Abstract
DISC1 influences susceptibility to psychiatric disease and related phenotypes. Intact functions of DISC1 and its binding partners, NDEL1 and NDE1, are critical to neurodevelopmental processes aberrant in schizophrenia (SZ). Despite evidence of an NDEL1–DISC1 protein interaction, there have been no investigations of the NDEL1 gene or the relationship between NDEL1 and DISC1 in SZ. We genotyped six NDEL1 single-nucleotide polymorphisms (SNPs) in 275 Caucasian SZ patients and 200 controls and tested for association and interaction between the functional SNP Ser704Cys in DISC1 and NDEL1. We also evaluated the relationship between NDE1 and DISC1 genotype and SZ. Finally, in a series of in vitro assays, we determined the binding profiles of NDEL1 and NDE1, in relation to DISC1 Ser704Cys. We observed a single haplotype block within NDEL1; the majority of variation was captured by NDEL1 rs1391768. We observed a significant interaction between rs1391768 and DISC1 Ser704Cys, with the effect of NDEL1 on SZ evident only against the background of DISC1 Ser704 homozygosity. Secondary analyses revealed no direct relationship between NDE1 genotype and SZ; however, there was an opposite pattern of risk for NDE1 genotype when conditioned on DISC1 Ser704Cys, with NDE1 rs3784859 imparting a significant effect but only in the context of a Cys-carrying background. In addition, we report opposing binding patterns of NDEL1 and NDE1 to Ser704 versus Cys704, at the same DISC1 binding domain. These data suggest that NDEL1 significantly influences risk for SZ via an interaction with DISC1. We propose a model where NDEL1 and NDE1 compete for binding with DISC1.
INTRODUCTION
Emerging evidence suggests that the gene disrupted in schizophrenia-1 (DISC1) confers an increased risk for a range of psychiatric illnesses (1,2), including major depression (3), bipolar disorder, schizoaffective disorder and schizophrenia (SZ) (4). Moreover, several recent studies have observed a link between DISC1 genotype and elements of neurocognitive function (5–9), as well as associations with other manifestations of illness such as reduced cerebral grey matter (5,8) and severity of positive psychotic symptoms (10). The mechanism by which DISC1 confers these effects on brain structure and function remains to be elucidated; however, the complexity of these phenotypes indicates that its action may be mediated by multiple loci within DISC1 and/or through interactions with critical binding partners.
The DISC1 protein is multifunctional and has at least 10 binding partners, many of which are involved in cell division and intracellular transport (2,7,11). Of particular interest are nuclear distribution element like (NDEL1) and its homolog, NDE1, centrosomal proteins involved in mitosis, neuronal migration and microtubule organization during brain development (12–14). Both NDEL1 and NDE1 were identified via their interactions with lissencephaly 1 (LIS1), a gene in which mutations cause human lissencephaly (15–19). These LIS1 mutations inhibit binding to NDEL1 and cause an abnormal pattern of cortical development resulting in the appearance of a ‘smoothed brain’ (16). NDEL1 has been demonstrated to act as a modulator of dynein function and as a critical component for centrosome–nucleus coupling during neuronal migration (12). A critical role of NDE1 in neurodevelopment was demonstrated in an NDE1-deficient mouse model (NDE1 knock out), in which the mice presented with microcephaly, reduced progenitor cell division and alterations in mitotic spindle formation and in chromosome segregation (16). Although NDE1 and NDEL1 share some common features, for example both interact with cytoplasmic dynein, a microtubule-dependent motor complex (18), they are distinct in their functions related to chromosomal alignment and segregation (20).
There is preliminary evidence that NDEL1’s function in neurodevelopment may be related to the pathophysiology of SZ (21,22). Biological data suggest that if NDEL1 plays a role in the etiology of SZ, it may do so via an interaction with the DISC1 protein. Lipska et al. (23) demonstrated that NDEL1 expression is decreased in the hippocampus (HC) of patients with SZ. Further, they reported an association between NDEL1 expression and three alleles influencing risk for SZ within the DISC1 gene, with these alleles consistently predicting reduced NDEL1 expression. Similarly, in a study by Kamiya et al. (24), results from yeast two-hybrid assays suggest that genetic variation at loci proximal to the DISC1–NDEL1 interaction domain modulates the binding affinity of NDEL1 to DISC1. Functionally, an intact NDEL1–DISC1 interaction has been shown to be critical to multiple neurodevelopmental processes that are abnormal in SZ including neural outgrowth (25). On the basis of this convergent evidence, we hypothesized that the gene that codes for NDEL1 (NDEL1 located on chromosome 17p13.1) may be involved in the pathophysiology of SZ, perhaps via interaction with DISC1.
Nuclear distribution gene E homolog 1 (NDE1), the homolog of NDEL1, is located on chromosome 16p13.11. There is early evidence that NDE1 may also play a role in susceptibility to SZ, and that its actions in this context may also be directly linked to DISC1 function. In a recent linkage study, Hennah et al. (26) identified a critical interaction between a DISC1 risk haplotype (HEP3) and NDE1 and risk for SZ in a Finnish sample. In an initial linkage analysis, the chromosomal region containing NDE1 showed no evidence for linkage; however, once conditioned on the presence of a previously identified risk haplotype in DISC1, significant linkage was detected in this region but only in carriers of the risk haplotype. Follow-up analyses revealed a significant association with a haplotype within the NDE1 gene and SZ in these subjects. A more recent study in a Japanese SZ population failed to replicate these findings in NDE1; however, DISC1 genotype was not taken into account in this cohort (27). Taken together, we hypothesized that variation in the NDE1 gene may not directly impact upon the risk for SZ but that its interaction with DISC1 may be critical to this relationship.
To date, there have been no studies examining the effect of NDEL1 genetic variation on SZ susceptibility. Moreover, most likely because of limitations in sample sizes, there has been a paucity of data on the relationship between NDEL1, NDE1 and DISC1 genotype and risk for SZ. Therefore, we conducted a case–control study in 275 Caucasian patients with SZ and 200 Caucasian healthy controls to assess the relationship between NDEL1 and SZ and to test for an epistatic interaction between NDEL1 genotype and DISC1 genotype on risk for SZ. We specifically focused on the DISC1 functional variant, Ser704Cys, as several lines of evidence converge to suggest that this locus may be of particular importance in increasing risk for SZ and in modifying the DISC1 protein interaction with NDEL1 (Table 1), including: (i) the Ser 704 allele at this single-nucleotide polymorphism (SNP) has previously been identified as a SZ risk allele (5); (ii) the Ser 704 allele has been associated in our sample with increased lifetime severity of delusions in patients with SZ (9); (iii) Ser704Cys has been shown to impact hippocampal structure and function in healthy controls (5); (iv) its location on DISC1 is proximal to the region that is known to interact with NDEL1 (28,29); (v) Ser704Cys has been shown to impact NDEL1 expression in patients with SZ (23) and (vi) the Ser704Cys locus directly impacts DISC1–NDEL1 protein binding (24). For epistatic analyses with DISC1, we utilized an NDEL1-tagged SNP that captured the majority of variance in the defined linkage disequilibrium (LD) structure of our sample. Finally, although not a primary focus of this study, we also tested for an effect of NDE1 genotype and its potential interaction with DISC1 on risk for SZ in a subset of the sample for which this information was available.
Table 1.
