Abstract
Recent years have seen unprecedented characterization of mammalian chromatin thanks to advances in chromatin assays, antibody development and genomics. Genome-wide maps of chromatin state can now be readily acquired using microarrays or next-generation sequencing technologies. These datasets reveal local and long-range chromatin patterns that offer insight into the locations and functions of underlying regulatory elements and genes. These patterns are dynamic across developmental stages and lineages. Global studies of chromatin in embryonic stem cells have led to intriguing hypotheses regarding Polycomb/trithorax and RNA polymerase roles in ‘poising’ transcription. Chromatin state maps thus provide a rich resource for understanding chromatin at a ‘systems level’, and a starting point for mechanistic studies aimed at defining epigenetic controls that underlie development.
Introduction
Once conceived as an inert scaffold to package feet of DNA into each cell, chromatin is now recognized to have a multiplicity of functions in genome regulation. The basic building block of chromatin, the nucleosome, comprises an octamer of histone proteins wrapped by ~146 basepairs of DNA. Histone proteins are subject to over one hundred known post-translational modifications, including acetylation, methylation, ADP-ribosylation, ubiquitination, and phosphorylation [1]. These modifications occur on the side chains of specific residues in the histone tails and cores and functionally impact transcription, replication, recombination, and repair. Accordingly, chromatin modifying proteins can show cell type specific phenotypes in gain- or loss-of-function studies [1–3]. Lysine acetylation and methylation are both reversible processes thanks to the activities of histone deacetylases and recently discovered demethylases [1,4]. Both of these enzyme families are implicated in a range of developmental and physiologic pathways as well as in malignancy [1,5,6].
Our ability to interrogate chromatin structure has been transformed in the past few years by a convergence of advances in chromatin antibody development and genomic technologies. Since its introduction 20 years ago, the chromatin immunoprecipitation (ChIP) assay has gradually become a mainstay for chromatin biology experimentation [7–9]. This procedure uses specific antibodies to enrich genomic DNA associated with a particular histone modification or chromosomal protein in vivo. The enriched DNA can then be interrogated by PCR (ChIP-PCR), microarray hybridization (ChIP-chip), or high-throughput sequencing (ChIP-Seq). PCR can be used to query a limited number of genomic positions – e.g., a small panel of known promoters. In contrast, high-density microarrays with probes placed at regular intervals (‘tiling arrays’) can be used to query large domains such as chromosomes or the whole genome, although the latter can be costly (reviewed in [10]). More recently, next-generation sequencing platforms have been applied to chromatin state mapping [11,12]. These new technologies can sequence ChIP DNA at sufficient depth to enable accurate and high-resolution whole genome analysis. Several technical advantages, including rapid throughput, high resolution, better genome coverage and the ability to interrogate comparatively small amounts of ChIP DNA, make the sequencing approach particularly powerful.
The power of genomic technologies to inform accurately on biology is contingent on high quality reagents. A vast number of chromatin modifications and structural proteins have been identified to date, and an immense number of antibody reagents have been raised against these epitopes. However, the specificity of these reagents and their efficacy in ChIP may vary widely depending on the different epitopes, antibody sources, and bleeds. As with any chromatin assay, precise standardization of these starting reagents is essential for effective genome-scale characterization.
Genome-scale chromatin assays also require large numbers of cells, and homogeneity of the input population is critical for clarity of data interpretation. Because of this requirement, most experiments to date have focused on cell lines that can be readily expanded in culture. However, recent studies have leveraged techniques in stem cell biology, in vitro differentiation and cell sorting to obtain populations of high biological interest. Genome-wide maps for a number of chromatin marks have now been reported for human and mouse embryonic stem (ES) cells, neural progenitor cells, fibroblasts, hepatocytes and T-cells [11–16].
Chromatin landscape of embryonic stem cells
Chromatin packaging in ES cells is of particular interest as increasing evidence suggests that it plays a critical role in maintaining pluripotency [17]. Since ES cells are derived from the inner cell mass of the blastocyst at a developmental time point that follows large-scale erasure and re-establishment of epigenetic modifications, chromatin state maps for ES cells may provide unique insights into the reprogramming process. The maps also offer a framework for understanding differentiation and lineage-specification, and how these processes alter and are altered by the chromatin state.
