another signaling pathway to modulate WRS expression. Studies are underway to decipher the mechanism underlying these phenomena.
In conclusion, our data reveal the interplay between a pro-inflammatory cytokine IFN-γ, and the anti-inflammatory cytokines IL-4 and IL-13, as well as a role of PI3K, in the differential regulation of IDO and WRS expression in microglia. The physiological benefit of inverse correlation between IDO and WRS in a pathological condition might favor tryptophan catabolism in infected cells, preventing their escape from immune attack due to IDO expression. Thus, these pathways may represent important therapeutic targets for treatment of neuroinflammatory diseases.
ACKNOWLEDGEMENTS
MCY and EMEB performed experiments with the assistance of CCF and DDW; MA performed the immunofluorescence studies. CML performed independent biostatistical analyses. MCY, EMEB and HSF interpreted the results. MCY and HSF wrote the manuscript, with input from the other authors. HSF was responsible for the overall research design and direction. We thank Drs. Cecilia Marcondes, Tricia Burdo, Gurudutt Pendyala, Salavador Huitron-Resendiz, Deepak Yadav and Sandrine Dabernat for their helpful suggestions in reviewing the manuscript. This study was supported by National Institute of Health Grants MH072477 and MH062261. This is manuscript #18478 from The Scripps Research Institute.
Footnotes
EMEB: International Livestock Research Institute, Nairobi, Kenya
REFERENCES
- Ahmed NN, Grimes HL, Bellacosa A, Chan TO, Tsichlis PN. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci U S A. 1997;94(8):3627–32. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–91. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- Bara H, Hainfellner JA, Kepplinger B, Mazal PR, Schmid H, Budka H. Kynurenic acid metabolism in the brain of HIV-1 infected patients. J Neural Transm. 2000;107(10):1127–38. doi: 10.1007/s007020070026. [DOI] [PubMed] [Google Scholar]
- Bialojan C, Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem J. 1988;256(1):283–90. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boasso A, Herbeuval JP, Hardy AW, Winkler C, Shearer GM. Regulation of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA-synthetase by CTLA-4-Fc in human CD4+ T cells. Blood. 2005;105(4):1574–81. doi: 10.1182/blood-2004-06-2089. [DOI] [PubMed] [Google Scholar]
- Braun D, Longman RS, Albert ML. A two-step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic-cell maturation. Blood. 2005;106(7):2375–81. doi: 10.1182/blood-2005-03-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S, Sui Y, Potula R, Pinson D, Adany I, Li Z, Huang M, Li S, Dhillon N, Major E. Role of interleukin-4 and monocyte chemoattractant protein-1 in the neuropathogenesis of X4 simian human immunodeficiency virus infection in macaques. J Neurovirol. 2004;10(Suppl 1):118–24. doi: 10.1080/753312763. [DOI] [PubMed] [Google Scholar]
- Burudi EM, Marcondes MC, Watry DD, Zandonatti M, Taffe MA, Fox HS. Regulation of Indoleamine 2,3-Dioxygenase Expression in Simian Immunodeficiency Virus-Infected Monkey Brains. J Virol. 2002;76(23):12233–12241. doi: 10.1128/JVI.76.23.12233-12241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves AC, Ceravolo IP, Gomes JA, Zani CL, Romanha AJ, Gazzinelli RT. IL-4 and IL-13 regulate the induction of indoleamine 2,3-dioxygenase activity and the control of Toxoplasma gondii replication in human fibroblasts activated with IFN-gamma. Eur J Immunol. 2001;31(2):333–44. doi: 10.1002/1521-4141(200102)31:2<333::aid-immu333>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Dhillon NK, Sui Y, Potula R, Dhillon S, Adany I, Li Z, Villinger F, Pinson D, Narayan O, Buch S. Inhibition of pathogenic SHIV replication in macaques treated with antisense DNA of interleukin-4. Blood. 2005;105(8):3094–9. doi: 10.1182/blood-2004-09-3515. [DOI] [PubMed] [Google Scholar]
