Abstract
Microtubules are a major structural component of the cytoskeleton and participate in cell division, intracellular transport, and cell morphogenesis. In the present study, 795 cotton tubulin expressed sequence tags were analysed and 19 β-tubulin genes (TUB) cloned from a cotton cDNA library. Among the group, 12 cotton TUBs (GhTUBs) are reported for the first time here. Transcription profiling revealed that nine GhTUBs were highly expressed in elongating fibre cells as compared with fuzzless-lintless mutant ovules. Treating cultured wild-type cotton ovules with exogenous phytohormones showed that individual genes can be induced by different agents. Gibberellin induced expression of GhTUB1 and GhTUB3, ethylene induced expression of GhTUB5, GhTUB9, and GhTUB12, brassinosteroids induced expression of GhTUB1, GhTUB3, GhTUB9, and GhTUB12, and lignoceric acid induced expression of GhTUB1, GhTUB3, and GhTUB12. When GhTUBs were transformed into the Saccharomyces cerevisiae inviable mutant, tub2, which is deficient in β-tubulin, one ovule-specific and eight of nine fibre-preferential GhTUBs rescued this lethality. This study suggests that the proteins encoded by cotton GhTUBs are involved during cotton fibre development.
Keywords: fuzzless-lintless mutant, Gossypium hirsutum (cotton) fibre, phytohormone, β-tubulin
Introduction
Upland cotton (Gossypium hirsutum) accounts for most of the world cotton fibre production for the textile industry. Cotton fibres are single-celled trichomes that are similar to Arabidopsis trichomes, differentiated from about 15–25% of the ovule epidermal cells (Basra and Malik, 1984; Kim and Triplett, 2001). Fibre cell development occurs through fibre initiation, cell elongation, cell wall deposition, and maturation, and is a highly regulated, fundamental biological process. Fibre cells are initiated on the day of anthesis and elongate up to several centimetres without cell division. This process provides a unique system for studying cell elongation and cell wall and cellulose biosynthesis (Kim and Triplett, 2001). Although microarray transcriptome profiling of cotton unique expressed sequence tags (EST) identified a number of fibre-specific/preferential genes involved in phytohormone biosynthesis, lipid biosynthesis, and cytoskeleton and cell wall structures (Ji et al., 2003; Arpat et al., 2004; Shi et al., 2006; Gou et al., 2007; Qin et al., 2007a), the molecular mechanism of fibre cell elongation is not fully understood.
Microtubules play an important role in plant cell morphology and development (Kopczak et al., 1992). For example, microtubule depolymerization by a specific antagonist causes loss of directionality of root hair growth (Bibikova et al., 1999). Microtubule reorientation is the key in changing the growth orientation leading to Arabidopsis trichome branching (Mathur and Chua, 2000). The principle component of microtubules is a heterodimer of highly conserved α- and β-tubulin, encoded by multigene families (Silflow et al., 1987; Goddard et al., 1994; Nogales et al., 1998). Microtubule nucleation at microtubule-organizing centres is mediated by γ-tubulin (Pastuglia et al., 2006), microtubule dynamics, organization, and establishment of polarity controlled by microtubule-associated proteins (Whittington et al., 2001; Sedbrook, 2004; Ambrose et al., 2007; Korolev et al., 2007; Wang et al., 2007). Arabidopsis has six α-tubulin genes (TUA), nine β-tubulin genes (TUB), and two γ-tubulin genes (Kopczak et al., 1992; Snustad et al., 1992; Liu et al., 1994). Seven TUAs and six TUBs are found in Zea mays (Montoliu et al., 1990; Villemur et al., 1994), eight TUBs are found in Oryza sativa (Yoshikawa et al., 2003), and eight TUAs and 20 TUBs are found in Populus (Oakley et al., 2007). Most tubulin genes show differential and tissue-specific expression patterns (Snustad et al., 1992; Cheng et al., 2001; Oakley et al., 2007), implying that they are regulated by developmental signals and play roles in specific tissues. In cotton, seven of nine TUAs are highly expressed in developing fibres (Dixon et al., 1994; Whittaker and Triplett 1999; Li et al., 2007). Among the seven TUBs previously reported, two accumulate in 20 d post-anthesis (dpa) fibre cells (Dixon et al., 1994). GhTUB1 and GhTUB9 are the only cotton β-tubulins to be characterized (Ji et al., 2002; Li et al., 2002).
To understand the overall contribution of tubulins in cotton fibre development, 19 tubulin genes were cloned, including 12 TUBs newly identified by in-depth analysis of cotton ESTs. The regulation of TUBs in response to different phytohormones and by very-long-chain fatty acids, molecules that are important for fibre cell elongation, was studied (Shi et al., 2006; Qin et al., 2007a). A complementation analysis was performed using a non-viable yeast mutant to examine the function of all TUBs that are preferentially expressed in fibres as well as certain non-specifically expressed TUBs. Several of the tubulins did not complement the yeast mutant, indicating that different tubulins have distinct functions despite the fact that the tubulin gene family is highly conserved.
Materials and methods
Plant materials
Upland cotton (G. hirsutum L. cv. Xuzhou142) and a fuzzless-lintless (fl) mutant (Zhang and Pan, 1992) were planted in the field. Ovules were excised from cotton bolls, and fibre cells were scraped from the epidermis of the ovule, frozen in liquid nitrogen, and stored at –80 °C before RNA extraction. Other tissues were obtained from cotton plants grown in the field or a fully automated greenhouse as described (Ji et al., 2003).
