Abstract
Floral transition in the obligate long-day (LD) plant sugar beet (Beta vulgaris ssp. vulgaris) is tightly linked to the B gene, a dominant early-bolting quantitative trait locus, the expression of which is positively regulated by LD photoperiod. Thus, photoperiod regulators like CONSTANS (CO) and CONSTANS-LIKE (COL) genes identified in many LD and short-day (SD)-responsive plants have long been considered constituents and/or candidates for the B gene. Until now, the photoperiod response pathway of sugar beet (a Caryophyllid), diverged from the Rosids and Asterids has not been identified. Here, evidence supporting the existence of a COL gene family is provided and the presence of Group I, II, and III COL genes in sugar beet, as characterized by different zinc-finger (B-box) and CCT (CO, CO-like, TOC) domains is demonstrated. BvCOL1 is identified as a close-homologue of Group 1a (AtCO, AtCOL1, AtCOL2) COL genes, hence a good candidate for flowering time control and it is shown that it maps to chromosome II but distant from the B gene locus. The late-flowering phenotype of A. thaliana co-2 mutants was rescued by over-expression of BvCOL1 thereby suggesting functional equivalence with AtCO, and it is shown that BvCOL1 interacts appropriately with the endogenous downstream genes, AtFT and AtSOC1 in the transgenic plants. Curiously, BvCOL1 has a dawn-phased diurnal pattern of transcription, mimicking that of AtCOL1 and AtCOL2 while contrasting with AtCO. Taken together, these data suggest that BvCOL1 plays an important role in the photoperiod response of sugar beet.
Keywords: Arabidopsis, Beta vulgaris, B gene, Caryophyllids, CONSTANS-LIKE gene, flowering time
Introduction
Improved knowledge of how floral induction pathways operate in crop plants is vitally important for both agronomy and quality. In sugar beet, Beta vulgaris ssp vulgaris, photothermal induction (long day photoperiods and exposure to cold temperatures) is required for reproductive growth (Owen et al., 1940). The transition is signified by rapid elongation of the stem (bolting) and is known to be tightly linked to the dominant early-bolting (B) gene locus (Munerati, 1931), although the underlying mechanisms are complex and poorly understood. Breeders still largely depend on phenotypic rather than genotypic selection and the development of high yielding cultivars has largely been achieved through the selection of late bolting genotypes (biennials) that are considered ‘recessive’ at the B gene locus (bb) and have an obligate requirement for vernalization in order to bolt. That these plants are still able to bolt indicates that the bb genotype might not represent a loss-of-function mutation. Expression of the bolting habit in both annual (BB, Bb) and biennial (bb) genotypes is positively regulated by long days (LD) (Bell and Bauer, 1942; Owen, 1954; Abe et al., 1997), thus photoperiod genes are considered to interact directly or indirectly with the B gene and may include candidates for the bolting gene itself. Furthermore, Abe et al. (1997) postulated that bolting is regulated by a gene complex that is formed by the B gene and a closely linked gene for LD requirement. Although there is a physiological and temporal distinction between bolting and flowering in sugar beet, such that plants can bolt without flowering, for example, under SDs (Fife and Price, 1953), bolting time is frequently used as a measure of flowering time because of the high correlation between induction of bolting and flowering. Hence, the important need for a molecular genetic dissection of potential B gene candidates in order to understand flowering time control in sugar beet.
The ability to perceive changes in daylength is dependent on a range of factors including light perception and photoperiodic regulation through the circadian clock. Variation in regulatory gene activation and transcript accumulation in different plant species ultimately results in the release of output signals that trigger flowering under different photoperiods. Regulatory genes in the photoperiod pathway were initially identified by screening for reversion to a late-flowering phenotype under long days (LD) in the rapid-cycling accessions of A. thaliana (Koornneef et al., 1991). GIGANTEA (GI) and CONSTANS (CO) were attributed to the molecular hierarchy of the long day (LD) response with GI activating transcription of CO which, in turn, positively affects transcription of two floral integrators, FLOWERING LOCUS T (FT) and the MADS-box gene SUPPRESSION OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Putterill et al., 1995; Park et al., 1999; Suarez-Lopez et al., 2001). The external coincidence model demonstrates the promotive effects of CO on FT expression and is typified by a distinctive pattern of oscillation for CO transcripts (Yanovsky and Kay, 2003; Hayama and Coupland, 2004; Valverde et al., 2004). CO homologues discovered thus far also exhibit a CO-like pattern of transcript accumulation which suggests that a similar mechanism of transcriptional and post-transcriptional regulation also exists in the photoperiod pathway of other plant species (Liu et al., 2001; Izawa et al., 2002; Nemoto et al., 2003; Turner et al., 2005). Functional equivalents of AtCO homologues have been demonstrated (Robert et al., 1998; Yano et al., 2000; Griffiths et al., 2003; Nemoto et al., 2003) and A. thaliana-like photoperiodic response pathways have been identified in other plant species through complementation studies (Hayama et al., 2002; Izawa et al., 2002; Kojima et al., 2002).
The downstream effects of CO activation are determined by two highly conserved domains on the protein. Two adjacent basic domains known as B-boxes at the amino-terminus bear similarities to the DNA-binding zinc-finger motifs of GATA-type transcription factors (Ramain et al., 1993; Tsuzuki et al., 2000) and may be involved in binding to the promoters of FT and SOC1 (Samach et al., 2000; Hepworth et al., 2002). In A. thaliana, mutations within these zinc-finger domains have resulted in a late-flowering phenotype (Redei, 1962; Putterill et al., 1995; Robson et al., 2001). The second conserved domain that is characteristic of CO proteins is located at the carboxyl terminus of CO. This CCT (CO, CO-like, TOC) domain contains about 45 amino acids with a nuclear localization signal present within the second half of the motif (Strayer et al., 2000; Robson et al., 2001). CCT domains are also common to other proteins including the circadian oscillator, TOC1 (TIMING OF CHLOROPHYLL A/B BINDING PROTEIN1) (Strayer et al., 2000) and may be important for mediating protein–protein interactions (Kurup et al., 2000; Yamashino et al., 2003). Based on domain structure and sequence information, genes encoding these characteristic motifs have been identified in almost all angiosperms. Extensive database searches and phylogenetic analyses have revealed up to 17 CO-like (COL) gene members in A. thaliana, 16 in rice, nine in barley, and three in wheat (Griffiths et al., 2003; Nemoto et al., 2003). Members from each of the COL multi-gene families can be loosely classified into four different groups (Group I to IV) based on conserved residues within domain structures (Robson et al., 2001). Functional studies on individual COL genes in A. thaliana have not been completed, although limited evidence exists for the function of AtCOL1, AtCOL2, AtCOL3, and AtCOL9. Cheng and Wang (2005) discovered that AtCOL9 (a Group III gene) represses AtCO transcription and hence delays flowering through indirect effects on the abundance of AtFT mRNA. It has also been reported that, despite about 67% identity to the AtCO protein, manipulation of transcriptional activity of the Group I genes AtCOL1 and AtCOL2 had no effect on flowering time (Ledger et al., 2001) whilst mutation in AtCOL3 (a Group I gene) had a significant effect on light signalling and root growth (Datta et al., 2006).