Molecular context of Ser704Cys variation of DISC1
| Genotype | Clinical phenotypes | References |
|---|---|---|
| Ser | The NDEL1 rs1391768 SNP on risk for SZ (Ser704/Ser704) | Current study |
| Increased lifetime severity of delusions in SZ (Ser704/Ser704) | (9) | |
| Poor cognitive performance in control as well as SZ (Ser704/Ser704) | (5) | |
| Cys | The NDE1 rs3784859 SNP on risk for SZ | Current study |
| Stronger cognitive decline in aged women (Cys704/Cys704) | (7) | |
| Molecular/anatomical phenotypes | ||
| Ser | Reduction of HC GM volume in control and lower HF NAA (Ser704/Ser704) | (5) |
| Altered engagement of HC during cognitive tasks (Ser704/Ser704) | (5) | |
| Lower expression of FEZ1, LIS1 and NDEL1 in HC, and FEZ1 in DLPFC of SZ (Ser704/Ser704) | (23) | |
| Less interaction of DISC1with NDEL1 | (24) | |
| Predicted phosphorylation by DNA-PKa (NetPhosK 1.0) | Current study | |
| Cys | Reduction of CC GM volume and decreased fractional anisotropy in prefrontal WM | (3) |
| Less phosphorylation of ERK1/2 | (3) | |
| Less interaction of DISC1 with NDE1 | Current study |
HF, hippocampal formation; NAA, N-acetylaspartate; DLPFC, dorsolateral prefrontal cortex; CC, cingulate cortex; GM, gray matter; WM, white matter; DNA-PK, DNA-dependent protein kinase.
aPossible phosphorylation of Ser704 is predicted by NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/).
The second stage of the current study was directed at extending the recently published finding of a gene–gene interaction between DISC1 and NDE1 (26). Although homologous at the gene level, the relationship between the NDEL1 and NDE1 proteins and their potential interactions with DISC1 has not yet been elucidated. To address the functional relationship among DISC1, NDEL1 and NDE1, we first examined whether NDE1 and NDEL1 share a common binding domain on DISC1. We then tested for an effect of DISC1 Ser704Cys on the NDE1–DISC1 protein interaction. Finally, we bring these data together with our previously published data (24) to compare the relative binding affinity of the DISC1 Cys704 versus Ser704 proteins to both NDEL1 and NDE1 and we propose a potential competitive binding mechanism for NDE1 and NDEL1 in relation to DISC1 Ser704Cys.
RESULTS
Stage one
NDEL1
In a cohort of 275 Caucasian patients with SZ and 200 Caucasian healthy controls, four SNPs formed two major yin–yang haplotypes, AGTC (57% frequency in SZ; 64% frequency in HCs) and GCCT (42% frequency in SZ; 36% frequency in HCs) as well as three additional haplotypes that were very rare; each <2.5% frequency. Because of the very low frequency of the three ‘rare’ haplotypes, the subjects were grouped by the status of only the two most common haplotypes (AGTC, GCCT). Thus, NDEL1 diplotype analyses focused on dichotomous groups comparing subjects who were homozygous for the common haplotype (AGTC HZs) versus subjects that carried at least one copy of the GCCT haplotype. This resulted in a complete sample of 268 patients with SZ and 196 healthy controls, in which further association analyses were conducted.
NDEL1 association with illness
χ2 analyses revealed that the AGTC haplotype was more common in healthy controls than in SZ patients, with a statistically significant haplotypic P-value of 0.041. In diplotype analyses, AGTC homozygosity was significantly more frequent in healthy controls (43%) versus SZ patients (34%) (χ2 = 3.90; P = 0.048; OR = 1.32; 95% CI 1.01–1.73).
As noted in Figure 1, the LD among the four SNPs comprising the haplotypes was very high (r2 ≥ 0.93). All SNPs were in HWE (data not shown) and, as expected on the basis of the high LD between the SNPs in the haplotype block, trend-level associations were found for each of the individual SNPs with disease (all P-values < 0.06). Using the same methodology as was used in the diplotype analysis (dominant/recessive model of common allele homozygosity versus risk allele carriers), each individual SNP was significantly associated with disease (all P-values < 0.05). The SNP that best tagged the AGTC haplotype was SNP rs1391768, with the G allele being over-represented in patients with SZ (42.1% frequency) versus healthy controls (35.8% frequency) (allelic χ2 = 3.67; P = 0.056; OR = 1.33, 95% CI 0.99–1.70). At SNP rs1391768, carriers of the G allele were at significantly greater risk for SZ than were subjects who were homozygous for the A allele (χ2 = 4.01; P = 0.045; OR = 1.47, 95% CI 1.01–2.15). Thus, to avoid redundancy in analyses and on the basis of very strong LD between markers, we utilized rs1391768 for all subsequent analyses.
Figure 1.
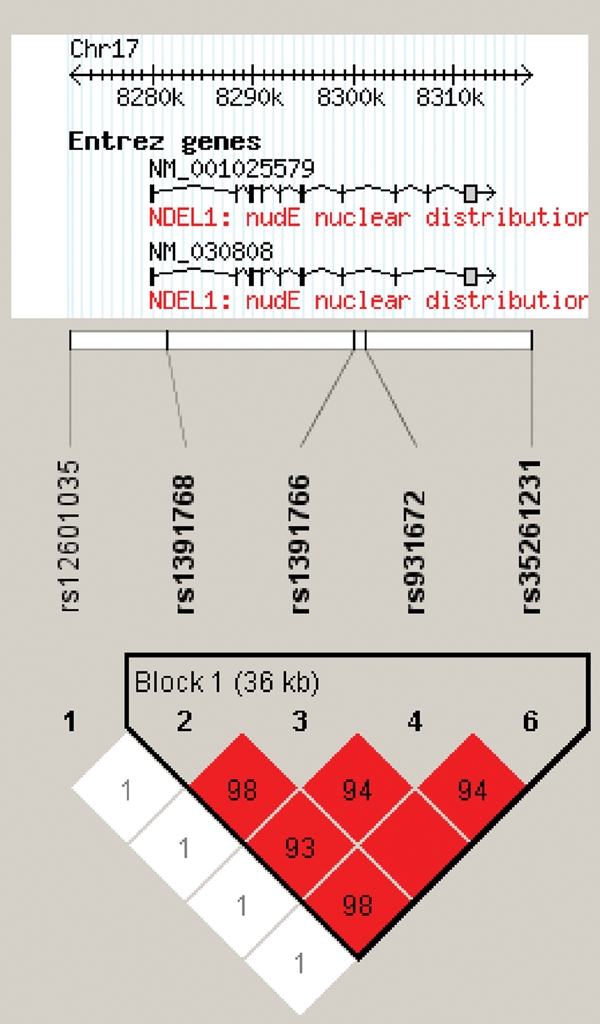
LD structure of NDEL1 haplotype block. LD (D′) structure using Haploview 3.32 (46). One SNP, rs 2012190, demonstrated extremely rare (<05%) minor allele frequency and was excluded from further analyses. Of the remaining SNPs, four formed a tight haplotype block spanning ∼36 kb, encompassing the NDEL1 gene.
Interaction between NDEL1 rs1391768 and DISC1 Ser704Cys
We tested for an epistatic interaction between the NDEL1 rs1391768 genotype and the DISC1 Ser704Cys polymorphism on risk for SZ first by unconditional logistic regression and second by conditioning the sample on DISC1 Ser704Cys. Owing to the low frequency of the Cys allele, DISC1 Ser704Cys genotypes were grouped to compare subjects carrying at least one copy of the Cys allele (Cys) with subjects homozygous for the Ser allele (SerSer). For NDEL1 SNP rs1391768, a dominant/recessive model of common allele homozygosity versus risk allele carriers was utilized as this was the strongest model for disease risk. Covariates included in the regression model were age, sex and estimated IQ.