The chromatin landscapes of human and mouse ES cells have been systematically characterized by a series of large-scale studies over the past few years [11,13–16,18,19]. Overall, the studies display a remarkable degree of concordance.
Chromatin marks of initiating and elongating RNA polymerase II: poised polymerase
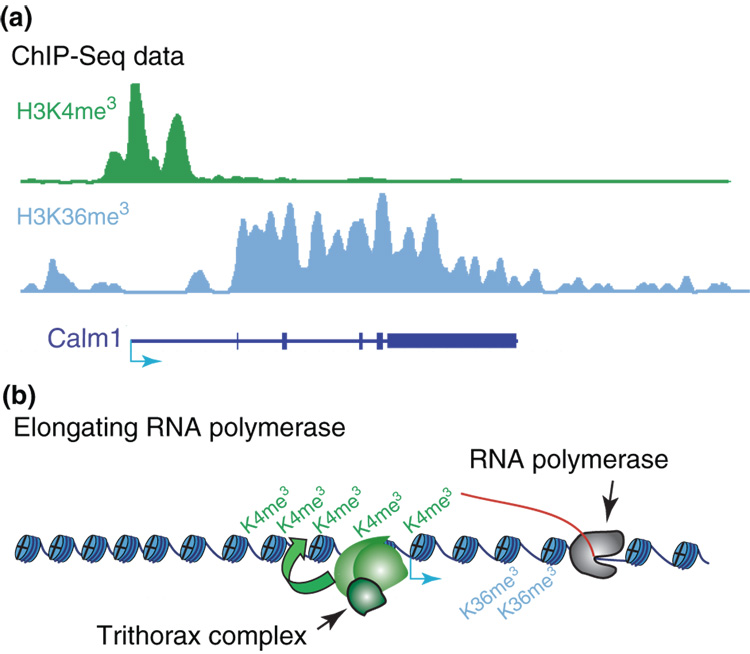
Studies in the model system S. cerevisiae correlated histone H3 lysine 4 (H3K4) tri-methylation with RNA polymerase II initiation, while H3 lysine 36 (H3K36) tri-methylation has been correlated with elongation [20]. Similar results have been obtained for mammalian cells where genome maps of H3K4 and H3K36 tri-methylation reveal the promoters and transcribed regions of protein coding genes as well as non-coding transcripts (Figure 1) [11–13,21,22].
Figure 1. Active Gene Chromatin Pattern.
Chromatin state maps reveal a stereotypical pattern at active genes. (A) In mouse ES cells, the transcription start site for the Calm1 gene (blue arrow) is marked by H3K4 tri-methylation, a trithorax-associated mark, while the remainder of the transcribed region is marked by H3K36 tri-methylation [11]. (B) Evidence from model systems supports a central role for initiating and elongating RNA polymerase II in recruiting the relevant histone methyltransferase enzymes [20].
In ES cells, H3K36 tri-methylation levels as determined by ChIP-Seq or ChIP-chip correlate strongly with RNA expression levels. H3K4 tri-methylation also correlates with expression, but to a lesser extent. This reflects the fact that in ES cells virtually all promoters with annotated CpG islands or otherwise high CG-density carry H3K4 tri-methylation [11,13–15]. Many CG-rich promoters regulate constitutive house-keeping genes, but others correspond to developmental regulators and signaling proteins that are not expressed in ES cells (see below). The roughly 25% of promoters that lack H3K4 tri-methylation have low CG content, frequently exist in clusters, and tend to correspond to clustered genes that encode tissue-specific proteins such as olfactory receptors or keratin proteins [11,13].