- Doherty TM, Kastelein R, Menon S, Andrade S, Coffman RL. Modulation of murine macrophage function by IL-13. J Immunol. 1993;151(12):7151–60. [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275(5300):661–5. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- Fleckner J, Martensen PM, Tolstrup AB, Kjeldgaard NO, Justesen J. Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine. 1995;7(1):70–7. doi: 10.1006/cyto.1995.1009. [DOI] [PubMed] [Google Scholar]
- Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101(2):151–5. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- Frumento G, Rotondo R, Tonetti M, Damonte G, Benatti U, Ferrara GB. Tryptophan derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J Exp Med. 2002;196(4):459–68. doi: 10.1084/jem.20020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Spitzer NC. In vivo regulation of axon extension and pathfinding by growthcone calcium transients. Nature. 1999;397(6717):350–5. doi: 10.1038/16927. [DOI] [PubMed] [Google Scholar]
- Grant RS, Naif H, Thuruthyil SJ, Nasr N, Littlejohn T, Takikawa O, Kapoor V. Induction of indolamine 2,3-dioxygenase in primary human macrophages by human immunodeficiency virus type 1 is strain dependent. J Virol. 2000;74(9):4110–5. doi: 10.1128/jvi.74.9.4110-4115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U, Fallarino F, Bianchi R, Orabona C, Vacca C, Fioretti MC, Puccetti P. A defect in tryptophan catabolism impairs tolerance in nonobese diabetic mice. J Exp Med. 2003;198(1):153–60. doi: 10.1084/jem.20030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin GJ, Brew BJ. Implications of the kynurenine pathway and quinolinic acid in Alzheimer’s disease. Redox Rep. 2002;7(4):199–206. doi: 10.1179/135100002125000550. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Smythe GA, Veas LA, Takikawa O, Brew BJ. A beta 1-42 induces production of quinolinic acid by human macrophages and microglia. Neuroreport. 2003;14(18):2311–5. doi: 10.1097/00001756-200312190-00005. [DOI] [PubMed] [Google Scholar]
- Heyes MP, Rubinow D, Lane C, Markey SP. Cerebrospinal fluid quinolinic acid concentrations are increased in acquired immune deficiency syndrome. Ann Neurol. 1989;26(2):275–7. doi: 10.1002/ana.410260215. [DOI] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Crowley JS, Davis LE, Demitrack MA, Der M, Dilling LA, Elia J, Kruesi MJ, Lackner A. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain. 1992;115(Pt 5):1249–73. doi: 10.1093/brain/115.5.1249. [DOI] [PubMed] [Google Scholar]
- Hulshof S, Montagne L, De Groot CJ, Van Der Valk P. Cellular localization and expression patterns of interleukin-10, interleukin-4, and their receptors in multiple sclerosis lesions. Glia. 2002;38(1):24–35. doi: 10.1002/glia.10050. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Jung JS, Lim SJ, Kim JY, Kim TH, Cho KH, Han IO. LY294002 inhibits interferon-gamma-stimulated inducible nitric oxide synthase expression in BV2 microglial cells. Biochem Biophys Res Commun. 2004;318(3):691–7. doi: 10.1016/j.bbrc.2004.04.082. [DOI] [PubMed] [Google Scholar]
- Iribarren P, Chen K, Hu J, Zhang X, Gong W, Wang JM. IL-4 inhibits the expression of mouse formyl peptide receptor 2, a receptor for amyloid betal-42, in TNF-alphaactivated microglia. J Immunol. 2005;175(9):6100–6. doi: 10.4049/jimmunol.175.9.6100. [DOI] [PubMed] [Google Scholar]
- Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300(5625):1527–8. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- Kwidzinski E, Bunse J, Aktas O, Richter D, Mutlu L, Zipp F, Nitsch R, Bechmann I. Indolamine 2,3-dioxygenase is expressed in the CNS and down-regulates autoimmune inflammation. Faseb J. 2005;19(10):1347–9. doi: 10.1096/fj.04-3228fje. [DOI] [PubMed] [Google Scholar]
- Levings MK, Schrader JW. IL-4 inhibits the production of TNF-alpha and IL-12 by STAT6-dependent and-independent mechanisms. J Immunol. 1999;162(9):5224–9. [PubMed] [Google Scholar]
- Ludowyke RI, Holst J, Mudge LM, Sim AT. Transient translocation and activation of protein phosphatase 2A during mast cell secretion. J Biol Chem. 2000;275(9):6144–52. doi: 10.1074/jbc.275.9.6144. [DOI] [PubMed] [Google Scholar]
- MacKenzie CR, Gonzalez RG, Kniep E, Roch S, Daubener W. Cytokine mediated regulation of interferon-gamma-induced IDO activation. Adv Exp Med Biol. 1999;467:533–9. doi: 10.1007/978-1-4615-4709-9_66. [DOI] [PubMed] [Google Scholar]
- Maffei CM, Mirels LF, Sobel RA, Clemons KV, Stevens DA. Cytokine and inducible nitric oxide synthase mRNA expression during experimental murine cryptococcal meningoencephalitis. Infect Immun. 2004;72(4):2338–49. doi: 10.1128/IAI.72.4.2338-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R, Zibert A, Laryea M, Gobel U, Daubener W, Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood. 2004;103(12):4619–21. doi: 10.1182/blood-2003-11-3909. [DOI] [PubMed] [Google Scholar]
- Mochizuki Y, Tsuda S, Kanetake H, Kanda S. Negative regulation of urokinase-type plasminogen activator production through FGF-2-mediated activation of phosphoinositide 3-kinase. Oncogene. 2002;21(46):7027–33. doi: 10.1038/sj.onc.1205736. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Sad S. The expanding universe of T-cell subsets: Thl, Th2 and more. Immunol Today. 1996;17(3):138–46. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189(9):1363–72. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Sharma MD, Hou D, Baban B, Lee JR, Antonia SJ, Messina JL, Chandler P, Koni PA, Mellor AL. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114(2):280–90. doi: 10.1172/JCI21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281(5380):1191–3. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- Murray MF. Tryptophan depletion and HIV infection: a metabolic link to pathogenesis. Lancet Infect Dis. 2003;3(10):644–52. doi: 10.1016/s1473-3099(03)00773-4. [DOI] [PubMed] [Google Scholar]
- Musso T, Gusella GL, Brooks A, Longo DL, Varesio L. Interleukin-4 inhibits indoleamine 2,3-dioxygenase expression in human monocytes. Blood. 1994;83(5):1408–11. [PubMed] [Google Scholar]
- Nguyen MH, Ho JM, Beattie BK, Barber DL. TEL-JAK2 mediates constitutive activation of the phosphatidylinositol 3′-kinase/protein kinase B signaling pathway. J Biol Chem. 2001;276(35):32704–13. doi: 10.1074/jbc.M103100200. [DOI] [PubMed] [Google Scholar]
- O’Keefe GM, Nguyen VT, Benveniste EN. Class II transactivator and class II MHC gene expression in microglia: modulation by the cytokines TGF-beta, IL-4, IL-13 and IL-10. Eur J Immunol. 1999;29(4):1275–85. doi: 10.1002/(SICI)1521-4141(199904)29:04<1275::AID-IMMU1275>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Oswald IP, Gazzinelli RT, Sher A, James SL. IL-10 synergizes with IL-4 and transforming growth factor-beta to inhibit macrophage cytotoxic activity. J Immunol. 1992;148(11):3578–82. [PubMed] [Google Scholar]
- Pahan K, Raymond JR, Singh I. Inhibition of phosphatidylinositol 3-kinase induces nitricoxide synthase in lipopolysaccharide- or cytokine-stimulated C6 glial cells. J Biol Chem. 1999;274(11):7528–36. doi: 10.1074/jbc.274.11.7528. [DOI] [PubMed] [Google Scholar]