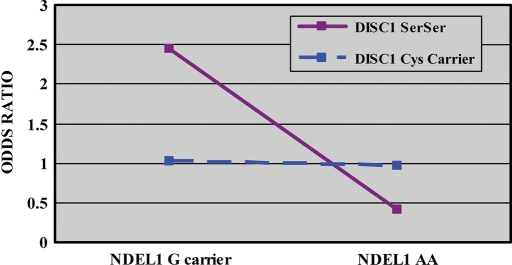
We used the likelihood ratio test in a backward stepwise regression to test for interaction. All terms were retained in the final best fit model. As predicted, the NDEL1 × DISC1 interaction term was significant (P = 0.024) after accounting for the main effects of NDEL1 rs1391768 (P = 0.008); DISC1 Ser704Cys (P = 0.047); age (P = 0.004); sex (P < 0.001) and estimated IQ (P < 0.001). The final model classified subject type with 73.7% accuracy with a χ2 = 111.98 (P < 0.001). To explicate the interaction effect, separate χ2 analyses for effects of NDEL1 on SZ risk were conducted, dependent upon DISC1 Ser704Cys status. These analyses revealed that the effect of the NDEL1 rs1391768 genotype on risk for SZ was significant in the context of a DISC1 SerSer background (χ2 = 8.86; df = 1; P = 0.003; OR = 2.44; 95% CI 1.35–4.41) but was not significant in patients carrying one or two copies of the Cys allele (χ2 = 0.01; df = 1; P = 0.907; OR = 1.03; 95% CI 0.61–1.73; Fig. 2).
Figure 2.
Effect of NDEL1 rs1391768 genotype in a DISC1 Ser704Cys background. Subjects are grouped by genotype at both NDEL1 rs1391768 and DISC1 Ser704Cys. The X-axis represents NDEL1 genotype group. The Y-axis represents OR. These data depict the DISC1–NDEL1 genotype interaction graphically. Sample sizes for each group are as follows: NDEL1 G carrier × DISC1 Ser704Ser (SZ = 74, HC = 43); NDEL1 AA × DISC1 Ser704Ser (SZ = 31, HC = 44); NDEL1 G carrier × DISC1 Cys carrier (SZ = 52, HC = 42) and NDEL1 AA × DISC1 Cys carrier (SZ = 83, HC = 65).
NDE1
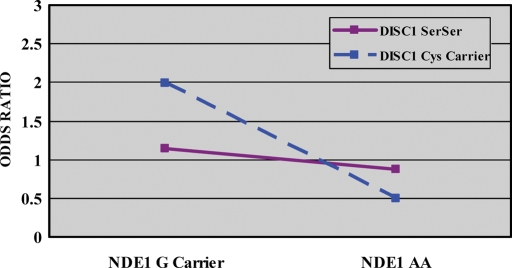
In a cohort of 267 Caucasian patients with SZ and 244 Caucasian healthy controls, overlapping with the sample genotyped for NDEL1, a total of six SNPs in the NDE1 gene were genotyped using methods described in what follows and in more detail elsewhere (30). Four SNPs formed a single haplotype block (Table 2; Fig. 3). Haploview analyses revealed that four haplotypes comprised this block, none of which was significantly associated with susceptibility to SZ; likewise, none of the single markers was significantly associated with disease (Table 2). As no significant associations was found with NDE1 and since we were primarily interested in the relationship between NDE1 and DISC1, we carried out exploratory interaction analyses with each individual NDE1 SNP and DISC1 Ser704Cys, as described earlier, using separate χ2 analyses for effects of NDE1 on SZ risk, dependent upon DISC1 Ser704Cys status. We did not utilize a regression model for NDE1–DISC1 analyses, as NDE1 genotype did not have a significant main effect in initial tests. In the subset of subjects for whom both NDE1 and DISC1 genotype were available (170 cases and 125 controls), χ2 analyses revealed that genotype at NDE1 rs3784859 had a significant effect on risk for SZ only in the context of a DISC1 Cys-carrying background (χ2 = 3.89; df = 1; P = 0.049; OR = 2.00; 95% CI 1.00–3.97), although this was not significant in patients with the SerSer genotype (χ2 = 0.15; df = 1; P = 0.698; OR = 1.15; 95% CI 0.57–2.30; Fig. 4). Although these results are quite modest and would not survive any level of correction for multiple testing, it is of note that this is the opposite pattern revealed in disease-associated risk relative to the NDEL1 SNP, at which increased susceptibility was present only in the context of a SerSer background. No other NDE1 SNPs were significant when conditioned on DISC1 Ser704Cys (data not shown).
Table 2.
NDE1 haplotype and SNP association with SZ
| Position (B35) | Frequency (total) | Frequency (cases) | Frequency (controls) | χ2-value | P-value | |
|---|---|---|---|---|---|---|
| Haplotype | ||||||
| GGAC | 0.55 | 0.54 | 0.56 | 0.46 | 0.50 | |
| TCGC | 0.29 | 0.29 | 0.28 | 0.13 | 0.72 | |
| GGGC | 0.11 | 0.11 | 0.11 | <0.01 | 0.95 | |
| GGGT | 0.04 | 0.05 | 0.03 | 2.32 | 0.13 | |
| SNP (allele) | ||||||
| rs8061376 (T)a | 15661969 | 0.30 | 0.29 | 0.16 | 0.69 | |
| rs4781679 (C)a | 15668934 | 0.29 | 0.28 | 0.13 | 0.72 | |
| rs3784859 (G)a | 15672904 | 0.45 | 0.43 | 0.56 | 0.45 | |
| rs12934645 (T)a | 15687868 | 0.05 | 0.03 | 2.77 | 0.10 | |
| rs8044738 (A) | 15688555 | 0.30 | 0.29 | 0.19 | 0.67 | |
| Rs881803 (G) | 15709835 | 0.31 | 0.28 | 0.76 | 0.38 | |
aSNP included in the haplotype.
Figure 3.
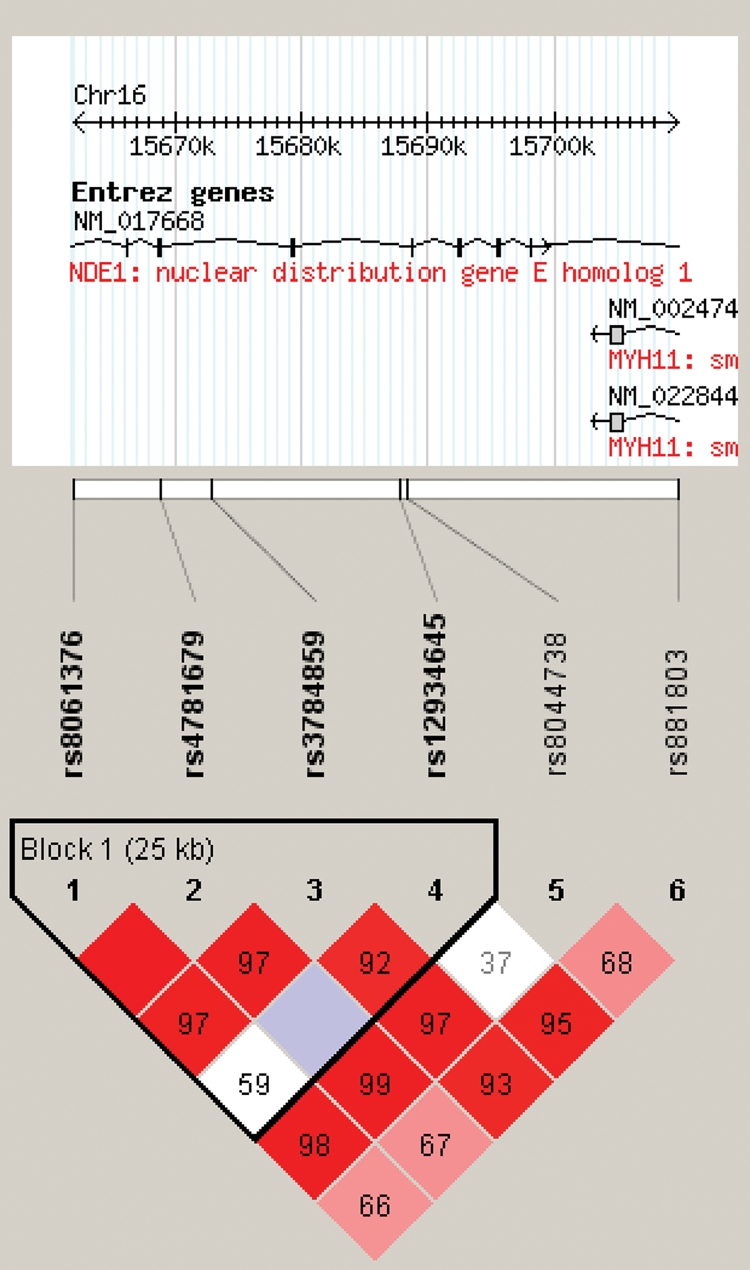
LD structure of NDE1 haplotype block. LD (D′) structure using Haploview 3.32 (24). Four of the six SNPs formed a tight haplotype block spanning ∼30 kb, within the NDE1 gene.