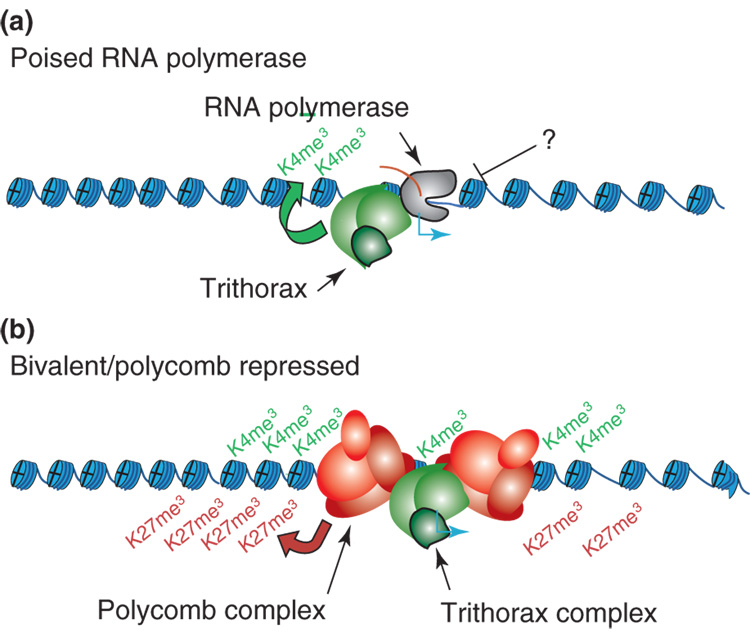
The disconnect between H3K4 tri-methylation and expression may reflect an unexpectedly widespread role for ‘poised polymerase’ at inactive promoters [13]. RNA polymerase II can be detected at the promoters of >50% of genes, including many that do not show evidence of elongation as judged by mRNA expression and H3K36 tri-methylation levels. Many of these ‘inactive’ promoters generate short abortive transcripts, suggesting they are initiating but are unable to transition to elongation [13]. Thus, regulation of the transition to elongation may play an important role in titrating gene expression and in keeping certain inactive genes poised for induction (Figure 2). Although the underlying mechanisms remain elusive, the phenomenon appears in other models, including B-cells and T-cells and Drosophila [12,13,23].
Figure 2. Inactive Gene Promoters in ES Cells.
Models of gene regulation in ES cells. (a) H3K4 tri-methylation and an initiating form of RNA polymerase II are evident at promoters of some inactive genes. These genes may be regulated at the level of the transition from initiation to elongation [13]. (b) A significant subset of inactive genes in ES cells, including many lineage-specifying transcription factor genes, are dually marked by tri-methylation at H3K4 (active) and at H3K27 (repressive). The relevant genes are largely silent in ES cells. The presence of the activating mark may keep them poised for activation upon ES cell differentiation [18,31].
So what is the functional relevance of H3K4 tri-methylation at active and inactive promoters? This mark may serve as a general means for recruiting the transcriptional machinery or ensuring its access to the promoter. Indeed, several components of transcriptional machinery contain domains that recognize the H3K4 tri-methylated histone tail [24–26]. Tissue specific expression of these genes would then be controlled by additional factors that titrate initiation rates or regulate the transition to elongation [13,15,20]. In addition H3K4 tri-methylation may protect inactive CG-rich promoters from DNA methylation by preventing engagement by the de novo DNA methyltransferase Dnmt3L [27,28].
Bivalent Chromatin Domains and Polycomb in Pluripotency
An extensive body of literature, largely focused on the Drosophila model system, has defined fundamental roles for trithorax and Polycomb protein complexes in the establishment and maintenance of lineage-specific gene expression [29,30]. Trithorax proteins are a diverse set of transcriptional activators, several of which catalyze or physically interact with H3K4 methylation. Polycomb proteins are transcriptional repressors that catalyze and bind Histone H3 lysine 27 (H3K27) methylation. The patterns of H3K27 tri-methylation and Polycomb protein binding in ES cells have been systematically characterized by a series of studies [11,14–16,18,19]. The repressive modification is evident at roughly 20% of promoters in ES cells. Its localization correlates with that of the Polycomb repressive complex 2, which catalyzes H3K27 tri-methylation [16,19]. Genes targeted by Polycomb and marked by H3K27 tri-methylation are transcriptionally silent in ES cells and include a large number that encode developmental regulators as well as key signaling proteins.
The unexpected observation that virtually all Polycomb targets in ES cells display the activating H3K4 tri-methylation mark in addition to the repressive H3K27 tri-methylation mark led to the hypothesis that a ‘bivalent’ chromatin state may poise these genes for subsequent activation (Figure 2) [18,31]. Consistent with this model, a high proportion of Polycomb targets, including key developmental regulators, are rapidly induced upon ES cell differentiation. The activated genes become selectively marked by H3K4 tri-methylation in differentiated populations. In contrast, non-induced genes tend to lose the activating mark and selectively acquire repressive H3K27 tri-methylation (Figure 3).