Understanding the photoperiodic response in sugar beet presents a new challenge not only because of the physiological complexity of the reproductive transition in beet but also because, unlike A. thaliana which is able to flower under both LD and SD (a facultative LD plant), sugar beet has an obligate LD requirement for flowering (Bell and Bauer, 1942). However, sequence information and functional analysis of sugar beet genes involved in flowering time as opposed to bolting time are limited and only just emerging. This includes a concerted effort towards obtaining sequence information for the B gene locus (Gaafar et al., 2005) and a systematic examination of candidate genes for relationships to B. Recently, an FLC-like gene, BvFL1 was isolated by Reeves et al. (2007) and was mapped to a different chromosome to the B gene. BvFL1 is alternatively spliced and showed classic responses to vernalization although epigenetic repression was not observed. One variant, BvFL1-v3 was found to delay flowering time significantly in complementation assays in A. thaliana (Atflc) mutants, but targets of this repression in sugar beet have not been demonstrated. Here, we have started to dissect the photoperiod pathway and describe the identification of a COL gene family in sugar beet. It is shown that one of the genes, BvCOL1 resembles Group I AtCO/AtCOL genes and is a putative homologue of AtCO and that it up-regulates transcription of AtFT and AtSOC1 in an A. thaliana co-2 mutant background. It is also shown that BvCOL1 does not map to the B locus.
Materials and methods
Plant material
Three sugar beet genotypes were used; (i) breeding line C600 (Lewellen, 1989), with an early bolting (annual) habit; (ii) commercial cultivar, Roberta (KWS Saatzucht GmbH, Einbeck, Germany) and (iii) breeding line NF (SESvanderHave, Tiennen, Belgium) which are both late bolting biennials. The annual and biennial habits were attributed to non-obligate and obligate requirements for vernalization, respectively. Arabidopsis thaliana wild-type ecotype Landsberg erecta (Ler-0) was obtained from the Nottingham Arabidopsis Stock Centre (NASC ID: NW20, http://arabidopsis.info/). co-2 mutant plants (Putterill et al., 1995) were a kind gift from Professor George Coupland (Max Planck Institute, Cologne, Germany).
Growth conditions and flowering time measurements
Sugar beet was grown in controlled growth chambers at 22 °C with a 16 h LD photoperiod. Photosynthetically active radiation (PAR) with a fluence rate of 220 μmol photons m−2 s−1 was provided by 125 W white fluorescent lamps (GE Lighting) and 40 W tungsten bulbs (Phillips) for 15 h. Plants were further illuminated for 1 h with tungsten light to enrich far-red light and prevent etiolation. Bolting and flowering were induced by imposing vernalizing conditions (8 weeks in annuals and 18 weeks in biennials) at 6 °C and under continuous low lights (10 μmol m−2 s−1). Vernalized plants were thermally buffered at 15 °C for 1 week to prevent de-vernalization before being returned to 22 °C and a 16 h photoperiod as described above. Plants initiated bolting within 2–3 weeks and flowered within 6 weeks of vernalization. Arabidopsis thaliana was grown under defined light regimes (LDs of 16 h and SDs of 8 h) in Sanyo Gallenkamp MLR 350 growth chambers at 20 °C. Lighting was supplied by 36 W fluorescent Daystar lamps (CEC Technology) providing 280 μmol m−2 s−1 of PAR. All leaves (vegetative and cauline leaves excluding young cotyledons) were counted at the first visible sign of budding as a measurement of flowering time.
Phylogenetic analysis
Sequences for BvCOL1 and CO-like family members were identified by BLAST searches of the Genbank database, the Beta vulgaris Gene Index, BvGI 1.0 (http://compbio.dfci.harvard.edu/tgi/), and the Sugar Beet EST Database at Michigan State University (http://genomics.msu.edu/sugarbeet/index.html). Sequences were aligned using the Clustal W-based alignment tool in Vector NTI, version 10.0.1 (Invitrogen). Computation of phylogenetic relationships by the Maximum Parsimony (MP) algorithm and bootstrap analysis was carried out using PHYLIP version 3.67 (Felsenstein, 1989).
Southern analysis
Total genomic DNA was extracted according to Smith et al. (1990). Aliquots (10 μg) of restriction enzyme digested DNA were analysed by Southern blot hybridization according to standard methods (Sambrook and Russell, 2001). BvCOL1 gene-specific probes were generated from a 787 bp fragment corresponding to the zinc-finger domains or from 470 bp of the inter-domain (middle) region of the cDNA. BvCO-like gene family-specific probes were generated from a 395 bp fragment corresponding to the CCT domain. Transgene-specific probes were generated either from the CaMV 35S promoter or BvCOL1 cDNA sequences.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from at least three individual plants using Plant RNAeasy Kit™ (Qiagen) and DNase treated. Two micrograms of RNA was reverse transcribed using AffinityScript (Stratagene) and diluted 5-fold for QPCR. Primers for QPCR were designed (see Supplementary Table S1 at JXB online) and optimized to 98–110% amplification efficiency with products verified by sequencing. Fluorescence of bound SYBR-GREEN (Stratagene) was detected in the MX3000 real-time PCR machine (Stratagene) over 40 cycles at 60 °C annealing temperature. BvCOL1 transcription levels were measured in triplicate and normalized against BvTubulin (Karetsou et al., 2005), a sugar beet alpha-tubulin gene with low variation in Ct values (18–19 cycles) for all tissues tested. QPCR in A. thaliana was normalized against AtUBC (At5g25760) (Czechowski et al., 2005).
Isolation of BvCOL1 sequences and generation of transgenic A. thaliana plants
Fragments of 1104 bp (cDNA) and 1771 bp (genomic DNA) were cloned using primers based on TC1427 (see Supplementary Table S2 at JXB online) and verified by custom sequencing (John Innes Centre Genome Laboratory, Norwich, UK). BvCOL1 promoter sequences were obtained by screening sugar beet BAC library filters (McGrath et al., 2004) and sequencing positive clones. To create transgenic plants overexpressing BvCOL1, the cDNA (1104 bp) was inserted into pEarleyGate 100 (Earley et al., 2006), mobilized into Agrobacterium tumefaciens strain GV2260 and used to transform the A. thaliana late-flowering co-2 mutant by the floral dip method (Clough and Bent, 1998).
Selection of transgenic A. thaliana plants
T1 progenies of Agrobacterium-dipped plants were selected by spraying 2-week-old plants with 0.3 mM glufosinate ammonium (Finale; supplied by Aventis Environmental Science, Essex, UK). Selfed T2 seeds and progenies were scored for herbicide resistance by painting rosette leaves of 2-week-old plants with a 0.3 mM Finale solution. Leaf necrosis was indicative of herbicide sensitivity and evident after four days whilst resistant plants remained healthy. The presence and expression of BvCOL1 in resistant lines were verified by PCR and RT-PCR, respectively. Homozygous T3 seeds were sown together with the wild type (Ler) and the late-flowering co-2 mutant under LD and SD conditions. Flowering time was recorded from a minimum of 20 individual plants for each T3 population and their controls. Statistical analysis of flowering time data was carried out by two-way ANOVA.