Figure 4.
Effect of NDE1 rs3784859 genotype in a DISC1 Ser704Cys background. Subjects are grouped by genotype at both NDE1 rs3784859 and DISC1 Ser704Cys. The X-axis represents NDE1 genotype group. The Y-axis represents OR. These data depict the DISC1–NDE1 genotype interaction graphically. Sample sizes for each group are as follows: NDE1 G carrier × DISC1 Ser704Ser (SZ = 45, HC = 35); NDE1 AA × DISC1 Ser704Ser (SZ = 28, HC = 25); NDE1 G carrier × DISC1 Cys carrier (SZ = 75, HC = 41) and NDE1 AA × DISC1 Cys carrier (SZ = 22, HC = 24).
Stage two
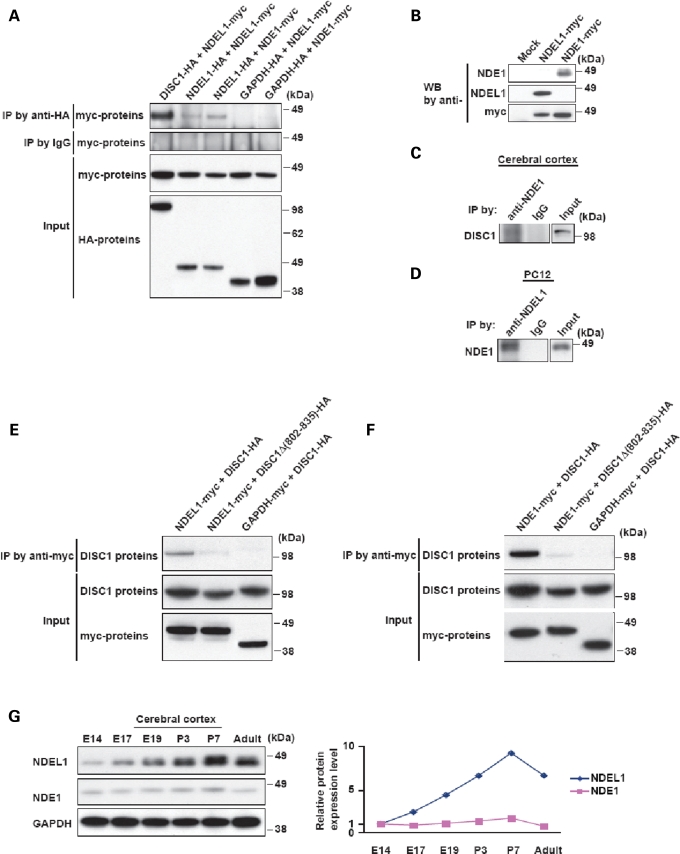
Data from stage one provide evidence of epistasis between the DISC1 and NDEL1 genes in SZ and weaker evidence of an interaction between DISC1 and NDE1, consistent with the previous report of genetic interaction between these genes in a Finnish cohort (26). Thus, we hypothesized that these three proteins (DISC1, NDEL1 and NDE1) may have functional relationships to one another. To address this question, we examined molecular interactions among these proteins biochemically. DISC1–NDE1 and DISC1–NDEL1 interactions as well as DISC1 and NDE1 self-associations have already been reported (15,25,28–29). Thus, to expand upon these previous data, we tested the potentials of NDEL1 self-association and NDE1–NDEL1 heterodimerization by co-immunoprecipitation. We observed specific NDE1–NDEL1 interaction and NDEL1 self-association, although their binding was significantly weaker than the noted DISC1–NDEL1 interaction (Fig. 5A). We confirmed protein interactions of NDE1–DISC1 and NDE1–NDEL1 at endogenous protein levels by using anti-NDE1 and NDEL1 antibodies that do not cross-react with the other molecules (Fig. 5C and D). Next, we explored the relationship between DISC1–NDEL1 and DISC1–NDE1 binding. By using a DISC1 deletion mutant that selectively lacks the NDEL1-binding domain (amino acids 802–835) (24) in co-immunoprecipitation, we observed that the same short domain was required for DISC1–NDE1 protein interaction (Fig. 5E and F). This result suggests that NDE1 and NDEL1 may compete with each other for DISC1. Feng et al. (15) studied expression of NDE1 at the messenger RNA level: its expression is the highest at embryonic day 11 (E11) and is still detectable in the ventricular zone, intermediate zone and cortical plate after E15.5. Expression level of NDEL1 is reportedly increased, according to brain development, and is prominent in the intermediate zone and cortical plate (18,31). Thus, we confirmed expression of NDE1 and NDEL1 at the protein level in the cerebral cortex from neurodevelopmental stages to adulthood (Fig. 5G). Consistent with the previous reports (15,18,31), we observed constant expression of NDE1 at low level, in contrast to time-course increase of NDEL1 during neurodevelopment. We observed one band for NDE1, versus two bands for NDEL1, which may reflect possible post-translational modification(s) on NDEL1. Taken together, although it is probable that NDE1 and NDEL1 interact with DISC1 in independent contexts, we propose an alternative notion that both NDE1 and NDEL1 can simultaneously associate with DISC1 at certain but key developmental time points.
Figure 5.
Protein interaction of DISC1, NDEL1 and NDE1. (A) Self-association of NDEL1 and hetero-oligomerization of NDEL1 and NDE1. HA-tagged DISC1, NDEL1 or GAPDH were transfected with myc-tagged NDEL1 or NDE1 in HEK293 cells. Immunoprecipitates with an anti-HA antibody were analyzed by western blotting with an anti-myc antibody. Myc-tagged NDEL1 is co-precipitated with HA-tagged DISC1 and NDEL1, but not with GAPDH (top panel). NDE1 is also co-precipitated with HA-tagged NDEL1, but not with GAPDH (top panel), suggesting that NDEL1 self-associates and forms a hetero-oligomer with NDE1. For a negative control in co-immunoprecipitation, we used a rabbit IgG and observed no precipitation of target proteins (second panel). The inputs of each protein in co-immunoprecipitation are also shown (bottom panels). (B) The specificities of monoclonal antibodies against NDE1 (see Materials and Methods section) and NDEL1 (24) were evaluated by western blotting. Anti-NDE1 antibody recognizes exogenous NDE1 protein but not exogenous NDEL1 protein. On the other hand, anti-NDEL1 antibody detects exogenous NDEL1 protein but not exogenous NDE1 protein. (C) Endogenous protein interaction of NDE1–DISC1 was confirmed by co-immunoprecipitation with the tissue extract of rat adult cerebral cortex. A monoclonal antibody against NDE1 was used for immunoprecipitation. (D) Endogenous protein interaction of NDE1–NDEL1 was confirmed by co-immunoprecipitation with the cell extract of undifferentiated PC12 cells. A monoclonal antibody against NDEL1 was used for immunoprecipitation. (E) Amino acids 802–835 of DISC1 are critical for DISC1–NDEL1 interaction. HEK293 cells were transfected with myc-tagged NDEL1 and HA-tagged DISC1 or DISC1 lacking amino acids 802–835 [DISC1Δ(802–835)]. Lysates were then immunoprecipitated with an anti-myc antibody and analyzed by western blotting with an anti-HA antibody. Deletion of amino acids 802–835 of DISC1, which is a critical domain for DISC1–NDEL1 interaction we previously reported, completely abolished the interaction of exogenous DISC1 [DISC1Δ(802–835)] with NDEL1 (top panel). The inputs of each protein are shown (bottom panels). (F) Amino acids 802–835 of DISC1 are critical for DISC1–NDE1 interaction. Myc-tagged NDE1 was transfected with HA-tagged DISC1 or HA-tagged DISC1Δ(802–835) in HEK293 cells. Protein extracts were immunoprecipitated with an anti-myc antibody. Deletion of amino acids 802–835 of DISC1 dramatically weakened the interaction with NDEL1 in co-immunoprecipitation (top panel). The inputs are shown (bottom panels). (G) Protein expression of NDE1 and NDEL1 in the cerebral cortex. NDE1 protein was expressed constantly at a low level from E14 to adulthood. Expression of NDEL1 is increased according to brain development, with the highest expression at P7.