Semi-quantitative reverse transcription (RT)-PCR and real-time quantitative (QRT)-PCR analysis of GhTUB expression in cotton tissues
The wild-type (wt) ovules were collected from –3, 0, 5, 10, 15, and 20 dpa cotton flowers. The fibres were stripped from ovules at different developmental stages. Total RNA was extracted from wt cotton roots, leaves, stems, ovules, fibres, and fl mutant ovules. The wt ovules collected from 0 dpa were used for in vitro cultures supplemented with different phytohormones or very-long-chain fatty acids. Cotton cDNA was reverse transcribed from 5 μg total RNA. Gene-specific primers were designed (Table 1), and QRT-PCR was carried out using the SYBR green PCR kit (Applied Biosystems) in a DNA Engine Opticon-Continuous Fluorescence Detection System (MJ Research). The cotton ubiquitin gene, UBQ7 (accession no. AY189972), was used as an internal control in each reaction. Samples were analysed in triplicate using independent RNA samples and were quantified by the comparative cycle threshold method (Wittwer et al., 1997).
Table 1.
Primers used in the current study
| Gene | Primers used for RT-PCR or QRT-PCR amplification and expression profiling |
| GhTUB1 | 5′-CACTGTTTGTGACATCCCTC-3′ |
| 5′-TCTCATCCATCCCTTCTCC-3′ | |
| GhTUB2 | 5′-TTTGTGACATCCCACCCACT-3′ |
| 5′-ATGGCAATAACTTTCCTTCTCC-3′ | |
| GhTUB3 | 5′-AAATGAGCACCAAGGAAGTT-3′ |
| 5′-AATCCCAGCACAAATGAAAA-3′ | |
| GhTUB4 | 5′-GAGATGGAGTTTACGGAGGCTGAG-3′ |
| 5′-TCTAACACCCAAACAAGGTATTCAG-3′ | |
| GhTUB5 | 5′-GAGTACCAGCAATACCAGG-3′ |
| 5′-AAGAAATCAATCCATCAAA-3′ | |
| GhTUB6 | 5′-GAGGAAGAGTACGAGGGGG-3′ |
| 5′-AAAGGAACGAACAGAGCAG-3′ | |
| GhTUB7 | 5′-ACAGAAGCGGAAAGTAACA-3′ |
| 5′-AATAAACAAGCCAAAGTGA-3′ | |
| GhTUB8 | 5′-ATCAGCAATACCAGGACGC-3′ |
| 5′-AAACACACCAATAGCCACA-3′ | |
| GhTUB9 | 5′-AACTCTTCCTACTTCGTCG-3′ |
| 5′-CCAAATCTTCCTCATCCTC-3′ | |
| GhTUB10 | 5′-GCTTTCTTGCACTGGTACAC-3′ |
| 5′-TAATCGCCAAACTTCGCTCT-3′ | |
| GhTUB11 | 5′-TCACAGAAGCAGAGAGCAAC-3′ |
| 5′-CAAAGGAACGAACAGAGCAG-3′ | |
| GhTUB12 | 5′-AAACCTTATTCCATTCCCTCG-3′ |
| 5′-TCACCCTCCTGAACATCTCTTG-3′ | |
| GhTUB13 | 5′-CACAGAAGCAGAAAGCAACA-3′ |
| 5′-ATACGAGCAAACCAGAAAAG-3′ | |
| GhTUB14 | 5′-ACGAGTATGAGGAAGGAGAG-3′ |
| 5′-AATGCAGAGACACAAGAAAT-3′ | |
| GhTUB15 | 5′-TACAAAGCCTTAACTGTCC-3′ |
| 5′-AACTCCATTTCATCCATCC-3′ | |
| GhTUB16 | 5′-GAATCTCATTCCCTTCCCC-3′ |
| 5′-ACTGTTCACTCACACGCCT-3′ | |
| GhTUB17 | 5′-AAAACAAGAACTCATCCTAC-3′ |
| 5′-ATACACATCAAAATAACTAA-3′ | |
| GhTUB18 | 5′-GAGGATTAGGAAGGAAAAAC-3′ |
| 5′-ATGAAAAGCTCAAGAAAAAA-3′ | |
| GhTUB19 | 5′-CAGTTTGTGACATTCCACCCA-3′ |
| 5′-TCCTAATCCTCGTACTCCGCT-3′ |
| Gene | Primers used for construction of all YCplac33-GhTUBs |
| GhTUB1 | 5′-CCGtctagaATGAGAGAAATCCTTCACATCCAA-3′ |
| 5′-CCGctcgagTTAAGCCTCTGCCTCGTATTCC-3′ | |
| GhTUB3 | 5′-CCGtctagaATGAGAGAAATCCTCCATGTTCAAGCC-3′ |
| 5′-CCGctcgagTCAATTTTCCTCAGCTTCATCTTCATAC-3′ | |
| GhTUB4 | 5′-CCGtctagaATGCGTGAGATTCTTCATATTCAGGC-3′ |
| 5′-CCGggtaccCTATTCCTGGAGTTCCTCCTCCTCA-3′ | |
| GhTUB5 | 5′-CCGtctagaATGAGAGAAATCTTGCACATCCAAGGT-3′ |
| 5′-CCGctcgagTCAAGCAGCTTCCTCTTCTTCTTCCT-3′ | |
| GhTUB7 | 5′-CCGtctagaATGAGAGAAATCCTCCACGTTCAAGC-3′ |
| 5′-CCGctcgagTTAATTTTCCATTGCCTCATCTTCATA-3′ | |
| GhTUB8 | 5′-CCGtctagaATGAGAGAAATCCTTCACGTTCAAG-3′ |
| 5′-CCGctcgagCTAGTTCTCTTCCACACCTTCCTCC-3′ | |
| GhTUB9 | 5′-CCGtctagaATGAGAGAGATCCTTCATGTTCAAGG-3′ |
| 5′-CCGctcgagTCACATGTGTTCTTCATCCAAATCT-3′ | |
| GhTUB10 | 5′-CCGtctagaATGAGGGAAATCCTTCACGTACAAG-3′ |
| 5′-CCGctcgagTTACATCTCATGAACAGCTTCCTCG-3′ | |
| GhTUB12 | 5′-CCGtctagagATGAGAGAAATCCTTCACATCCAA-3′ |
| 5′-CCGctcgagTTAAGCCTCTGCCTCGTATTCCT-3′ | |
| GhTUB16 | 5′-CCGgctagcATGCGTGAAATCCTTCACATCC-3′ |
| 5′-CCGcccgggTTAGTCTTGATACTCCTCCTCTTCCTC-3′ |
Identification and clustering analysis of cotton tubulin ESTs
The tubulin ESTs were identified from 110 812 G. hirsutum ESTs (Shi et al., 2006; Gou et al., 2007; Y X Zhu et al., unpublished data) with an expectation value (E) of <0.01 and an identity score >50%. They were clustered into contigs using the stackPACK™v2.1 program (http://stackpack2.cbi.pku.edu.cn/). One cDNA clone from a cotton cDNA library (Shi et al., 2006) was chosen to represent a GhTUB EST contig.