Genetic mapping of BvCOL1
To map BvCOL1 on a genetic map of sugar beet, two parents (K1P1 and K1P2) and an F1 plant (K1F1) of the F2 mapping population K1 (kindly provided by Dr B Schulz, KWS Saat AG, Einbeck, Germany) (Mohring et al., 2004; Schneider et al., 2007) were screened for polymorphisms by PCR amplification of genomic DNA and subsequent sequencing using an ABI PRISM™ 3700 DNA Analyzer. Primers A242 and A245 (see Supplementary Table S2 at JXB online) are located approximately 0.6 kb upstream and 0.1 kb downstream of the start codon, respectively. The PCR fragments of 97 F2 individuals of the mapping population were sequenced using primer A242 followed by co-dominant scoring of the electropherograms at the sites of four single nucleotide polymorphism (SNP) markers. Marker scores were added to the genotypic data for 305 SNP and RFLP markers in K1 that were generated previously by Schneider et al. (2007). The map position of BvCOL1 was calculated using the Kosambi mapping function (Kosambi, 1944) in JoinMap® 3.0 (Van Ooijen and Voorrips, 2001), as described by Schneider et al. (2007).
BvCOL sequence accession numbers
Three full-length BvCOL genomic sequences cloned from biennial sugar beet, cultivar Roberta were deposited into GenBank. Accession numbers are EU437782 (BvCOL1), EU437783 (BvCOL2), and EU437784 (BvCOL3).
Results
A CO-like (COL) multi-gene family in sugar beet
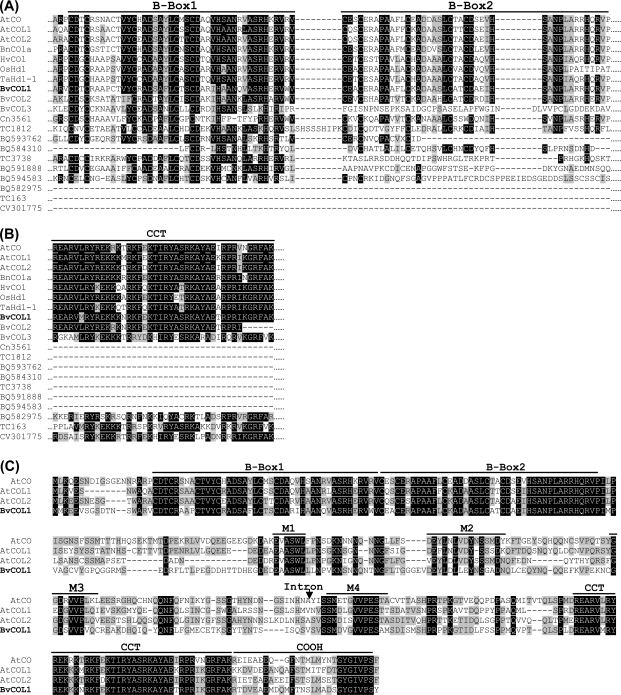
Currently, about 26 887 ESTs for Beta vulgaris are available through the dbEST division of Genbank. Stringent cluster analyses have further aligned these ESTs into longer sequences thereby allowing ‘Tentative Consensuses’ (TC) to be annotated and assigned putative functions (Quackenbush et al., 2000). From the collection of ESTs, 13 618 unigenes or TCs may be identified and are presently available through the Beta vulgaris Gene Index, BvGI 1.0 (http://compbio.dfci.harvard.edu/tgi/). BvCOL genes were identified by first searching this database and confirming the results by querying the sugar beet EST database (http://genomics.msu.edu/sugarbeet/index.html). A TBLASTN search with AtCO and CO-like protein sequences resulted in 13 hits with Expect (E) values <0.5 (Table 1). The nucleotide sequences were first translated to identify conserved domain residues that typify COL genes (B-box and CCT) and then compared by alignment against AtCO and AtCOL proteins and homologues from other plant species (Fig. 1A, B).
Table 1.
Beta vulgaris CONSTANS-like (BvCOL) expressed sequence tag/tentative consensus (EST/TC) sequences
| Name | ESTs in BvGI 1.0 | TCs in BvGI 1.0 | ESTs in Bv database at MSU | Domains | E-value | |
| 1 | BvCOL1 | BI543628, BI543722 | TC1427 | Cn2349 | B1, B2, CCT | 1.1e-41 |
| BI543724, BQ488270 | ||||||
| BQ588630, BQ589119 | ||||||
| 2 | BvCOL2 | BQ583937, BQ588069 | TC879 | Cn2600 | B1, B2, CCT | 4.4e-45 |
| 3 | BvCOL3 | BQ487825, BQ487842, | TC2903 | Cn3652 | B1, CCT | 2.2e-10 |
| BQ583909, BQ583972 | ||||||
| 4 | BQ584191, BQ584479 | Cn3561 | B1, B2 | 9.5e-22 | ||
| 5 | BQ489825, BQ589815 | TC1812 | Cn1811 | B1, B2 | 1.5e-16 | |
| BQ591614 | ||||||
| 6 | BQ593762 | B1, B2 partial | 1.9e-13 | |||
| 7 | BQ584310 | B1 partial, B2 | 7.8e-05 | |||
| 8 | BQ489587, BQ489817 | TC3738 | Cn3977 | B1 | 5.0e-11 | |
| 9 | BQ591888 | Cn1730 | B1 | 5.1e-09 | ||
| 10 | BQ594583 | B1 | 5.7e-08 | |||
| 11 | BQ582975 | CCT | 0.035 | |||
| 12 | BQ588535, BQ589202 | TC163 | Cn2315 | CCT | 0.46 | |
| BQ589272, BQ589370 | ||||||
| 13 | CV301775 | CCT | 0.49 |
Gene sequences were identified by TBLASTN of BvGI 1.0 (http://compbio.dfci.harvard.edu/tgi/) and the sugar beet EST database (http://genomics.msu.edu/sugarbeet/index.html) using AtCO, AtCOL1 to AtCOL16 protein sequences. The presence of CO-like domains is denoted by B1 (B-box 1), B2 (B-box 2) and CCT. Accession numbers with a ‘TC’ (Tentative Consensus), ‘BQ’ and ‘CV’ prefix were obtained from BvGI, and those with a ‘Cn’ prefix were obtained from the sugar beet EST database at MSU. The E-values given in the table refer to similarity to AtCO.
Fig. 1.
Multiple sequence alignment of putative CO-like EST/TC protein sequences in sugar beet. Comparison of putative sugar beet CO-like EST/TCs against Group Ia (AtCO/AtCOL1/AtCOL2) proteins of A. thaliana and CO/Hd1 homologues for (A) only the B-box domains of 10 EST/TCs and (B) only the CCT domain of six EST/TCs. (C) Full-length protein sequence of BvCOL1 aligned against Group Ia proteins of A. thaliana. The position of the intron within the BvCOL1 sequence is indicated by an arrow. Characteristic domains of CO-like proteins are marked by lines over sequences. Identical and similar residues are highlighted in black and grey, respectively. Accessions with a ‘TC’ or ‘BQ’ prefix are obtained from BvGI 1.0. Other accession numbers are AtCO (At5g15840; NM_121589) (Putterill et al., 1995), AtCOL1 (At5g15850; AY065001) (Putterill et al., 1995), AtCOL2 (At3g02380; NM_111105) (Ledger et al., 1996), BnCOa1 (AY280868) (Robert et al., 1998), OsHd1 (AB041838) (Yano et al., 2000), HvCO1 (AF490469) (Griffiths et al., 2003), TaHd1-1 (AB094490) (Nemoto et al., 2003).