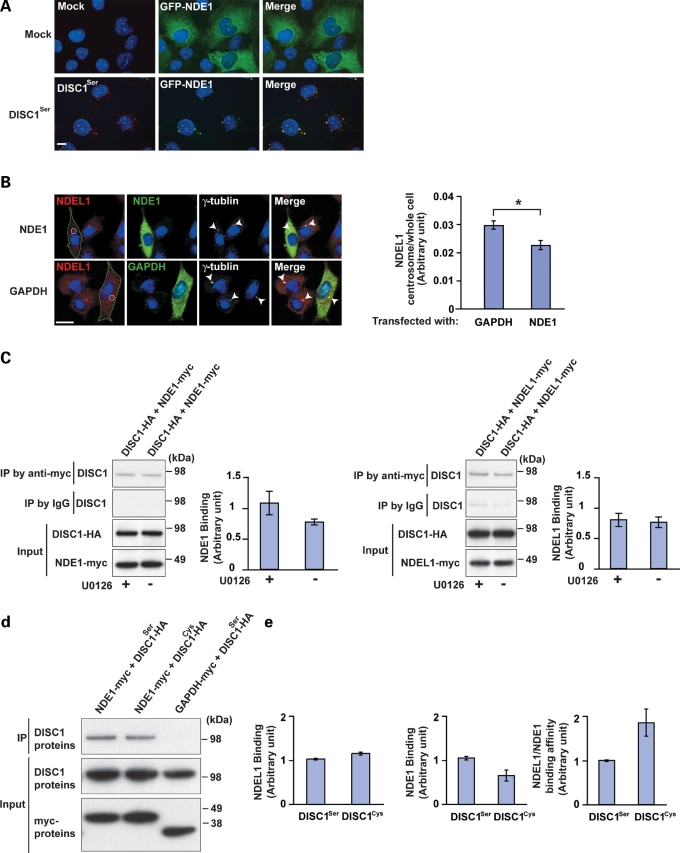
To examine the interaction of NDE1 to DISC1 in cell cultures, green fluorescent protein (GFP)-tagged NDE1 was exogenously expressed together with mock or HA-tagged DISC1 in COS7 cells. GFP-NDE1 becomes co-localized with DISC1 in punctuated patterns after expression of exogenous DISC1 (Fig. 6A), similar to the observation of DISC1 and NDEL1 (32). We previously reported that DISC1 is required for the maintenance at the centrosome of the dynein motor complex in which both NDE1 and NDEL1 are involved. We also observed that DISC1 increases the accumulation of NDEL1 to the centrosome (25). To address a competitive relationship of NDE1 and NDEL1 for DISC1, we tested whether overexpression of NDE1 affects the centrosomal accumulation of endogenous NDEL1 in PC12 cells. Expression of NDE1, but not an unrelated protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH), led to decreased accumulation of NDEL1 at the centrosome (Fig. 6B). These data also suggest that NDEL1 and NDE1 may have a competitive role at the centrosome, probably in association with DISC1. Our previous report indicated that DISC1 is, at least in part, required for proper maintenance of ERK1/2 activity in cultured neurons (3). Thus, we tested whether NDE1–DISC1 or NDEL1–DISC1 protein interactions are affected by ERK1/2. U0126 is known to block NGF-induced activation of ERK1/2 in PC12 cells (33). Protein interactions of NDE1–DISC1 or NDEL1–DISC1 were not altered by the addition of an ERK1/2 inhibitor U0126, suggesting that ERK1 signaling may be independent of these two protein interactions (Fig. 6C). Finally, we tested whether the SZ-associated genetic variation at DISC1 Ser704Cys may affect DISC1–NDEL1 and DISC1–NDE1 protein interactions. We followed up on our previous report that NDEL1 binds to DISC1-Cys704 more strongly than DISC1-Ser704 (24) by carrying out a series of parallel binding experiments with NDE1. Interestingly, we observed the opposite binding pattern for NDE1 and NDEL1 with regard to DISC1 Ser704Cys, with a stronger affinity of NDE1 to DISC1-Ser704 than to DISC1-Cys704 (Fig. 6D and E).
Figure 6.
Possible competition of NDE1 and NDEL1 for DISC1. (A) Change in subcellular distribution of NDE1 by overexpression of DISC1. Exogenous expression of DISC1 (DISC1-Ser704) (red) leads to punctuated distribution of GFP-tagged NDE1 (green) in COS7 cells. Scale bar, 10 µm. (B) NDE1 affects the accumulation of NDEL1 to the centrosome in PC12 cells. Endogenous NDEL1 (red) is localized in the perinuclear region, including the centrosome, in undifferentiated PC12 cells. Overexpression of NDE1 (green, upper panels), but not GAPDH (green, lower panels), redistributes endogenous NDEL1 (red) from the perinuclear regions (lower panels). To semi-quantify the localization change, immunointensity of NDEL1 in the centrosome area (white circle) relative to that in the whole cell region surrounded by green line was examined. Bars represent averages of each group of cells in blinded experiments (*P < 0.005). Error bars represent SEM. Representative images are shown. Blue, nucleus; red, NDEL1; green, NDE1 or GAPDH; white, γ-tubulin (indicated by arrowheads). Scale bar, 10 µm. (C) Protein interactions of DISC1–NDE1 and DISC1–NDEL1 were not affected in the presence of U0126, which is known to block NGF-induced activation of ERK1/2 in PC12 cells. Error bars represent SEM. (D) HEK 293 cells were transfected with HA-tagged DISC1Ser or DISC1Cys together with myc-tagged NDE1. Lysates were then immunoprecipitated with an anti-myc antibody and western-blotted with an antibody to HA to analyze the DISC1–NDE1 interaction. Interaction of DISC1Ser with NDE1 is slightly stronger than that with DISC1Cys as shown in co-immunoprecipitation with an anti-myc antibody (top panel). The inputs are also shown (bottom panels). (E) Influence of the Ser704Cys variation of DISC1 on the DISC1–NDEL1 and DISC1–NDE1 protein interactions. NDE1 binds to DISC1Ser more tightly than to DISC1Cys (left graph and D), whereas NDEL1 interacts with DISC1Ser less tightly than with DISC1Cys as published previously (middle graph). The opposite binding pattern for NDE1 and NDEL1 with regard to DISC1 Ser704Cys results in about 1.6-fold difference between DISC1Ser and DISC1Cys (right graph) (P = 0.054). Error bars represent SEM.