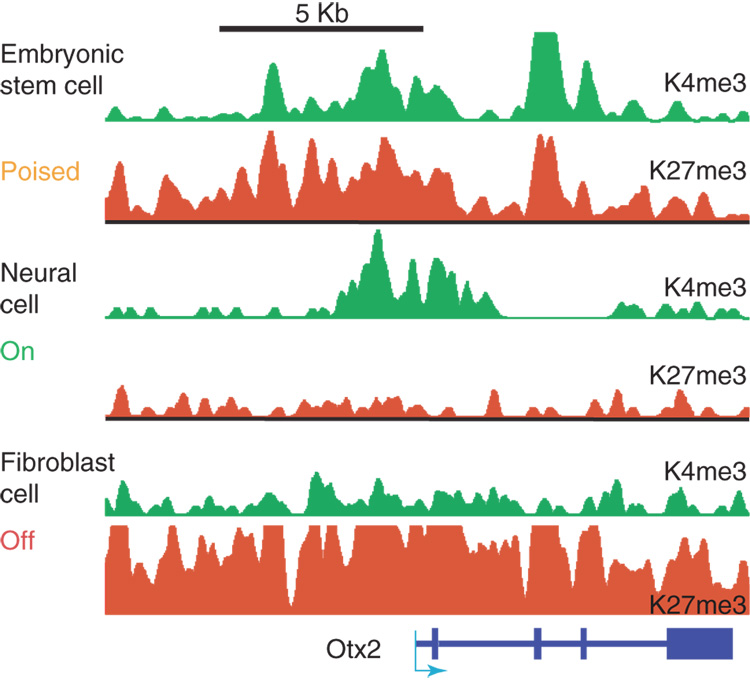
Figure 3. Bivalent Domain Resolution.
Resolution of a bivalent chromatin domain during neural differentiation of mouse ES cells. Chromatin state for an 18 KB region at the developmental locus encoding Otx2 (a neural specific transcription factor). In ES cells, the gene is expressed at very low levels and its promoter region is organized into chromatin that carries both H3K4 and H3K27 tri-methylation. In neural progenitors Otx2 is expressed and is associated with H3K4 tri-methylation. In embryonic fibroblasts, the gene is inactive and marked by a broad domain of H3K27 tri-methylation [11].
Role of bivalent domains in differentiated cells
Studies on multipotent and committed cells offer insight and generate new questions regarding the bivalent domain hypothesis [11,18]. In ES cell–derived neural progenitors, most bivalent domains resolve in accord with expression changes (Figure 3). A small subset of bivalent promoters are retained in neural progenitors, and many correspond to genes with glial or neuronal functions whose fate is yet to be determined. A key issue remains whether the retention of H3K4 tri-methylation indeed renders these promoters more readily activated than those that retain only H3K27 tri-methylation is an open question. Such a finding could suggest a broad role for bivalent domains in hierarchical differentiation. However, other studies have revealed potential bivalent promoters in differentiated T-cells as well as evidence for de novo formation of bivalent chromatin during ES cell differentiation (e.g., at the OCT4 promoter) [12,14]. These findings are not readily explained by such a hierarchical model.
An open question remains as to why bivalent chromatin is evident at so many promoters in ES cells. Estimates of the number of bivalent promoters from various studies are fairly consistent at around two to three thousand genes [11,14,15]. In contrast, differentiated cells contain far fewer examples [11]. The complete set of bivalent genes could represent key regulatory targets of Polycomb and trithorax that are inactive in ES cells but important for downstream differentiation pathways. However, an alternate explanation is that only a subset of the bivalent genes are subject to such epigenetic controls, while others reflect distinct phenomena. Notably, those targets that encode key developmental regulators are marked by particularly expansive regions containing robust signals for the opposing histone modifications [11,16]. Furthermore, only a few ‘bivalent’ targets have been rigorously confirmed by sequential ChIP to have concomitant H3K4 and H3K27 tri-methylation on the same chromatin segment. Identification of the most relevant set of gene targets and the functional import of the bivalent chromatin state in ES cells awaits further investigation.
Although clearly distinct, the stalled polymerase and bivalent chromatin domain models are not mutually exclusive. A recent study found evidence for poised polymerase at a panel of bivalent gene promoters in ES cells [32].