Three transcripts (TC1427, TC879, TC2903) were found to encode putative full-length coding sequences with predicted B-box and CCT domains (Table 1; Fig. 1A, B). These have been designated BvCOL1, BvCOL2, and BvCOL3, respectively. Seven other EST/TCs appear to be truncated at the 3′ end but encode distinguishable B-boxes. Of these putative transcripts, four (Cn3561, TC1812, BQ593762, and BQ584310) contained B-box 1 and 2. However, incomplete 3′ (BQ593762) and 5′ (BQ584310) sequences meant that only one complete and one partial B-box could be identified reliably for two of these proteins. The three other ESTs (TC3738, BQ591888, BQ594583) contained only a single B-box. Three remaining EST/TCs (BQ582975, TC163, CV301775) were truncated at the 5′ end and encoded only a characteristic CCT domain. To verify expression of all 13 sequences in sugar beet vegetative leaf tissues, RT-PCR (see Supplementary Table S2 at JXB online) was carried out and the resultant cDNA fragments were cloned and confirmed by sequencing (data not shown).
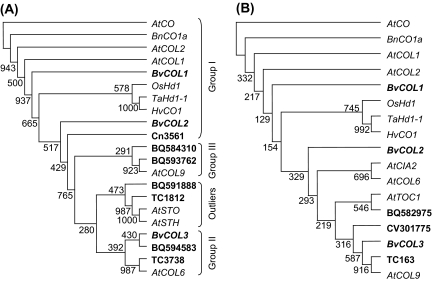
The encoded B-boxes have a high level of identity (up to 82%) with the classic zinc-finger motif commonly found in COL proteins (Omichinski et al., 1993; Putterill et al., 1995) and hence it is predicted that these truncated ESTs are likely to derive from CO-like genes in sugar beet. However, zinc binding motifs also exist in other plant proteins (Torok and Etkin, 2001) such as salt tolerance (STO) and salt tolerance homologue (STH) (Lippuner et al., 1996; Song et al., 1998; Holm et al., 2001). Similarly, ESTs with predicted CCT domains are equally likely also to belong to genes that are not part of the COL family, such as TOC1 (Strayer et al., 2000), CHLOROPLAST IMPORT APPARATUS (CIA2) (Sun et al., 2001), and other ‘unknown/hypothetical’ proteins. To identify true COL gene members and to understand the relationship of the 13 sugar beet CO-like genes, estimation of genetic distance by maximum parsimony (MP) was carried out on B-box and CCT domain nucleotides only. CO/Hd1 homologues (Group I: B-box 1, B-box 2), AtCOL6 (Group II: B-box 1), AtCOL9 (Group III: B-box 1, diverged B-box 2), and genes encoding B-box and CCT domains that are outside the COL gene family were included in the phylogenetic analysis of sugar beet genes (Fig. 2).
Fig. 2.
Phylogenetic analysis of sugar beet CO-like genes by maximum parsimony (MP). (A) MP tree of CO-like DNA sequences encoding only the B-box domains and two other zinc-binding genes that are not part of the CONSTANS family from A. thaliana (AtSTO, At1g06040 and AtSTH, At2g31380). (B) MP tree of CO-like DNA sequences encoding only the CCT domain and two other CCT-containing genes that are not part of the CONSTANS family from A. thaliana (AtTOC1, At5g61380 and AtCIA2, At4g25990). Bootstrap values from 1000 replicates were used to assess robustness of the tree. Group I genes are represented by CO/Hd1 homologues from different species (see Fig. 1 legend for accession numbers.). AtCOL6 (At1g68520; NM_105523) and AtCOL9 (At3g07650; NM_111644) (Cheng and Wang, 2005) were included as representatives of Group II and Group III genes, respectively.
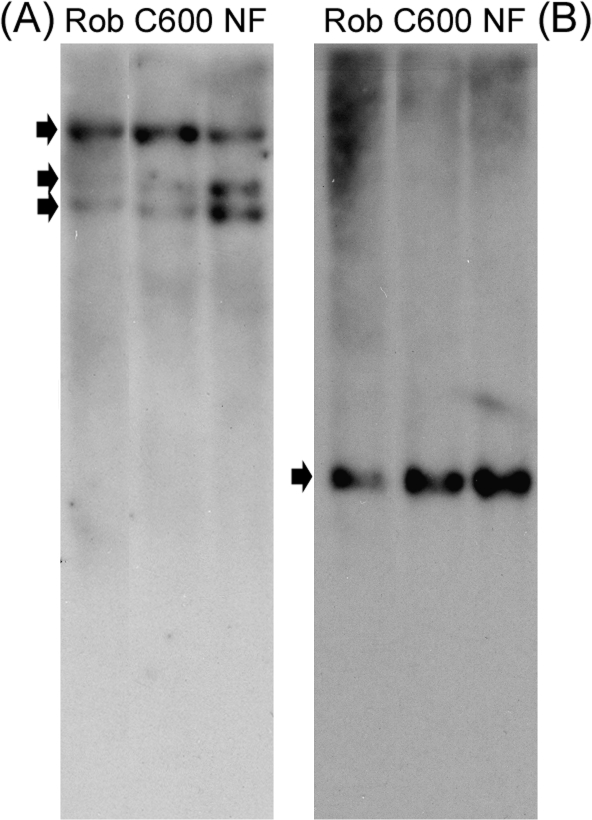
The 10 sugar beet EST/TCs encoding B-boxes fell into four clades/groups that are consistent with the predicted number of zinc-finger domains. BvCOL1, BvCOL2, and Cn3561 clustered with Group I CO-like/Hd1 genes with two B-boxes. The remaining EST/TCs with two B-boxes (BQ584310 and BQ593762) clustered with AtCOL9, a Group III gene with a diverged B-Box 2. Three EST/TCs (BvCOL3, BQ594583, TC3738) encoding only a single B-box were allocated as Group II genes together with AtCOL6. Interestingly, the phylogenetic tree placed two ESTs (BQ591888 and TC1812) in a cluster with the salt tolerance genes, AtSTO and AtSTH. TC1812 is 60% and BQ591888 is 45% identical to AtSTO at the nucleotide level and hence could be related to this class of genes. Analysis of the six sugar beet EST/TCs encoding CCT domains placed BvCOL1 and BvCOL2 in the same cluster as Group I CO/Hd1 genes. Three ESTs/TCs (CV301775, BvCOL3, and TC163) were clustered with AtCOL9, a Group III gene. However, one EST (BQ582975) showed 57% identity with AtTOC1 and is hence likely not to belong to the CONSTANS gene family. MP analysis of corresponding protein sequences also consistently allocated each EST/TCs within the same major clusters (data not shown). The presence of a sugar beet multi-gene family was supported further by Southern analysis using a fragment corresponding to the CCT domain of BvCOL1. At low stringency, three hybridized bands were present in both biennial and annual sugar beet genotypes suggesting that there are at least three Group I COL genes (Fig. 3A). This concurs with the above phylogentic cluster analysis of existing sugar beet ESTs.
Fig. 3.