DISCUSSION
We report a significant, although modest, association between a four-locus NDEL1 haplotype (AGTC) and SZ. Subjects carrying one or more copies of the second most common haplotype (GCCT) were at increased risk for developing SZ. Because of very high LD among the SNPs tested, analyses of the best tagged SNP (NDEL1 rs1391768) captured most of the effect noted in haplotype analyses. In addition, we provide preliminary evidence of an epistatic interaction between the functional DISC1 polymorphism, Ser704Cys and variation in NDEL1, on SZ susceptibility. Subjects carrying the G allele at NDEL1 rs1391768 demonstrated a more than two-fold increased risk of developing SZ in the context of DISC1 Ser704 homozygosity; however, in the background of DISC1 Cys704, no increased risk was evident. These interaction data are both biologically plausible and consistent with previous data, as the Ser704 allele at this polymorphism is associated with increased severity of positive symptoms in our cohort (9), has been identified as a risk allele for SZ (5) and is associated with reduced NDEL1 expression in the HC of SZ patients (23).
We further report no direct association between NDEL1's homolog, NDE1, and risk for SZ; however, our analyses did reveal initial evidence for an interaction between NDE1 and DISC1 on SZ susceptibility, although this was a much weaker relationship than that noted between NDEL1 and DISC1 and not statistically indicative of epistasis. Of note, the pattern of risk associated with NDE1 genotype relative to DISC1 Ser704Cys was the opposite of that seen with NDEL1 genotype; specifically, an increased risk due to NDE1 variation was present only in DISC1 Cys carriers and not in DISC1 Ser homozygotes.
Both NDEL1 and NDE1 are essential components of the dynein motor at the centrosome which is required for normal neurodevelopmental processes, including neuronal migration in the cerebral cortex and adult hippocampal neurogenesis (12,34,35). DISC1 also has a parallel role in the dynein complex, playing a key regulatory role in neurodevelopmental processes implicated in the pathophysiology of SZ (24,25,29,35–38). In addition, NDEL1 is labeled by its enzyme activity in vitro as an endo-oligopeptidase (39) and may be a protein of particular interest as a potential treatment target for novel antipsychotic medications which act as neurotensin agonists (40,41). Interaction of DISC1 and NDEL1 may be important, at least as evidenced by its crucial role in neurite outgrowth of differentiating PC12 cells (24).
Our results are consistent with both expression data, in which the DISC1 Ser704 allele predicted reduced NDEL1 expression in SZ patients (23), and protein-binding data, which suggest that the Ser704 protein binds less readily to NDEL1 than does the Cys704 protein (24). Our interaction data of DISC1, NDEL1 and NDE1 are compatible with a recent report of significant linkage between SZ and the chromosomal region containing the NDEL1 homolog, NDE1, in a subset of families conditioned on the presence of a DISC1 risk haplotype (26). As the relationship between the protein products of these homologous genes (NDEL1 and NDE1) has not yet been elucidated, these findings form an important link to our results from the second stage of the current study.
Specifically, we have now demonstrated an epistatic interaction between DISC1 and NDEL1 at the gene level, which follows logically from the previous biological evidence supporting an interaction at the protein level. Likewise, Hennah et al. (26) provided evidence of gene–gene interaction between DISC1 and NDE1, supported by our association results, and in our stage two analyses we provide initial data describing a biological association between NDE1 and DISC1, with an opposite pattern of binding as that reported by our group for NDEL1 (24). These data may indicate that both NDEL1 and NDE1 are independently implicated in the pathophysiology of SZ, or rather, this may suggest that imbalanced interaction of NDEL1 and NDE1 with DISC1 disturbs coordinated functions of the DISC1-associated protein complex. Although additional studies will be required to fully understand the implications of our hypothesized competitive binding model of NDEL1 and NDE1 with regard to DISC1 Ser704Cys, disturbance of this pattern may disrupt normal neurodevelopment. Specifically, expression profiles of NDEL1 versus NDE1 differ during the developmental period marked by neuronal maturation, with NDEL1 more prominently expressed. Although speculative, a perturbation of this balance may result in abnormal binding of these proteins to DISC1 and interfere with normal neuron growth, offering a possible explanation for the observation in brain imaging that the DISC1 Ser704 allele is associated with reduced gray matter in the HC (5).
In summary, we present preliminary evidence indicating an epistatic interaction between NDEL1 and DISC1, as the effect of NDEL1 on risk for SZ appears to be reliant on a background of DISC1 Ser704Ser genotype. Further, we report new evidence in support of a shared binding domain for NDEL1 and NDE1 to DISC1, with opposite effects of the DISC1 Ser704Cys mutation on binding patterns. Finally, we hypothesize a potential competitive binding mechanism to explain these results. These data support the importance of assessing the interactions between putative risk genes and proteins located within known molecular networks implicated in the pathophysiology of SZ.
MATERIALS AND METHODS
Stage one
Subjects: NDEL1–DISC1 association
The study group included 275 Caucasian patients with SZ or schizoaffective disorder (30.9% female) with a mean age of 45.0 ± 10.5 years, mean education of 13.0 ± 2.2 years and an estimated IQ (based on WRAT-3 Reading) of 96.1 ± 11.8. All subjects provided written informed consent to an Institutional Review Board of the North Shore–Long Island Jewish Health System (NSLIJHS)-approved protocol. Patients were recruited from the inpatient units of the Zucker Hillside Hospital (ZHH), a division of the NSLIJHS, in Glen Oaks, NY, USA.
All patients were clinically stable at the time of assessment. Clinical characteristics that were collected included duration of illness 17.7 ± 10.6 years, age of onset (first medicated) 21.4 ± 6.3 years and global assessment of function 38.1 ± 15.2.
Caucasian healthy control subjects (n = 200) were recruited from the general population via word of mouth, newspaper and internet advertisements and posted flyers. Subjects were excluded if they had an Axis I diagnosis, active or recent substance abuse or if they had a first-degree relative with a known or suspected Axis I disorder, based on family history questionnaire. Controls were 62.0% female, had a mean age of 47.7 ± 15.7 years, mean education of 15.8 ± 2.4 years and an estimated IQ (based on WRAT-3 Reading) of 105.6 ± 9.2.
All subjects were Caucasian by self-report and drawn from a single geographic location (Glen Oaks, NY area, USA). Although population stratification is a potential confound in any case–control study, we have recently demonstrated that undetected substructure is not present in our geographically homogeneous population. In our recent whole-genome association study (30) of a case–control cohort collected by the same methods described earlier, we tested for stratification using 210 ancestry informative markers selected for maximal informativeness and observed no differences between patients and controls beyond chance levels. Moreover, in the same cohort, none of the subjects deviated from a single population as assessed by the STRUCTURE program (42).
Diagnostic measures
Patient diagnosis was established through structured interview (Structured Clinical Interview—DSM-IV; SCID-IV) (43) and confirmed by diagnostic consensus conference, which utilizes expert clinical opinion alongside SCID-IV data and corroborating medical record information. Healthy controls for the project were assessed using the Structured Clinical Interview for DSM-IV, Non-Patient Edition (43), specifically designed for healthy subjects to rule out Axis I diagnoses. In addition to the structured diagnostic interview, potential subjects are screened to rule out any history of CNS trauma, neurological disorder or previously diagnosed learning disability.