In vitro ovule culture and treatments with exogenous phytohormones or very-long-chain fatty acids
Cotton ovules were collected at 1 dpa. They were sterilized and cultured in medium with or without the following compounds: 5 μM brassinosteroid (BR), 5 μM gibberellin (GA), and 5 μM lignoceric acid (C24:0). Supplementation with ethylene (final concentration: 0.1 μM) was carried out as described (Shi et al., 2006). C24:0 was dissolved in methyl tert-butyl ether to a stock concentration of 5 mM.
Functional complementation of the yeast tub2Δ strain by cotton GhTUBs
Saccharomyces cerevisiae diploid strain W1536 TUB2/tub2Δ was generated by amplifying the tub2::kanMX4 cassette by PCR using template genomic DNA extracted from BY4743 TUB2/tub2Δ (Mat a/a; his3D1/his3D1; leu2D0/leu2D0; lys2D0/LYS2; MET15/met15D0; ura3D0/ura3D0;tub2::kanMX4/TUB2, EUROSCARF). The PCR product was transformed into W1536, and selection of W1536 TUB2/tub2Δ followed the described protocol (Qin et al., 2007b). As a control, S. cerevisiae TUB2 was amplified with forward primer, 5′ cccgggTGGAGTGACATAGCAGCTACTACAAC-3′ and reverse primer 5′ gggcccGCTCGGAAGGTTAAAGGTTGT-3′ (lower-case letters indicate restrictions sites for cloning), and cloned into TRP1-marked pYADE4 behind the same ADH promoter with SmaI/ApaI sites, resulting in pYADE4-ScTUB2, which was transformed into W1536 TUB2/tub2Δ. The transformants were selected on Sc-Trp (synthetic complete medium lacking tryptophan) plates, and sporulated on plates containing 0.25% (wt/v) yeast extract, 1.5% (wt/v) potassium acetate, and 0.05% (wt/v) D-glucose supplemented with amino acids. After sporulation, ascospores were digested with zymolyase (Seikagaku), and the tetrads were dissected using a Singer MSM manual dissection microscope (Singer Instruments). Separated ascospores were grown on YPD [1% (wt/v) yeast extract, 2% (wt/v) peptone, and 2% (wt/v) D-glucose] for 5 d. The mutant complemented by ScTUB2 was selected by replica plating on YPD-G418 (YPD supplemented with 300 μg ml−1 of geneticin) plates and 2-amino-5-flurobenzoic acid (FAA) plates [synthetic complete medium containing 2% (wt/v) D-glucose and 0.05% (wt/v) FAA] simultaneously. Positive candidates, tub2Δ carrying pYADE4-ScTUB2, grew on YPD-G418 plates but not on FAA plates. YCplac-GhTUB was transformed into haploid tub2Δ carrying pYADE4-ScTUB2 and was selected on Sc-Trp or Sc-Ura plus FAA plates.
Sequence analysis
All cotton β-tubulin sequences were aligned by ClustalW (http://www.ebi.ac.uk). A neighbor-joining tree was constructed in MEGA3.1 (Kumar et al., 2004). A neighbor-joining bootstrap tree was constructed from the alignments of GhTUBs using Molecular Evolutionary Genetics Analysis (MEGA) software 3.1 with 1000 bootstrap replicates.
Results
Identification and cloning of 19 cotton TUB genes
A total of 795 putative ESTs containing cotton α-, β-, and γ-tubulin sequences were identified by aligning Arabidopsis tubulin cDNAs with 110 812 G. hirsutum ESTs. In accordance with their highly fibre-preferential expression pattern (Ji et al., 2003; Arpat et al., 2004; Shi et al., 2006; Gou et al., 2007), 1540 UBQ ESTs, 674 E6 ESTs, 414 expansin ESTs, and 501 ESTs encoding lipid transfer proteins were identified. The tubulin ESTs were further clustered into 46 contigs. All 17 previously reported tubulin genes (Table 2; Dixon et al., 1994; Li et al., 2007) were represented in 26 contigs, implying that the assembly used in this study should reflect a relatively complete G. hirsutum tubulin family.