DNA gel blot analysis of CO-like (COL) gene copies in sugar beet. Genomic DNA from two biennial (Roberta and NF) and one annual variety of sugar beet (C600) was digested with HindIII and probed with fragments derived from BvCOL1 (TC1427). (A) 395 bp fragment corresponding to CCT domain under low stringency, and (B) 787 bp fragment corresponding to B-box 1 and 2 domains under high stringency.
BvCOL1 shares common motifs with A. thaliana Group I CO and CO-like proteins
A single locus of BvCOL1 is present in both annual and biennial genotypes (Fig. 3B) and has highest homology to Group I CO/Hd1 genes compared to the other CO-like ESTs in sugar beet. An alignment of BvCOL1 protein sequence (Fig. 1C) against Group I CO-like proteins of A. thaliana revealed that the closest similarities were to Group I, sub-group Ia, proteins (Griffiths et al., 2003). AtCO, AtCOL1, and AtCOL2 are characteristic sub-group Ia proteins due to the presence of four conserved motifs (M1 to M4) in the more diverse inter-domain (middle) region of the peptides. BvCOL1 is highly conserved at these motifs with the exception of M2 where it diverges from the M2a motif (L-V-D-Y) of sub-group Ia proteins. Another variant of the M2 motif (M2c) has been reported in Physcomitrella patens ssp. patens PpCOL genes (Zobell et al., 2005). Comparison of sequence identity at both nucleotide and protein level further revealed that BvCOL1 is more similar to AtCOL1 (66% and 59%) and AtCOL2 (67% and 62%) than AtCO (64% and 55%) (data not shown).
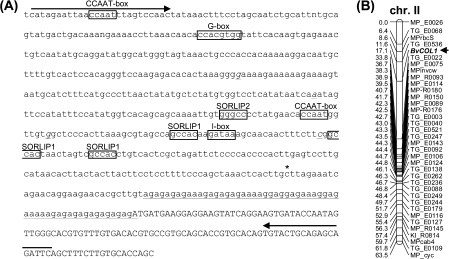
The genomic region spanning the BvCOL1 coding sequence was amplified and found to contain two exons of similar sizes to Group Ia genes in A. thaliana (746 bp and 358 bp) and an intron located between the M3 and M4 motifs as in the Group I COL genes (Griffiths et al., 2003; Zobell et al., 2005). To investigate cis-regulatory elements of BvCOL1, upstream promoter sequences were isolated by using a short fragment comprising the B-box 1 domain to screen a sugar beet BAC library (McGrath et al., 2004) under high stringency. Six positive BACs were identified (consistent with the expected library size), and a 600 bp fragment upstream of the initiation start codon was isolated (Fig. 4A). Multiple sequence alignment with promoters of A. thaliana Group Ia CO-like genes revealed 44% nucleotide conservation to AtCO, 45% to AtCOL1 and 50% to AtCOL2 (data not shown). Cis-regulatory DNA sequence motifs that are commonly found in circadian- and/or light-regulated genes in A. thaliana were also identified: G-box (Martinez-Garcia et al., 2000; Michael and McClung, 2003), I-box (Terzaghi and Cashmore, 1995), and SORLIP (‘sequences over-represented in light-induced promoters’) 1 and 2 (Hudson and Quail, 2003). The region containing the three SORLIP 1 motifs and the I-box is largely conserved in AtCOL1, AtCOL2 and, to a lesser extent, AtCO (data not shown). CCAAT boxes recently implicated in regulation of flowering (Wenkel et al., 2006) and a 57 bp GA-rich tract that includes several GAGA or GAGAG sites and a GA octodinucleotide repeat motif [(GA)8; (Santi et al., 2003)] were also identified upstream of the translation start codon (Fig. 4A).
Fig. 4.
BvCOL1 promoter/5′ UTR sequence and genetic map position. (A) BvCOL1 promoter/5′ UTR sequence. The 600 bp sequence upstream of the ATG start codon is shown in lower case letters. The positions of four single nucleotide polymorphisms in the K1 mapping population that were used for genetic mapping are underlined and in italics. The positions of primers used for SNP detection are indicated by arrows above the sequence. The asterisk indicates the 5′ end of the sequence corresponding to EST TC1427. Cis-regulatory DNA sequence motifs that are commonly found in circadian- and/or light-regulated genes are indicated as G-box (Martinez-Garcia et al., 2000; Michael and McClung, 2003), I-box (Terzaghi and Cashmore, 1995), SORLIP1 and 2 (Hudson and Quail, 2003) and CCAAT boxes (Wenkel et al., 2006). A 57 bp GA-rich tract that includes several GAGA sites and a GA octodinucleotide repeat motif (GA)8, (Santi et al., 2003) just upstream of the translation start codon is underlined. (B) Using the four polymorphic sites indicated in (A), the position of BvCOL1 (arrow) was mapped on a SNP-based genetic map of expressed genes (Schneider et al., 2007). BvCOL1 is located on chromosome II between markers TG_E0536 and TG_E0022.
BvCOL1 maps to sugar beet chromosome II and is not the bolting gene B
BvCOL1 was mapped to chromosome II of B. vulgaris on a SNP-based genetic map of expressed genes (Schneider et al., 2007). PCR amplification yielded a single PCR product and sequencing gave rise to unambiguous sequence electropherograms that revealed four single nucleotide polymorphisms between K1P1 and K1P2 in the genomic region upstream of the start codon (Fig. 4A). At two other positions, the BvCOL1 BAC sequence differed from K1P1 and K1P2 (Fig. 4A). These two sites are located within the GA-rich simple sequence repeat region upstream of the start codon and are likely to reflect the sites of polymorphisms between the parent plants of the mapping population and the genotype USH20 used for BAC library construction. The BvCOL1 gene is located between markers TG_E0536 and TG_E0022, at a cumulative genetic distance of 17.1 cM (Fig. 4B). To examine the possibility that BvCOL1 corresponds to the bolting gene, which was previously also mapped to chromosome II (Boudry et al., 1994; El-Mezawy et al., 2002), a small subset of the F2 mapping population that was used by El-Mezawy et al. (2002) for genetic linkage between BvCOL1 and B was analysed. Among 20 F2 plants that are phenotypically and genotypically well characterized for the early bolting character (including phenotypic data for >20 F3 plants each and extensive molecular marker data), six plants showed recombination between polymorphic sites in BvCOL1 and the B locus (data not shown). This high frequency of recombination indicates that BvCOL1 is not a constituent of the bolting gene locus.