DNA procedures
Genomic DNA was extracted from venous blood samples, and six SNPs (rs12601035, rs1391768, rs1391766, rs931672, rs2012190 and rs35261231) were genotyped by 5′ exonuclease assay, using the primer-probe sets available as Taqman® Assays-on-Demand (Applied Biosystems, Foster City, CA, USA). SNPs were selected, based on available Taqman inventories at the time of genotyping, to span the entirety of NDEL1, including 5′ and 3′ flanking regions. Call rates exceeded 99% across the six SNPs. Allele frequencies, Hardy–Weinberg equilibrium (HWE) and LD structure were examined using Haploview 3.32 (44). One SNP, rs2012190, demonstrated extremely rare (<0.5%) minor allele frequency and was excluded from further analyses. Of the remaining SNPs, four formed a tight haplotype block spanning ∼36 kb and encompassing the gene, as shown in Figure 1. One SNP (rs12601035), located ∼8 kb upstream of NDEL1, was not part of the NDEL1 haplotype block and was also excluded from further analyses. For the four SNPs in the NDEL1 haplotype block, phase and diplotype assignment were estimated using PHASE 2.1.1 (45). Genotyping for DISC1 Ser704Cys was conducted as described previously (4).
NDEL1 association with illness
None of the four SNPs in the haplotype deviated from HWE (P-values > 0.05). As described earlier, alleles at these four SNPs formed a yin–yang haplotype. Since analyses were designed to examine DISC1 status at the level of the individual subject, and to maximize cell size for these comparisons, phased diplotype and individual genotype associations were examined under a dominant/recessive model, using df = 1 χ2 test statistics, comparing homozygotes of the most common variant versus carriers of risk variant. Odds ratios (ORs) were calculated as a measure of effect size for all association analyses.
NDEL1 and DISC1 epistasis
We tested for a genotype interaction between the best tagged SNP for the most common NDEL1 haplotype (rs1391768) and DISC1 Ser704Cys by utilizing a likelihood ratio test in an unconditional logistic regression model, with subject type (SZ versus healthy controls) as the dependent measure and entering NDEL1 rs1391768 genotype, DISC1 Ser704Cys genotype and the interaction term NDEL1 × DISC1 in a backward stepwise model. In addition, χ2 analyses were conducted to determine ORs for the significant interaction, by conditioning the sample on DISC1 Ser704Cys.
It should be noted that the secondary analyses with the NDE1 gene utilized a patient sample directly overlapping the sample used in the primary analyses (n = 267) and a slightly larger healthy control sample (n = 244) recruited and diagnosed by the same methods described earlier. These samples were comparable with the NDEL1–DISC1 sample in both demographics and illness features but were genotyped by different methods. Briefly, genomic DNA was extracted from whole blood and hybridized to two chips containing ∼262 000 and ∼238 000 SNPs on the basis of the manufacturer’s (Affymetrix, Santa Clara, CA, USA) specifications. Patients and controls were proportionally distributed on each 96-well plate. Genotype calls were made using Bayesian robust linear model with Mahalanobis distance classifier algorithm threshold at 0.5 applied to batches of 100 samples. Mean call rates <90% on both chips (or <85% on one chip) were rejected, resulting in a mean call rate for the retained sample of 97%. For additional detail on genotyping methodology, see the work of Lencz et al. (30). Statistical procedures to test for association between NDE1 and SZ and interactions between NDE1 and DISC1 were carried out identically to those described earlier for NDEL–DISC1 analyses.
Stage two
Plasmids and antibodies
The deletion DISC1 expression construct was made by PCR-based mutagenesis protocol (24). The following antibodies were also used: mouse monoclonal antibody against γ-tubulin (Sigma-Aldrich, St Louis, MO, USA); mouse monoclonal antibodies against HA-tag and myc-tag (BAbCO, Berkeley, CA, USA); rabbit polyclonal antibody against HA-tag (Clontech, Mountain View, CA, USA); rabbit polyclonal antibody against myc-tag (Santa Cruz, Santa Cruz, CA, USA). Rabbit polyclonal anti-GAPDH antibody was prepared as described previously (46). Rabbit polyclonal anti-DISC1 antibody (D27) was a gift from Dr Nicholas J. Brandon. Rat monoclonal antibody against NDEL1 was characterized in the previous report (24). Rat monoclonal antibody against NDE1 was newly prepared for this study, according to the established protocol (47).
Cell culture and transfection
HEK293 and COS7 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (PS). PC12 cells were maintained in DMEM with 10% FBS, 5% horse serum (HS) and 1% PS. Transfection of expression constructs was carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for PC12 cells, with PolyFect Transfection Reagent (Qiagen, Valencia, CA, USA) for HEK293 cells, and FuGENE6 (Roche Applied Science, Indianapolis, IN, USA) for COS7 cells.
Co-immunoprecipitation and cell extraction
Immunoprecipitation: Cells were lysed in a RIPA buffer (50 mm Tris–HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm ethylenediaminetetraacetic acid, 1% Triton X-100 and protease inhibitor mixture) (Roche, Basel, Switzerland). Rat adult cerebral cortexes were homogenated in the RIPA buffer. Supernatant fractions obtained after centrifugation at 14 000g for 10 min were incubated with primary antibodies (rabbit polyclonal antibody against HA-tag or against myc-tag; rat monoclonal antibody against NDE1 or against NDEL1) overnight, followed by the addition of Protein G Plus/Protein A agarose (Calbiochem, Darmstadt, Germany) for 1 h. The immunoprecipitates were analyzed with SDS–PAGE followed by western blotting after extensive washing.
Immunofluorescent staining
Cells were fixed with ice-cold methanol at –20°C 1 day after transfection. After blocking with 1.5% bovine serum albumin and 0.5% normal goat serum in PBS, cells were treated with primary antibodies for 1 h followed by the reaction with secondary antibodies conjugated to Rhodamine Red-X, Cy2 and Cy5 (Jackson ImmunoResearch, West Grove, PA, USA) for 1 h. Hoechst 33258 (Molecular Probes) was used at 1:500 dilution for 3 min to visualize nuclei. Confocal microscopy (Zeiss LSM 510 Meta, Grottingem, Germany) was used for epifluorescent image collection. To quantify the distribution of NDEL1 at the centrosome in the cells, a circle with 3 µm diameter was drawn centering on the γ-tubulin and defined as the area, including the centrosome. In all experimental groups, the immunointensity of NDEL1 in the whole cell area versus centrosome area was quantified with Image J (http://rsb.info.nih.gov/ij/). The intensity ratio of the signal of more than 30 cells per group was analyzed in three independent experiments in a blinded manner. Statistical analyses were conducted by using a one-way ANOVA followed by post hoc testing. Values depicted are means ± SEM.
Quantitative and statistical analyses
Quantitative densitometric measurement of western blotting was performed using Image J. Statistical analyses were conducted by Student’s t-test or Welch’s t-test. Values depicted are mean ± SEM.
FUNDING
This work is financially supported by grants to K.E.B., A.K., A.S. and A.K.M. from the National Institutes of Health, Stanley Medical Research Institute, and NARSAD. We gratefully acknowledge additional support from the Donald and Barbara Zucker Foundation.