Table 2.
Renaming cotton TUBs and comparison with their former names
| Rename | cDNA gene accession no. | Former name |
| GhTUB1 | AF484959 | Tubulin β-1 (Tub1) |
| AF487511 | β-Tubulin (TUB1) gene | |
| AY345610 | β-Tubulin 9 | |
| GhTUB3 | AF521240 | Xu-142 β-tubulin 1 |
| GhTUB5 | AY345607 | β-Tubulin 5 |
| GhTUB6 | AY345608 | β-Tubulin 6 |
| GhTUB7 | AY345609 | β-Tubulin 7 |
| GhTUB9 | AY345606 | β-Tubulin 3 |
| GhTUB12 | DQ023526 | Cultivar CIM707 tubulin |
The poly(A) tail of each GhTUB was obtained by re-sequencing the cDNA insert in the library, and the 5′-untranslated regions of most GhTUB genes were obtained by 5′-RACE using the SMART RACE cDNA amplification kit (Clontech Laboratories, Inc.). Full-length cDNA was obtained by PCR amplification using primers that included 5′- and 3′-untranslated regions to ensure that the 5′-RACE products were not the result of cross-reactions of homologous transcripts derived from different subgenomes. All GhTUB cDNAs were cloned into the pGEM-T vector, and the sequences were verified by DNA sequencing from both directions. The open reading frames of GhTUBs were further amplified with proofreading pfu DNA polymerase using gene-specific primers (Table 1) and were cloned into URA3-marked YCplac33 behind the ADH promoter. YCplac33-GhTUB was verified by DNA sequencing from both directions using the CEQ dye terminator cycle sequencing quick start kit and the CEQ8000 analysis system (Beckman Coulter). Sixteen full-length tubulin cDNAs were obtained from the remaining 20 contigs, such that a total of 33 full-length cDNAs including 12 α-, 19 β-, and two γ-tubulin genes were isolated. Seven of these β-tubulin genes, including GhTUB1, 3, 5, 6, 7, 9, and 12, have been described previously (Table 2).
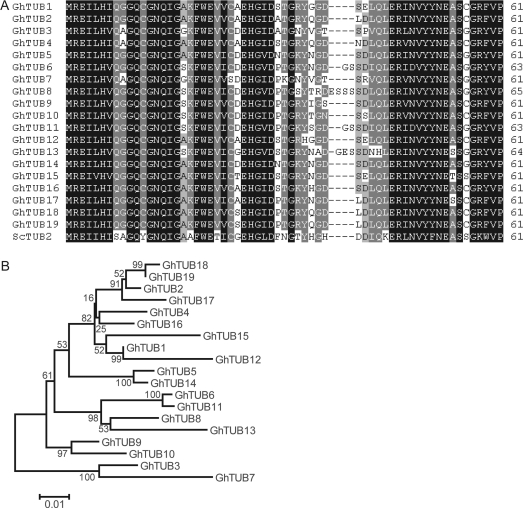
All 12 new TUB genes, designated as GhTUB2, 4, 8, 10, 11, 13, 14, 15, 16, 17, 18, and 19, were submitted to GenBank with accession numbers EU375992–EU376003. The predicted open reading frames ranged in size from 1335 bp to 1359 bp and shared 74–99% nucleotide identity (Fig. 1A). The putative GhTUB protein sequences varied from 444 to 452 amino acid residues in length and were highly conserved (85–99% overall sequence identity). There were large variations in the C termini, and there was an insertion of two to four residues at position 39 in the N termini of GhTUB6, GhTUB8, GhTUB11, and GhTUB13 when aligned with ScTUB2 (Fig. 1A). A phylogenetic tree of full-length GhTUBs is shown in Fig. 1B. The 19 GhTUBs were divided into four groups. GhTUB3 and GhTUB7 occupied a distinct branch that is basal to the clades containing the other GhTUBs. GhTUB9 and GhTUB10 formed one subgroup with the remaining 15 GhTUBs forming two sister subgroups. No δ-, ε-, ζ-, or τ-tubulins were found in the cotton assembly used in this study.
Fig. 1.
Alignments and neighbor-joining tree of the predicted amino acid sequences encoded by cotton GhTUB genes. (A) Amino acid sequences at the N termini of 19 cotton TUBs are aligned. The conserved residues are shaded in black. (B) A neighbor-joining tree was constructed in MEGA3.1 from 1000 bootstrap replicates. The scale bar corresponds to 0.1 estimated amino acid substitutions per site. GhTUB1, GhTUB3, GhTUB5, GhTUB6, GhTUB7, GhTUB9, and GhTUB12 were from GenBank (respective accession numbers: AY345610, AY345606, AY345607, AY345608, AY345609, AF521240, DQ023526). The other GhTUBs were from this study.