BvCOL1 transcription is under circadian regulation
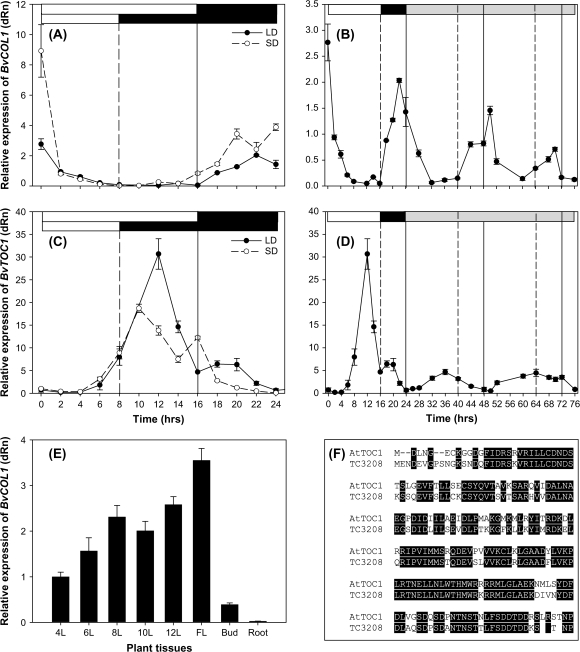
Diurnal regulation of BvCOL1 expression was profiled by QRT-PCR in vegetative leaves of sugar beet plants grown under LD and SD photoperiods. Under both light regimes, BvCOL1 transcript level was highest at the end of the dark period (time 0 h) which is immediately before the beginning of the light period. At dawn, transcript levels dropped rapidly within the first two hours and remained at low or undetectable levels until night occurred (Fig. 5A). In the dark, BvCOL1 transcription began immediately in LD-grown plants but SD-grown plants only began transcription after 6 h before they both reached a maximum level at the end of the night. This pattern of diurnal regulation closely mimics that of AtCOL1 and AtCOL2 (Ledger et al., 2001) and not AtCO where transcription peaks in the light under LDs and decreases in the dark (Suarez-Lopez et al., 2001). Except for the last 8 h in the dark and the first 2 h in the light, there was little difference between BvCOL1 transcription levels in plants grown under both photoperiods (Fig. 5A). To further determine if BvCOL1 is regulated by the endogenous circadian clock, plants were entrained under LD before transfer to continuous light where BvCOL1 transcription was followed for 48 h (Fig. 5B). Continuous oscillation of transcript levels with a 24 h rhythm was maintained but gradual decrease in peak amplitude was observed. Robust cycling of transcripts under a free-running period suggests that BvCOL1 is indeed under regulation by the circadian clock.
Fig. 5.
Expression profiles of BvCOL1 and a putative BvTOC1 in biennial sugar beet. Quantified transcription of BvCOL1 and BvTOC1 (TC3208) (Boxall et al., 2005) by QRT-PCR. BvCOL1 (A, B) and BvTOC1 (C, D) transcription in leaf tissues from 1-month-old plants (with eight fully-expanded leaves) over a 24 h period (A, C) and from plants moved to continuous light following entrainment under LDs (B, D). BvCOL1 transcription in developing leaves (4L to 12L) and in different organs (leaves of elongating stems; FL, floral bud; bud; and roots) sampled in LDs (E). Alignment of protein sequence of BvTOC1 against AtTOC1 showing a high degree of homology (76% amino acid identity) in the signal-receiver domain (F). Bars above represent the light (white), dark (black) or subjective light (grey) periods of the day. Solid and dashed lines represent expression levels for plants grown under LD and SD, respectively. Error bars represent standard error of three biological and three technical repetitions.
To confirm diurnal and circadian control of gene expression in the same tissues, we followed transcription of a sugar beet EST (TC3208) with homology to AtTOC1 (Boxall et al., 2005). It was demonstrated that this sequence is 76% identical to the amino-terminus signal-receiver domain of AtTOC1 at the protein level (Fig. 5F) and 70% identical at the nucleotide level. TC3208 mRNA transcripts oscillated over a 24 h period and peaked 4 h before the end of the day (LD) or at the beginning (after 2 h) of the night (SD), respectively (Fig. 5C) as has been observed for AtTOC1 (Strayer et al., 2000). Robustness of clock regulation was also examined under continuous light and maintenance of the rhythmic cycling of this putative BvTOC1 transcript was consistent with circadian-clock regulated genes albeit at reduced amplitude (Fig. 5D).
BvCOL1 transcription is developmentally regulated
The presence of BvCOL1 transcripts in vegetative leaf, cauline leaf (on elongating reproductive stems), and in fibrous roots of plants at the 10-leaf stage under LD was determined using QRT-PCR. Figure 5E shows that the amount of BvCOL1 mRNA increased as the plants matured until the 12-leaf stage when we normally consider the plants to have reached the adult phase. Following vernalization-induced bolting (reproductive transition), BvCOL1 transcription was also found to be high in cauline leaves but low in the first emerging floral bud and in fibrous roots (Fig. 5E). Expression profiles of the remaining BvCOL genes (see Supplementary Table S2 at JXB online) were also examined in these tissues but only BvCOL2 (another Group I gene) matched that of BvCOL1. By contrast, the salt-tolerant gene homologues, TC1812 and BQ591888 were preferentially expressed in roots compared to vegetative leaves (data not shown).
BvCOL1 complements the late-flowering phenotype of the A. thaliana co-2 mutant
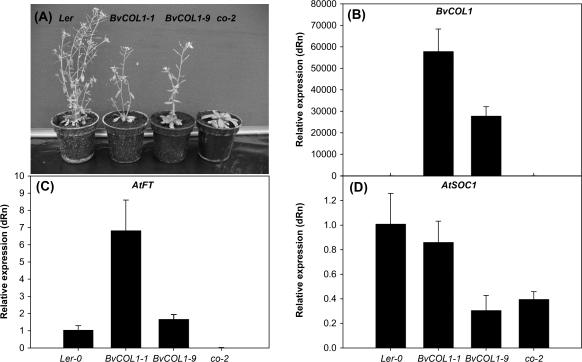
To assess a possible role of BvCOL1 in flowering control, the BvCOL1 cDNA driven by the strong CaMV 35S promoter was transformed into the late-flowering co-2 mutant. Sixteen T1 herbicide-resistant and transgene positive transformants exhibited a range of advanced flowering time phenotypes compared to the co-2 mutant. Two lines that flowered earliest (BvCOL1-1) and latest (BvCOL1-9) were selected for further characterization. Following segregation in the T2 generation, flowering time of homozygous T3 lines were compared to the co-2 mutant and wild-type (Ler) under both LDs (16 h light) and SDs (8 h light). Under our conditions, the vegetative phase of co-2 mutants was prolonged with an almost 5-fold increase in leaf number before flowering was initiated under both photoperiods compared to Ler (Table 2). This vegetative phase is significantly reduced when BvCOL1 is over-expressed in the mutant background. Under LD, lines BvCOL1-1 and BvCOL1-9 flowered significantly earlier than untransformed co-2 mutants, with flowering time being close to but not completely restored to wild-type numbers (P < 0.01) (Table 2; Fig. 6A). However, the acceleration of flowering time by BvCOL1 was most obvious under non-inductive SD where line BvCOL1-1 not only flowered earlier than the co-2 mutant, but was also slightly earlier than wild-type plants grown under the same conditions (P < 0.05) (Table 2). By contrast, line BvCOL1-9 only partially restored flowering time under SD. Southern blot analysis revealed that line BvCOL1-1 contained two transgene copies but only one copy was found in line BvCOL1-9 (data not shown). Consistent with this difference in transgene copy number, the level of BvCOL1 transcript accumulation was found to be two-fold higher in transgenic line BvCOL1-1 than in line BvCOL1-9 (Fig. 6B). Thus it is possible that gene dosage and/or positional effects may have contributed to the observed differences in the degree of phenotypic complementation.
Table 2.