ACKNOWLEDGEMENTS
We would like to thank Drs Bernice Morrow and Raju Kucherlapati for thier assistance with DNA preparation. We would like to thank Yukiko Lema for assistance in organizing the figures. We appreciate Drs Yuanyi Feng and Nick J. Brandon for GFP-tagged/myc-tagged NDE1 constructs and D27 antibody, respectively. Funding to pay the Open Access publication charges for this article was provided by The Zucker Hillside Hospital, Glen Oaks, NY.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Porteous D.J., Thomson P., Brandon N.J., Millar J.K. The genetics and biology of DISC1—an emerging role in psychosis and cognition. Biol. Psychiatry. 2006;60:123–131. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Ishizuka K., Paek M., Kamiya A., Sawa A. A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol. Psychiatry. 2006;59:1189–1197. doi: 10.1016/j.biopsych.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto R., Numakawa T., Ohnishi T., Kumamaru E., Yagasaki Y., Ishimoto T., Mori T., Nemoto K., Adachi N., Izumi A., et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum. Mol. Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkinson C.A., Goldman D., Jaeger J., Persaud S., Kane J.M., Lipsky R.H., Malhotra A.K. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 2004;75:862–872. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callicott J., Straub R.E., Pezawas L., Egan M.F., Mattay V.S., Hariri A.R., Verchinski B.A., Meyer-Lindenberg A., Balkissoon R., Kolachana B., et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2004;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdick K.E., Hodgkinson C.A., Szeszko P.R., Lencz T., Ekholm J.M., Kane J.M., Goldman D., Malhotra A.K. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- 7.Thomson P.A., Harris S.E., Starr J.M., Whalley L.J., Porteous D.J., Deary I.J. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci. Lett. 2005;389:41–45. doi: 10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Cannon T.D., Hennah W., van Erp T.G., Thompson P.M., Lonnqvist J., Huttunen M., Gasperoni T., Tuulio-Henriksson A., Pirkola T., Toga A.W., et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 9.DeRosse P., Hodgkinson C.A., Lencz T., Burdick K.E., Kane J.M., Goldman D., Malhotra A.K. Disrupted in schizophrenia 1 genotype and positive symptoms in schizophrenia. Biol. Psychiatry. 2007;61:1208–1210. doi: 10.1016/j.biopsych.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 10.Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. Disrupted in schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2006;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 11.Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol. Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 12.Shu T., Ayala R., Nguyen M.D., Xie Z., Gleeson J.G., Tsai L.H. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Toyo-Oka K., Sasaki S., Yano Y., Mori D., Kobayashi T., Toyoshima Y.Y., Tokuoka S.M., Ishii S., Shimizu T., Muramatsu M., et al. Recruitment of katanin p60 by phosphorylated NDEL1, an LIS1 interacting protein, is essential for mitotic cell division and neuronal migration. Hum. Mol. Genet. 2005;14:3113–3128. doi: 10.1093/hmg/ddi339. [DOI] [PubMed] [Google Scholar]
- 14.Hirohashi Y., Wang Q., Liu Q., Li B., Du X., Zhang H., Furuuchi K., Masuda K., Sato N., Greene M.I. Centrosomal proteins Nde1 and Su48 form a complex regulated by phosphorylation. Oncogene. 2005;25:6048–6055. doi: 10.1038/sj.onc.1209637. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Walsh C.A. Protein–protein interactions, cytoskeletal regulation and neuronal migration. Nat. Rev. Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y., Walsh C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Niethammer M., Smith D.S., Ayala R., Peng J., Ko J., Lee M.S., Morabito M., Tsai L.H. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 19.Soukoulis V., Reddy S., Pooley R.D., Feng Y., Walsh C.A., Bader D.M. Cytoplasmic LEK1 is a regulator of microtubule function through its interaction with the LIS1 pathway. Proc. Natl Acad. Sci. USA. 2005;102:8549–8554. doi: 10.1073/pnas.0502303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vergnolle M.A., Taylor S.S. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- 21.Rapoport J.L., Addington A.M., Frangou S. The neurodevelopmental model of schizophrenia: update 2005. Mol. Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 22.Lewis D.A., Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu. Rev. Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 23.Lipska B.K., Peters T., Hyde T.M., Halim N., Horowitz C., Mitkus S., Weickert C.S., Matsumoto M., Sawa A., Straub R.E., et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 24.Kamiya A., Tomoda T., Chang J., Takaki M., Zhan C., Morita M., Cascio M.B., Elashvili S., Koizumi H., Takanezawa Y., et al. DISC1-NDEL1/NDEL1 protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum. Mol. Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- 25.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell. Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 26.Hennah W., Tomppo L., Hiekkalinna T., Palo O.M., Kilpinen H., Ekelund J., Tuulio-Henriksson A., Silander K., Partonen T., Paunio T., et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum. Mol. Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 27.Numata S., Ueno S., Iga J., Nakataki M., Tanahashi T., Itakura M., Sano A., Ohi K., Hashimoto R., Takeda M., et al. No association between the NDE1 gene and schizophrenia in the Japanese population. Schiz. Res. 2008;99:367–369. doi: 10.1016/j.schres.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Brandon N.J., Handford E.J., Schurov I., Rain J.C., Pelling M., Duran-Jimeniz B., Camargo L.M., Oliver K.R., Beher D., Shearman M.S., et al. Disrupted in Schizophrenia 1 and NDEL1 form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol. Cell. Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Ozeki Y., Tomoda T., Kleiderlein J., Kamiya A., Bord L., Fujii K., Okawa M., Yamada N., Hatten M.E., Snyder S.H., et al. Disrupted in schizophrenia (DISC-1): mutant truncation prevents binding to NDE1-like (NDEL1) and inhibits neural outgrowth. Proc. Natl Acad. Sci.USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lencz T., Morgan T.V., Athanasiou M., Dain B., Reed C.R., Kane J.M., Kucherlapati R., Malhotra A.K. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., Hirotsune S.A. LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 32.Morris J.A., Kandpal G., Ma L., Austin C.P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 33.Park S.Y., Lee J.Y., Choi J.Y., Park M.J., Kim D.S. Nerve growth factor activates brain-derived neurotrophic factor promoter IV via extracellular signal-regulated protein kinase 1/2 in PC12 cells. Mol. Cells. 2006;21:237–243. [PubMed] [Google Scholar]
- 34.Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. NDEL1 contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 2006;17:680–689. doi: 10.1091/mbc.E05-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan X., Chang J.H., Ge S., Faulkner R.L., Kim J.Y., Kitabatake Y., Liu X.B., Yang C.H., Jordan J.D., Ma D.K., et al. Disrupted-in-schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taya S., Shinoda T., Tsuboi D., Asaki J., Nagai K., Hikita T., Kuroda S., Kuroda K., Shimizu M., Hirotsune S., et al. DISC1 regulates the transport of the NDEL1/LIS1/14-3-3epsilon complex through kinesin-1. J. Neurosci. 2007;27:15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyoshi K., Honda A., Baba K., Taniguchi M., Oono K., Fujita T., Kuroda S., Katayama T., Tohyama M. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol. Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 38.McGlashan T.H., Hoffman R.E. Schizophrenia as a disorder of developmentally reduced synaptic connectivity. Arch. Gen. Psychiatry. 2000;57:637–648. doi: 10.1001/archpsyc.57.7.637. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi M.A., Portaro F.C., Bastos M.F., Guerreiro J.R., Oliveira V., Gorrao S.S., Tambourgi D.V., Sant’Anna O.A., Whiting P.J., Camargo L.M., et al. Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc. Natl Acad. Sci. USA. 2005;102:3828–3833. doi: 10.1073/pnas.0500330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinkead B., Nemeroff C.B. Novel treatments of schizophrenia: targeting the neurotensin system. CNS Neurol. Disord. Drug Targets. 2006;5:205–218. doi: 10.2174/187152706776359655. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q., Jaaro-Peled H., Sawa A., Brandon N.J. How has DISC1 enabled drug discovery? Mol. Cell. Neurosci. 2008;37:187–195. doi: 10.1016/j.mcn.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.First M.B., Spitzer R., Williams J.B.W., Gibbon M. Structured Clinical Interview for Axis I DSM IV Disorders(SCIDI/P, Version 20) Patient Edition. New York: Biometric Research Department; 1998. [Google Scholar]
- 44.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 45.Stephens M., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hara M.R., Agrawal N., Kim S.F., Cascio M.B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J.H., Tankou S.K., Hester L.D., et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell. Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi N., Takanezawa Y., Koizumi H., Umezu-Goto M., Aoki J., Arai H. Expression of NUDEL in manchette and its implication in spermatogenesis. FEBS Lett. 2004;566:71–76. doi: 10.1016/j.febslet.2004.04.009. [DOI] [PubMed] [Google Scholar]