Nine of 19 GhTUB genes are preferentially expressed in cotton fibre cells
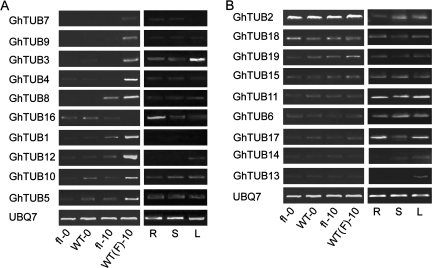
To identify TUBs differentially expressed in fibres, an fl mutant that fails to initiate fibre cells was used as a control (Ji et al., 2003; Shi et al., 2006). The transcript levels of all GhTUBs were examined by RT-PCR (Fig. 2). The transcripts of GhTUB3, 4, 7, and 9 predominantly accumulated in 10 dpa fibres stripped from ovules (Fig. 2A). The transcripts of GhTUB1, 5, 10, and 12 were detected at higher levels in 10 dpa wt fibres than in wt-0 and 10 dpa fl mutant ovules (Fig. 2A), whereas GhTUB8 transcripts were detected with similar expression levels in both 10 dpa wt fibres and fl mutant ovules (Fig. 2A). Low transcript levels were observed in roots, stems, and leaves for these GhTUBs, except that GhTUB3 was highly expressed in leaves (Fig. 2A). GhTUB16 transcripts were not detected in 10 dpa wt fibres but had a higher expression level in roots (Fig. 2A). The transcript levels of other GhTUBs did not vary between wt and fl mutants (Fig. 2B). These data suggest that GhTUB1, 3, 4, 5, 7, 8, 9, 10, and 12 are preferentially expressed in wt fibres, in which GhTUB16 is not expressed. The expression patterns of these GhTUBs in various fibre developmental stages were further determined by QRT-PCR (Table 3). The GhTUB1, 8, 9, and 12 transcripts reached peak expression levels of >1.0 at 10 dpa relative to that of the GhUBQ7 (Table 3). The GhTUB16 transcripts accumulated predominantly in 0 dpa ovules and rapidly decreased to low levels in later stages (Table 3), indicating that it is not a fibre-specific tubulin gene.
Fig. 2.
RT-PCR analysis of 19 GhTUB transcripts in wild-type and fl mutants. (A) RT-PCR analysis of nine GhTUB transcripts expressed preferentially or differentially in fibres. (B) RT-PCR analysis of nine GhTUB transcripts that were not preferentially expressed in fibres. Dpa, Days post-anthesis; WT-0, 0 dpa wild-type ovules; WT(F)-10, 10 dpa wild-type fibres stripped from ovules; fl-0, 0 dpa fl mutant ovules; fl-10, 10 dpa fl mutant ovules; R, roots; S, stems; L, leaves. UBQ7 (GenBank accession no. AY189972) was included as a control.
Table 3.
Relative expression levels of GhTUB compared with GhUBQ7 by QRT-PCR during different cotton fibre growth stages
| –3 dpa | 0 dpa | 5 dpa | 10 dpa | 15 dpa | 20 dpa | |
| GhTUB12 | 0.05±0.01 | 0.15±0.03 | 1.66±0.11 | 5.54±0.16 | 5.25±0.25 | 1.25±0.25 |
| GhTUB1 | 0.08±0.01 | 0.08±0.04 | 0.32±0.08 | 3.71±0.13 | 2.97±0.17 | 0.85±0.13 |
| GhTUB9 | 0.05±0.01 | 0.07±0.01 | 0.43±0.11 | 1.89±0.16 | 1.45±0.12 | 0.13±0.03 |
| GhTUB8 | ND | ND | 0.25±0.03 | 1.52±0.05 | 1.25±0.02 | 1.13±0.03 |
| GhTUB7 | ND | ND | 0.08±0.02 | 0.25±0.03 | 1.13±0.06 | 0.43±0.04 |
| GhTUB16 | 0.75±0.06 | 1.01±0.09 | 0.42±0.04 | 0.04±0.01 | 0.03±0.00 | ND |
| GhTUB3 | 0.03±0.01 | 0.08±0.03 | 0.25±0.02 | 0.87±0.03 | 0.83±0.04 | 0.85±0.03 |
| GhTUB5 | 0.25±0.01 | 0.31±0.02 | 0.82±0.03 | 0.86±0.03 | 0.76±0.03 | 0.71±0.03 |
| GhTUB10 | 0.26±0.01 | 0.46±0.02 | 0.52±0.02 | 0.71±0.03 | 0.46±0.03 | 0.37±0.03 |
| GhTUB4 | 0.24±0.01 | 0.27±0.01 | 0.46±0.03 | 0.45±0.02 | 0.54±0.02 | 0.56±0.02 |
The values (mean ±SD; n=3) for expression levels >0.5 relative to that of GhUBQ7 are in bold. In the upper section, GhTUB1, 9, and 8 with expression levels greater than 1.0 relative to that of GhTUB7. ND, Not detected; dpa, days post-anthesis.