Flowering time of transgenic 35S::BvCOL1 A. thaliana plants
Effect of BvCOL1 driven by the CaMV 35S promoter on the flowering time of A. thaliana co-2 mutants under LD (16/8 h light/dark) and SD (8/16 h light/dark) conditions. Data were collected from at least 20 individual homozygous T3 plants selected from 16 original transformants and are presented as the mean ±SE.
Student's t test showed significant difference (P < 0.01).
Fig. 6.
Flowering time and QRT-PCR analysis of gene expression in transgenic and untransformed A. thaliana. (A) Phenotype of 6-week-old wild-type, transgenic, and co-2 mutant lines. (B–D) Quantification of the BvCOL1 transgene and endogenous AtFT and AtSOC1 gene transcription in 2-week-old A. thaliana plants grown under LD by QRT-PCR. Error bars represent standard error of three biological and three technical repetitions.
BvCOL1 up-regulates AtFT and AtSOC1 transcription
The effect of BvCOL1 transcription in transgenic A. thaliana lines was investigated by QRT-PCR using tissues from plants grown under inductive LD. In the untransformed co-2 mutant, endogenous AtFT transcription was undetectable compared with basal levels in the wild type (Ler). By comparison, high transgene expression observed in line BvCOL1-1 correlated with a significantly higher up-regulation of AtFT expression compared to the lower expresser, BvCOL1-9. Furthermore, AtFT transcripts detected in line BvCOL1-1 exceeded levels detected in the wild-type (Ler) background (Fig 6C). In addition to direct effects on levels of AtFT expression, AtCO has also been shown to up-regulate AtSOC1 expression. In accordance with previous findings (Samach et al., 2000), it was found that endogenous AtSOC1 transcription in the co-2 mutant was down-regulated compared to the wild type (Fig. 6D). By over-expressing BvCOL1, AtSOC1 transcription was almost fully restored only in line BvCOL1-1 and not in line BvCOL1-9 (Fig. 6D).
Discussion
The effects of the environment on floral initiation in sugar beet have been observed for years but the molecular mechanisms that underlie its control remain to be fully elucidated. We have started the dissection of the obligate LD response in biennial beet by identifying a family of CO-like genes that play an important role in mediating between external light signals and flowering time. An extensive search of publicly available EST sequences and phylogenetic analysis has identified at least 10 genes (partial and full-length) that bear homology to the CO gene family of Arabidopsis with characteristic zinc-finger and/or CCT domains. Southern blot analysis and expression in vegetative leaves further verified the presence of a multigene family in sugar beet, as had also been shown to exist in monocots, dicots, and even in the moss, Physcomitrella (Robert et al., 1998; Yano et al., 2000; Liu et al., 2001; Griffiths et al., 2003; Kim et al., 2003; Nemoto et al., 2003; Hecht et al., 2005; Zobell et al., 2005).
Among the BvCOL genes that were identified, the closest homologue to Group 1a CO-like genes is BvCOL1 with promoter and coding regions most similar to AtCOL2 at 50% and 62%, respectively. In common with the COL genes, we found that BvCOL1 was under circadian control. It is estimated that the circadian clock controls between 2% to 36% of plant gene expression and plays an important intermediary role between light regulation and plant gene expression (Harmer et al., 2000; Schaffer et al., 2001; Michael and McClung, 2003). Some common cis-regulatory motifs have been attributed to clock response such as the CCA1-binding site (CBS, AAAAATCT) and evening element (EE, AAAATATCT) (Harmer et al., 2000). CBS and a morning element (ME, AACCACGA) (Harmer and Kay, 2005) are present in genes exhibiting a dawn-phased rhythmic expression pattern such as LHY (Piechulla et al., 1998; Michael and McClung, 2002, 2003). In the BvCOL1 promoter fragment identified in this study, none of these circadian-clock response elements (CCRE) are present, but circadian regulation of transcript oscillation was observed, indicating that other CCREs governing morning-phased rhythmic expression are yet to be identified. However, several light-response motifs are present in our BvCOL1 promoter fragment such as G-boxes which are known to be targets of the light-transducing factor, PIF3 (Martinez-Garcia et al., 2000). PIF3s are known to bind G-boxes located in the promoters of the MYB transcription factors LHY and CCA1, thereby activating their transcription.
It was observed that, when sugar beet plants are grown under different photoperiods, the diurnal regulation of BvCOL1 was strikingly different from the characteristic late evening (dusk) peak of AtCO and Hd1 homologues. Instead, BvCOL1 transcripts peak just before dawn, thereby closely resembling AtCOL2 (and AtCOL1) but AtCOL2 (and AtCOL1) does not control flowering as mis-expression had no effect on flowering time in A. thaliana (Ledger et al., 2001). This raises the possibility that BvCOL1 is not the true homologue of CO as the diurnal profile of mRNA accumulation in existing CO homologues consistently supports the external coincidence model of photoperiod-dependent activation. However, It was observed that overexpressed BvCOL1 can complement the loss-of-function co-2 mutant thereby demonstrating a functional equivalence with the AtCO/Hd1 homologues. It may be argued that the disparity in the apparent degree of conservation in gene sequence and regulation between A. thaliana and B. vulgaris on the one hand and gene function on the other hand is due to duplication of an ancestral gene bearing similarities to AtCOL1/AtCOL2 followed by subsequent functional diversification, and/or gene loss (e.g. in the lineage leading to B. vulgaris).
Under both LD and SD photoperiods, BvCOL1 was also found to be equally transcribed in the 14 h period that immediately followed the first 2 h after dawn. The differences in transcription under different day-lengths were in the last 8 h when BvCOL1 mRNA is more abundantly expressed under SD than LD. This is surprising as CO homologues in the long-day plants Arabidopsis (AtCO) and wheat (TaHd1-1) are more highly expressed under LD (Putterill et al., 1995; Nemoto et al., 2003) and raises another possibility that BvCOL1 may also act as a repressor of flowering under SDs. Such a lack of significant differences in mRNA abundance under different photoperiods is not unusual as OsHd1 in the SD-rice plant is also equally transcribed under both photoperiods (Yano et al., 2000). Further, phylogenetic analyses consistently placed BvCOL1 in a separate group to AtCOL9, a known repressor of flowering (Cheng and Wang, 2005) and we observed that BvCOL1 advances flowering time under SD and LD but with a more pronounced effect under SD in transgenic Arabidopsis. This suggests that BvCOL1 is unlikely to be a repressor of flowering. Certainly, the level of transcription under inductive photoperiods does not directly indicate functional activation nor does it define the period length at which the plant is responsive to reproductive switches. Instead, the activation of CO/Hd1 in response to light signals is more likely due to the balance of endogenous trans-activating/repressive factors induced by exposure to inductive and non-inductive photoperiods. This notion was supported by complementation analysis of the short-day rice Hd1 mutant line with a genomic fragment of the long-day wheat gene, TaHd1-1 (Nemoto et al., 2003). In addition, the rice Hd1 homologue is able to not only promote heading (flowering) under SD but also to inhibit flowering in LD, termed a bifunctional response (Yano et al., 2000). To understand the upstream regulatory domains necessary for LD-responsive activation in sugar beet will require further work such as expression under the BvCOL1 promoter and mis-expression in transgenic plants.