- Popov A, Abdullah Z, Wickenhauser C, Saric T, Driesen J, Hanisch FG, Domann E, Raven EL, Dehus O, Hermann C. Indoleamine 2,3-dioxygenase-expressing dendritic cells form suppurative granulomas following Listeria monocytogenes infection. J Clin Invest. 2006;116(12):3160–3170. doi: 10.1172/JCI28996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo BI, Alvarez JI, Castano JA, Arias LF, Restrepo M, Trujillo J, Colegial CH, Teale JM. Brain granulomas in neurocysticercosis patients are associated with a Th1 and Th2 profile. Infect Immun. 2001;69(7):4554–60. doi: 10.1128/IAI.69.7.4554-4560.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Shirey KA, Carlin JM. Synergistic transcriptional activation of indoleamine dioxygenase by IFN-gamma and tumor necrosis factor-alpha. J Interferon Cytokine Res. 2003;23(8):413–21. doi: 10.1089/107999003322277829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BY, Anderson SL, Xing L, Powell RJ, Tate WP. Interferon induces tryptophanyl-tRNA synthetase expression in human fibroblasts. J Biol Chem. 1991;266(36):24245–8. [PubMed] [Google Scholar]
- Sakurai K, Zou JP, Tschetter JR, Ward JM, Shearer GM. Effect of indoleamine 2,3-dioxygenase on induction of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2002;129(12):186–96. doi: 10.1016/s0165-5728(02)00176-5. [DOI] [PubMed] [Google Scholar]
- Sedgwick JD, Schwender S, Imrich H, Dorries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci U S A. 1991;88(16):7438–42. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin WH, Lee DY, Park KW, Kim SU, Yang MS, Joe EH, Jin BK. Microglia expressing interleukin-13 undergo cell death and contribute to neuronal survival in vivo. Glia. 2004;46(2):142–52. doi: 10.1002/glia.10357. [DOI] [PubMed] [Google Scholar]
- Stone TW. Kynurenines in the CNS: from endogenous obscurity to therapeutic importance. Prog Neurobiol. 2001;64(2):185–218. doi: 10.1016/s0301-0082(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Szczepanik AM, Funes S, Petko W, Ringheim GE. IL-4, IL-10 and IL-13 modulate A beta(1--42)-induced cytokine and chemokine production in primary murine microglia and a human monocyte cell line. J Neuroimmunol. 2001;113(1):49–62. doi: 10.1016/s0165-5728(00)00404-5. [DOI] [PubMed] [Google Scholar]
- Terness P, Bauer TM, Rose L, Dufter C, Watzlik A, Simon H, Opelz G. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J Exp Med. 2002;196(4):447–57. doi: 10.1084/jem.20020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253(1):239–54. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Widner B, Ledochowski M, Fuchs D. Interferon-gamma-induced tryptophan degradation: neuropsychiatric and immunological consequences. Curr Drug Metab. 2000;1(2):193–204. doi: 10.2174/1389200003339063. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Imanishi J, Oku T, Kishida T, Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by interferon. Proc Natl Acad Sci U S A. 1981;78(1):129–32. doi: 10.1073/pnas.78.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Collado-Hidalgo A, Yufit T, Taylor M, Varga J. Modulation of cellular tryptophan metabolism in human fibroblasts by transforming growth factor-beta: selective inhibition of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA synthetase gene expression. J Cell Physiol. 1998;177(1):174–86. doi: 10.1002/(SICI)1097-4652(199810)177:1<174::AID-JCP18>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Zganiacz A, Santosuosso M, Wang J, Yang T, Chen L, Anzulovic M, Alexander S, Gicquel B, Wan Y, Bramson J. TNF-alpha is a critical negative regulator of type 1 immune activation during intracellular bacterial infection. J Clin Invest. 2004;113(3):401–13. doi: 10.1172/JCI18991. [DOI] [PMC free article] [PubMed] [Google Scholar]