GhTUB genes are up-regulated by phytohormones or very-long-chain fatty acids
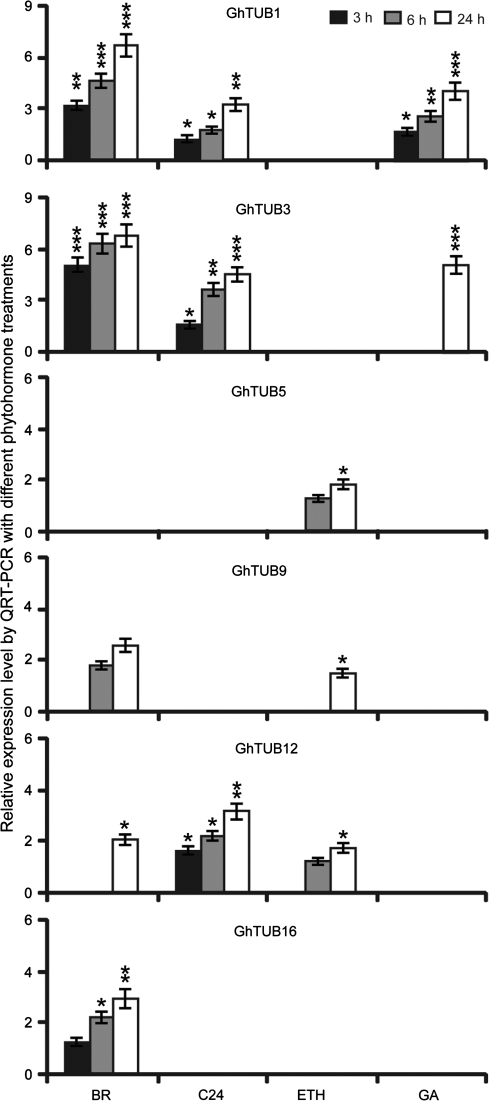
Plant hormones and lignoceric acid are important regulators of fibre development (Lee et al., 2007; Qin et al., 2007a). Exogenous treatment of wt ovules with ethylene, GA, BR, or C24:0 were performed to identify cotton TUBs regulated by phytohormones. QRT-PCR results showed that the expression of GhTUB1 or GhTUB3 was significantly increased 3-fold in 3 h to 5-fold in 24 h by BR, whereas the expression of GhTUB9 and GhTUB12 transcripts increased in response to BR at 6 h and 24 h, respectively (Fig. 3). C24:0 induced the expression of GhTUB1, 3, and 12 with a similar pattern, starting at 3 h (Fig. 3). GA increased GhTUB1 and GhTUB3 transcript levels at 3 h and 24 h, respectively, and ethylene induced an ∼2-fold increase in the expression of GhTUB5, 9, and 12 (Fig. 3). Non-fibre-specific GhTUB16 was up-regulated after BR treatment (Fig. 3).
Fig. 3.
Exogenous phytohormones and lignoceric acid promote the accumulation of GhTUB transcripts. RNA samples were prepared from three independent ovule cultures in the presence or absence of 5 μM brassinosteroid (BR), 0.1 μM ethylene (ETH), 5 μM gibberellin (GA), and 5 μM lignoceric acid (C24:0) for the indicated times. QRT-PCR experiments used the gene-specific primers reported in Table 1. The relative expression level of each GhTUB transcript is expressed relative to the level of the same transcript in the medium with or without ∼0.8 μM methyl tert-butyl ether for lignoceric acid supplementation at each indicated time. Values <0.1 are not shown. Statistically significant differences were determined using one-way ANOVA combined with Tukey's test. ***, P <0.001; **, P <0.01; *, P <0.05.
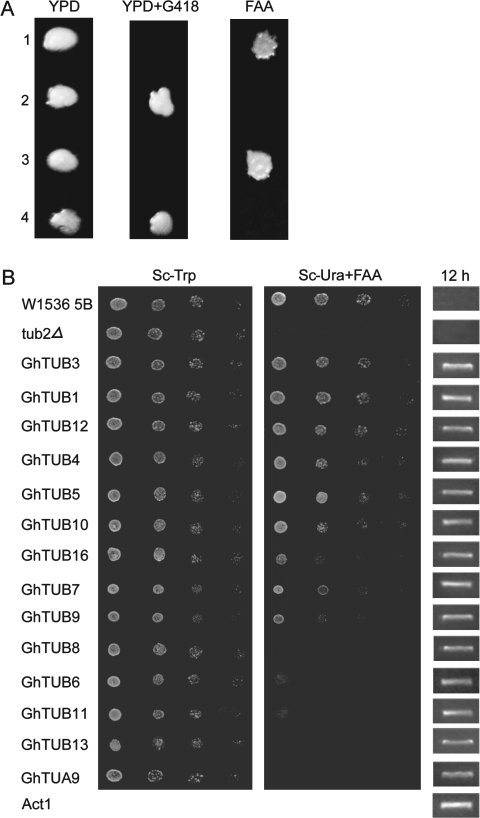
Functional characterization of cotton TUB genes in a yeast mutant
Saccharomyces cerevisiae ScTUB2 encodes β-tubulin (Thomas et al. 1985; Reijo et al., 1994). The yeast tub2Δ deletion mutant is lethal to cell growth (Thomas et al. 1985). To elucidate the essential biological function of the cotton TUB genes, the viability of the tub2Δ mutant cells complemented by individual GhTUBs was examined. The wt ScTUB2 was transformed into the diploid W1536 TUB2/tub2Δ and two viable spores were dependent on the presence of TUB2 for survival. The cells were unable to grow on medium containing FAA (Fig. 4A). The open reading frames of GhTUBs were separately cloned downstream of the ADH promoter in URA3-marked YCplac33 vectro, which were subsequently transformed into tub2Δ mutant cells carrying ScTUB2. The transformants were able to lose the pYADE4-ScTUB2 plasmid, as indicated by their survival on FAA-containing medium, confirming that GhTUBs, but not the plasmid backbone, were required for survival (Fig. 4B). The data showed that eight GhTUBs, excluding GhTUB6, 8, 11, and 13, and GhTUA9, encoding a cotton α-tubulin, were able to complement tub2Δ. These cotton TUBs were transcribed in yeast cells (Fig. 4B), indicating that the genetic complementation correlated with the cellular functions of GhTUBs. Amino acid sequence alignment of GhTUBs with ScTUB2 revealed that there are an additional two to four residues in the N termini of GhTUB6, 8, 11, and 13 (Fig. 1A), which may explain why they failed to rescue the lethality of tub2Δ (Fig. 4B).
Fig. 4.