Chromatin domains, boundaries and insulators
The uninterrupted, high resolution maps of chromatin modification patterns across genomic loci achieved with tiling arrays and ChIP-Seq methodology have revealed remarkably expansive regions with continuous chromatin markings. Domains of H3K27 methylation, H3K4 methylation and/or histone acetylation extending 10 or even 100 kilobases are evident at the mammalian Hox clusters and other evolutionarily conserved developmental loci [16,21,33]. The domain patterns vary among different cell types and likely function in the maintenance of gene expression or repression. Chromatin domains are attractive models of ‘epigenetic’ memory as their large size could ensure robust transmission through cell division [34,35].
Studies in model organisms have identified specific sequence elements capable of halting the propagation of a given chromatin state along the chromosomal DNA [36]. These boundary elements play an important role in defining chromatin domain structure in such organisms. The mechanisms of boundary formation in mammalian cells remain poorly understood, but some insights have emerged in recent publications.
Non-coding RNAs, well known for their roles in silencing the inactive X chromosome and other loci, are potential candidates for regulating the establishment of chromatin domains and their boundaries [37–40]. A recent microarray study identified many non-coding RNAs in the mammalian Hox clusters [40]. Remarkably, antisense knock-down of a non-coding RNA expressed at the edge of a chromatin domain in HoxC caused expansion of a chromatin domain in HoxD. The authors proposed that an interaction between the non-coding RNA and the Polycomb protein Suz12 mediates silencing of the HoxD locus in trans [40]. Global studies are documenting an increasing repertoire of RNAs without apparent coding potential [41]. Although speculative, the potential for non-coding RNAs to impact the chromatin landscape is considerable.
Chromatin insulators may also demarcate chromatin boundaries. In Drosophila, the gypsy insulator element can block the silencing influence of a Polycomb response element and halt the spread of associated H3K27 tri-methylation [42]. In vertebrates, the major insulator protein identified is CTCF, which binds to a highly conserved DNA sequence motif [43,44]. Studies using ChIP-Chip and ChIP-Seq found that CTCF binding is frequently invariant across tissues [12,43]. CTCF often binds between genes that are in close genomic proximity, but have distinct expression patterns and opposing chromatin modifications [12,43]. Additional CTCF binding sites are evident between alternative promoters of a single gene, and may allow tissue specific promoter usage [43]. A functional role for CTCF in influencing the locations and boundaries of mammalian chromatin domains, though intriguing, has yet to be documented.
Chromatin signatures of functional genomic elements
State maps have also revealed chromatin signatures associated with a variety of other functional genomic elements. By mapping an array of histone modifications and chromatin proteins across a common 30 MB portion of genome, a common signature of enhancer elements has been identified [45,46]. It was found that the presence of the histone acetylase p300 along with mono-methylated and di-methylated H3K4 could effectively predict novel enhancers and distinguish them from promoters, which are marked instead by tri-methylated H3K4. This chromatin signature was used to identify a novel enhancer element for the SLC22A5 gene [45]. Notably, the enhancer elements also tend to show hypersensitivity to DNase in genomic assays, but lack the nucleosome-free region shown to be a general characteristic of transcription start sites [47,48].
More broadly, chromatin state maps may be generally applicable for identifying the locations and cell type-specificities of functional DNA elements. An example includes the combined use of H3K4 and H3K36 tri-methylation to identify promoters and primary transcripts of microRNAs and other small RNAs [11]. These entities are otherwise difficult to characterize due to rapid processing in the nucleus. In addition, the repressive histone modification H3K9 tri-methylation marks certain repeat classes as well as imprinting control regions, and may thus also have potential for identifying novel sites of epigenetic regulation [11,49].
Future directions
Technological developments are transforming our ability to interrogate chromatin state and introducing exciting opportunities as well as formidable challenges. It is becoming both routine and cost-effective to acquire genome-wide maps for a rapidly expanding panel of chromatin epitopes in any cell type that can be obtained in sufficient numbers. State maps will provide a powerful and increasingly routine means for characterizing novel chromatin marks; i.e., by comparing their localization to known modifications and genome annotations, and/or following their dynamics upon perturbation of cell state. Alternatively, the phenotype and differentiation potential of a stem cell population might be ‘read out’ by mapping informative chromatin modifications and comparing the chromatin patterns against in vivo tissues. Once a critical mass of these ‘normal’ cell populations has been characterized, the chromatin states could be compared against cell models of human disease to potentially identify underlying defects.