Certain GhTUBs restore the viability of S. cerevisiae tub2Δ haploid cells. (A) Tetrad from diploid cells W1536 TUB2/tub2Δ transformed with pYADE4-ScTUB2. The ascospores grown on the YPD plate were replicated to G418 and FAA plates. (B) Complementation of tub2Δ mutant cells by individual GhTUBs. 12 h, RT-PCR analysis of yeast mutant cells expressing different GhTUBs harvested at 12 h from Sc-Trp culture medium. One S. cerevisiae actin gene (Act1) was used as the template control of RT-PCR analysis.
Discussion
Higher plants have greater numbers of TUA and TUB genes compared with mammals (Sullivan, 1988). The present current study identified 12 new β-tubulin genes, bringing the total known GhTUBs to 19 (Fig. 1), which is comparable with the recently identified 20 Pupulus TUBs (Oakley et al., 2007). The present findings suggest that there must be more TUAs than reported previously (Li et al., 2007), as plant cells cannot tolerate an imbalance in the ratio of α-tubulin to β-tubulin within the cytoplasm (Anthony and Hussey, 1998).
Comprehensive QRT-PCR analyses showed that nine of 19 GhTUBs were highly expressed at 10–15 dpa (Table 3), a period of fast elongation and primary cell wall synthesis, pointing to their roles in fibre development. Among these genes, GhTUB4, 8, and 10 were newly identified here, and the transcript level of GhTUB8 ranked third highest behind that of GhTUB1 and 12 (Table 3). GhTUB1, 5, 7, and 9 were highly expressed in fibre cells, as reported (Dixon et al., 1994; Ji et al., 2002; Li et al., 2002). Interestingly, the phylogenetic location of GhTUB3 and 7 is basal to the clade containing the other 17 GhTUBs (Fig. 1B), suggesting that GhTUB3 and 7 may have diverged early from the other GhTUBs. The GhTUB3/7, GhTUB9/10, and GhTUB1/12 pairs reside on distinct branches (Fig. 1B), implying that the genes encoding proteins that are preferentially expressed in fibres diverged during evolution. The expression levels of GhTUBs in developing fibre cells are in accordance with the need for fast assembly of microtubule arrays to meet rapid cellular expansion. Previous work also indicated that plant TUBs were expressed primarily in rapidly dividing cells or growing tissues such as root tips and elongating stems (Creelman and Mullet, 1991; Joyce et al., 1992).
The need for increased tubulin biosyntheses during the accelerated elongation process was verified by quantitative analysis of various GhTUB transcript levels after exogenous application of BR, ethylene, GA, and C24:0 to cultured cotton ovules. Interestingly, many TUBs, such as GhTUB1, 3, 9, 12, and 16 responded to BR treatment, whereas GhTUB1, 3, and 12 were also up-regulated by C24:0. Ethylene induced expression of GhTUB5, 9, and 12, and GA was only able to induce expression of GhTUB1 and 3 (Fig. 3), indicating that different chemicals promote fibre cell elongation via different mechanisms.
Plant cells contain ordered cortical microtubules, which may guide the movement of the cellulose synthase complex in the plasma membrane and regulate deposition of cellulose (Paredez et al., 2006). Tubulins are involved in microtubule assembly and function, and disruption of microtubule structure upon a reduction of α-tubulin expression causes abnormal cell expansion (Bao et al., 2001). Complementation of the S. cerevisiae tub2Δ mutant by cotton TUB genes (Fig. 4) provides evidence that these genes, functionally equivalent to ScTUB2, are essential for cell growth. Interestingly, GhTUB8 or the phylogenetically related GhTUB6, 11, and 13, which are poorly expressed in cotton fibres, contain amino acid insertions at position 39 (Fig. 1A) and were unable to complement the S. cerevisiae tub2Δ mutant (Fig. 4B). This region is important for interactions between the tubulin dimers (Chène et al., 1992), and mutations in this region are lethal to cells (Reijo et al., 1994). In Populus, additional residues at this position are also present in TUB19 and TUB20, which may be involved in pollen development (Oakley et al., 2007). These data suggest that the insertion region in plant TUBs may interfere with the tubulin–tubulin interactions in yeast cells but may perform some plant-specific functions. The existence of TUB genes in higher plants supports the notion that specialized tubulins are required for the growth and development of a plant cell and specifically for fibre cells and pollen tubes. Extensive arrays of microtubules are essential for the assembly of transversely oriented cellulose microfibrils to accommodate fast elongation.
Acknowledgments
This work was supported by grants from the China National Basic Research Program (grant no. 2004CB117302) and the Ministry of Science and Technology, People's Republic of China (grant nos 2006AA10A109-1 and 2007AA10Z136).