The next few years will see the generation of a vast number of chromatin state maps. Effective interpretation of this data windfall will require major investment in computational infrastructure. A single state map consists of millions of contiguous datapoints (depending on resolution) that describe enrichment as a function of genomic position. Enriched regions, domains and chromatin boundaries must be defined systematically – suitable algorithms are under development, although they will need to be streamlined in order to handle the sheer mass of data and made generally accessible. Maps for multiple modifications and different cell types must then be integrated and evaluated in the context of existing data and genome annotations.
Significant challenges remain at the levels of cell biology, reagent standardization and computational analysis before the full potential of emerging chromatin state mapping technologies can be realized. However, as each of these issues is addressed, we will be rewarded with systems-level views that greatly enhance our understanding of chromatin structure, its role in normal development, and how its dysfunction contributes to disease pathology.
Acknowledgements
We thank Richard Koche, Andrew Chi and Mezher Adli for critical reading of the manuscript. E.M. is supported by an institutional training grant from the National Cancer Institute. Research by the authors is supported by funds from the National Human Genome Research Institute, the Burroughs Wellcome Fund, the Culpeper Foundation, the Harvard Stem Cell Institute, Massachusetts General Hospital, and the Broad Institute of MIT and Harvard.
Glossary
- ChIP
Chromatin Immunoprecipitation
- ChIP-chip
Chromatin Immunoprecipitation followed by microarray (chip) analysis of the DNA
- ChIP-Seq
Chromatin Immunoprecipitation followed by deep sequencing analysis
- H3K4
Histone H3 lysine 4, which when tri-methylated is associated with CG-rich gene promoters, additionally, mono- and di-methylated forms can be associated with enhancers
- H3K27
Histone H3 lysine 27, which when methylated by Polycomb group proteins is associated with transcriptional repression
- Suz12
Suppressor of zeste 12 homologue, a polycomb group protein that is involved with H3K27 methylation and chromatin silencing
- CTCF
CCCTC-binding factor protein, a sequence-specific DNA binding protein with 11 zinc fingers domains that can block (insulate) enhancer activity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 3.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 6.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 8.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 9.Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 10.Mockler TC, Chan S, Sundaresan A, Chen H, Jacobsen SE, Ecker JR. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- ** 11.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ** 12.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009.The Mikkelsen et al and Barski et al. papers report the first applications of next-generation sequencing to acquire genome-wide maps of histone modifications. Mikkelsen et al. focused on embryonic and differentiating stem cells, and identified new bivalent chromatin marks as well as specific chromatin signatures for a number of additional genomic features. Barski et al. mapped 20 different chromatin epitopes in purified human T Cells, and identified chromatin modifications associated with active or repressed genes, as well as insulators, enhancers and transcripts.
- ** 13.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042.This study describes RNA polymerase at a surprising number of inactive gene promoters, many of which appear to be initiating despite the absence of full length transcripts. The finding proposes an unexpectedly wide role for poised RNA polymerase in gene regulation.
- * 14.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- * 15.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung W-K, Shahab A, Kuznetsov VA, et al. Whole-Genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004.The studies by Pan et al. and Zhao et al. investigated chromatin patterns in human ES cells. Their findings are highly consistent, despite the use of distinct methodologies. Overall, the data suggest that the chromatin state of ES cells, including the prevalence and distribution of bivalent domains, is highly conserved between human and mouse.
- 16.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 19.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007 doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 25.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of Trimethylated Histone H3 Lysine 4 Facilitates the Recruitment of Transcription Postinitiation Factors and Pre-mRNA Splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 28.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 32.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 33.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;105:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 36.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 37.Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 38.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. Rna. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 42.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 43.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048.This study defines a consensus CTCF binding motif and describes over 13,000 CTCF binding sites in the human genome. This analysis suggests that CTCF binding is largely invariant across tissues.
- * 44.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104.A computational analysis of evolutionarily-conserved non-coding DNA segments identified numerous long DNA sequence motifs, including a novel CTCF binding motif that appears to partition genomic regions with distinct regulation.
- ** 45.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- ** 46.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874.The ENCODE Consortium used tiling arrays covering 30 megabases of the human genome to gather over 200 data sets, including profiles of transcription factor binding, histone modifications, chromosomal proteins and DNase hypersensitivity. They identify , among many other things, chromatin signatures of enhancers and promoters. Heintzman et al. applied a similar approach to identify discriminatory signatures for these elements.
- 47.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genomescale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 48.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 49.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]