References
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. The Plant Cell. 2007;19:2763–2775. doi: 10.1105/tpc.107.053777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RG, Hussey PJ. Suppression of endogenous α and β tubulin synthesis in transgenic maize calli overexpressing α and β tubulins. The Plant Journal. 1998;16:297–304. doi: 10.1046/j.1365-313x.1998.00296.x. [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. Functional genomics of cell elongation in developing cotton fibers. Plant Molecular Biology. 2004;54:911–929. doi: 10.1007/s11103-004-0392-y. [DOI] [PubMed] [Google Scholar]
- Basra A, Malik CP. Development of the cotton fiber. International Review of Cytology. 1984;89:65–113. [Google Scholar]
- Bao Y, Kost B, Chua NH. Reduced expression of α-tubulin genes in Arabidopsis thaliana specifically affects root growth and morphology, root hair development and root gravitropism. The Plant Journal. 2001;28:145–157. doi: 10.1046/j.1365-313x.2001.01142.x. [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. The Plant Journal. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Chène P, Mazarguil H, Wright M. Microtubule assembly protects the region 28–38 of the β-tubulin subunit. Cell Motility and the Cytoskeleton. 1992;22:25–37. doi: 10.1002/cm.970220104. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Snustad DP, Carter JV. Temporal and spatial expression patterns of TUB9, a β-tubulin gene of Arabidopsis thaliana. Plant Molecular Biology. 2001;47:389–398. doi: 10.1023/a:1011628024798. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Water deficit modulates gene expression in growing zones of soybean seedlings: analysis of differentially expressed cDNAs, a new β-tubulin gene, and expression of genes encoding cell wall proteins. Plant Molecular Biology. 1991;17:591–608. doi: 10.1007/BF00037046. [DOI] [PubMed] [Google Scholar]
- Dixon DC, Seagull RW, Triplett BA. Two-dimensional gels: an easy method for large quantities of proteins. Plant Physiology. 1994;105:1347–1353. doi: 10.1104/pp.105.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard RH, Wick SM, Silflow CD, Snustad DP. Microtubule components of the plant cell cytoskeleton. Plant Physiology. 1994;104:1–6. doi: 10.1104/pp.104.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu ML, Chen XY. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Research. 2007;17:422–434. doi: 10.1038/sj.cr.7310150. [DOI] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX. Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Research. 2003;31:2534–2543. doi: 10.1093/nar/gkg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Li J, Wei G, Liang X, Zhu YX. A β-tubulin-like cDNA expressed specifically in elongating cotton fibers induces longitudinal growth of fission yeast. Biochemistry Biophysics Research Communication. 2002;296:1245–1250. doi: 10.1016/s0006-291x(02)02069-7. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Willemur R, Snustad DP, Silflow CD. Tubulin gene expression in maize (Zea mays L.): change in isotype expression along the developmental axis of seedling root. Journal of Molecular Biology. 1992;227:97–107. doi: 10.1016/0022-2836(92)90684-c. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiology. 2001;127:1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis thaliana contains at least six expressed α-tubulin genes. The Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev AV, Buschmann H, Doonan JH, Lloyd CW. AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. Journal of Cell Science. 2007;120:2241–2247. doi: 10.1242/jcs.007393. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Annals of Botany. 2007;100:1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang XL, Huang GQ, Li XB. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. Journal of Experimental Botany. 2007;58:3227–3238. doi: 10.1093/jxb/erm167. [DOI] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiology. 2002;130:666–674. doi: 10.1104/pp.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. The Plant Cell. 1994;6:303–314. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua NH. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. The Plant Cell. 2000;12:465–477. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènech P. A tandem of α-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Molecular Biology. 1990;14:1–15. doi: 10.1007/BF00015650. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Oakley RV, Wang YS, Ramakrishna W, Harding SA, Tsai CJ. Differential expansion and expression of α- and β-tubulin gene families in Populus. Plant Physiology. 2007;145:961–973. doi: 10.1104/pp.107.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- Pastuglia M, Azimzadeh J, Goussot M, Camillen C, Belcram K, Evrard JL, Schmit AC, Guerche P, Bouchez D. γ-Tubulin is essential for microtubule organization and development in Arabidopsis. The Plant Cell. 2006;18:1412–1425. doi: 10.1105/tpc.105.039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu YX. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. The Plant Cell. 2007a;19:3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Pujol FM, Hu CY, Feng JX, Kastaniotis AJ, Hiltunen JK, Zhu YX. Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. Journal of Experimental Botany. 2007b;58:473–481. doi: 10.1093/jxb/erl218. [DOI] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Molecular Biology of the Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC. MAPs in plant cells: delineating microtubule growth dynamics and organization. Current Opinion in Plant Biology. 2004;7:1–6. doi: 10.1016/j.pbi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. The Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Oppenheimer DG, Kopczak SD, Ploense SE, Ludwig SR, Haas NA, Snustad DP. Plant tubulin genes: structure and differential expression during development. Developmental Genetics. 1987;8:435–460. [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. The Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF. Structure and utilization of tubulin isotypes. Annual Review of Cell Biology. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Neff NF, Botstein D. Isolation and characterization of mutations in the β-tubulin gene of Saccharomyces cerevisiae. Genetics. 1985;111:715–734. doi: 10.1093/genetics/111.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R, Haas NA, Joyce CM, Snustad DP, Silflow CD. Characterization of four new β-tubulin genes and their expression during male flower development in maize (Zea mays L.) Plant Molecular Biology. 1994;24:295–315. doi: 10.1007/BF00020169. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M. Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. The Plant Cell. 2007;19:877–889. doi: 10.1105/tpc.106.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DJ, Triplett BA. Gene-specific changes in alpha-tubulin transcript accumulation in developing cotton fibers. Plant Physiology. 1999;121:181–188. doi: 10.1104/pp.121.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–131. doi: 10.2144/97221bi01. 134–138. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Yang G, Kawaguchi K, Komatsu S. Expression analyses of β-tubulin isotype genes in rice. Plant Cell Physiology. 2003;44:1202–1207. doi: 10.1093/pcp/pcg150. [DOI] [PubMed] [Google Scholar]
- Zhang T, Pan J. Genetic analysis of a fuzzless-lintless mutant in Gossypium hirsutum L. Jiangsu Journal of Agriculture Sciences. 1992;7:13–16. [Google Scholar]