Although complementation of null mutants may be regarded with caution as a tool for identifying gene homologues, it does nevertheless provide useful information about protein function. Thus, functional equivalence between BvCOL1 and AtCO/Hd1 proteins was supported by two observations. One, the differences in flowering time of lines BvCOL1-1 and BvCOL1-9 indicate that gene dosage and/or positional effects may have significantly influenced complementation of the A. thaliana mutant. Southerns and qPCR showed that the degree of phenotypic complementation and the level of BvCOL1 expression correlate with copy number of the transgene (one versus two copies) thereby suggesting that CO activity is a limiting factor in flowering induction. This has been observed by Putterill et al. (1995) in A. thaliana where over-expression of AtCO in wild-type plants further advanced flowering time. The same conclusion may be inferred from the intermediate flowering phenotype of heterozygous plants expressing any of the seven classical co mutations (Robson et al., 2001). It appears therefore that the genetic composition of BvCOL1 functions similarly to the endogenous AtCO in its ability quantitatively to determine flowering time by copy number and expression level. Two, the expressed BvCOL1 protein in A. thaliana is able to interact positively with the two downstream genes AtFT and AtSOC1 in the absence of the endogenous AtCO activator. Heterologous interaction between BvCOL1 with AtFT and AtSOC1 may be due in part to functional domains within BvCOL1. Despite a higher degree of homology to AtCOL2, BvCOL1 is also highly similar to the AtCO protein, with 77% amino acid identity within the B-box domains and 86% within the CCT domain. It may be that BvCOL1 can substitute for the absence of AtCO by being recruited to promoters of downstream genes to affect transcription, but an indirect effect of the transgene cannot be ruled out at this point. It is interesting also to note that the conservation of domain residues between group Ia proteins in A. thaliana are between 85–89% (B-box) and 88–93% (CCT) and yet, only AtCO has been demonstrated to control flowering time. It is, however, possible that like BvCOL1, AtCOL1 and AtCOL2 may play a role in flowering time in the absence of AtCO although complementation analysis of the classical co mutants has yet to be done.
So far it has been shown that BvCOL1 is a light-responsive gene that is regulated by the circadian clock and encodes a protein that is able to mediate the LD response in A. thaliana. Molecular mapping has demonstrated that BvCOL1 is located on chromosome II, but is not incorporated in the early bolting B gene locus. Preliminary haplotype analysis of the BvCOL1 gene from different Beta accessions (including wild species) has only detected synonymous substitutions resulting in silent mutations (data not shown). Hence, the difference in bolting response of annuals and biennials could not be attributed to BvCOL1. However, data from Abe et al. (1997) and Owen (1954) suggested that the annual bolting habit conferred by B is modified by LD-responsive genes. Therefore, the quantitative effects of CO-like or other photoperiod genes on bolting time, rather than the bolting response per se, cannot be excluded and this is, therefore, currently still under investigation. The question remains as to whether BvCOL1 is the main determinant of the LD photoperiod response in sugar beet and we are working to address this by transgenic manipulation of BvCOL1 in sugar beet and by screening of TILLING populations for mutations in BvCOL1. In order to interpret results from this work correctly, the molecular hierarchy of GI – CO – FT in governing LD response in sugar beet must be proven. We are therefore also engaged in studies to identify these genes and examine their relationships and physiological significance in sugar beet.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table 1. Sequences of primers for QPCR.
Supplementary Table 2. Sequences of primers for PCR and RT-PCR of BvCOL EST/TCs and BvCOL1 promoter.
Supplementary Material
Acknowledgments
This work is funded by Biotechnology and Biological Sciences Research Council Core Strategic Grant 4668. ES Mutasa-Gottgens is supported by The British Beet Research Organization (BBRO grant No. 06/20). We thank Andrea Jennings and Cay Kruse for technical assistance, Professor George Coupland (Max Planck Institute, Cologne, Germany) for providing the co-2 mutants, Dr Britta Schulz (KWS Saat AG, Einbeck, Germany) for providing the K1 mapping population and Professor Eberhard Weber (Martin-Luther-University Halle-Wittenberg, Germany) for providing the SNP and RFLP marker data set for K1. A Müller is financially supported by the German research foundation (DFG, grant No. Ju 205/14-1).
References
- Abe J, Guan GP, Shimamoto Y. A gene complex for annual habit in sugar beet (Beta vulgaris L.) Euphytica. 1997;94:129–135. [Google Scholar]
- Bell GDH, Bauer AB. Experiments on growing sugar beet under continous illumination. I. Developmental studies. Journal of Agricultural Science. 1942;32:112–141. [Google Scholar]
- Boudry P, Wieber R, Saumitoulaprade P, Pillen K, Vandijk H, Jung C. Identification of RFLP markers closely linked to the bolting gene B and their significance for the study of the annual habit in beets (Beta vulgaris L.) Theoretical and Applied Genetics. 1994;88:852–858. doi: 10.1007/BF01253996. [DOI] [PubMed] [Google Scholar]
- Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J. Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiology. 2005;137:969–982. doi: 10.1104/pp.104.054577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. The Plant Journal. 2005;43:758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. The Plant Cell. 2006;18:70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- El-Mezawy A, Dreyer F, Jacobs G, Jung C. High-resolution mapping of the bolting gene B of sugar beet. Theoretical and Applied Genetics. 2002;105:100–105. doi: 10.1007/s00122-001-0859-z. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fife JM, Price C. Bolting and flowering of sugar beets in continuous darkness. Plant Physiology. 1953;28:475–480. doi: 10.1104/pp.28.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaafar RM, Hohmann U, Jung C. Bacterial artificial chromosome-derived molecular markers for early bolting in sugar beet. Theoretical and Applied Genetics. 2005;110:1027–1037. doi: 10.1007/s00122-005-1921-z. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiology. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. The Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiology. 2004;135:677–684. doi: 10.1104/pp.104.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Izawa T, Shimamoto K. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant and Cell Physiology. 2002;43:494–504. doi: 10.1093/pcp/pcf059. [DOI] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiology. 2005;137:1420–1434. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO Journal. 2002;21:4327–4337. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO Journal. 2001;20:118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiology. 2003;133:1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes and Development. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetsou K, Koutita O, Skaracis G. Development of a quantitative analysis protocol of heat shock protein 70 in heat stressed sugar beet (Beta vulgaris L.) plants. Plant Molecular Biology Reporter. 2005;23:195a–195j. [Google Scholar]
- Kim SJ, Moon J, Lee I, Maeng J, Kim SR. Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil. Journal of Experimental Botany. 2003;54:1879–1887. doi: 10.1093/jxb/erg217. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annuals Eugenetics. 1944;12:172–175. [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. The Plant Journal. 2000;21:143–155. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Ledger S, Dare A, Putterill J. COL2 (Accession no. L81119 and L81120) is a homologue of the Arabidopsis flowering time gene CONSTANS (PGR 96-081) Plant Physiology. 1996;112:862. [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. The Plant Journal. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Lewellen RT. Regulation of cytoplasmic male sterile sugar beet germplasm C600 CMS. Crop Science. 1989;29:246. [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. Journal of Biological Chemistry. 1996;271:12859–12866. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu J, McIntosh L, Kende H, Zeevaart JA. Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiology. 2001;125:1821–1830. doi: 10.1104/pp.125.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Shaw RS, de los Reyes BG, Weiland JJ. Construction of a sugar beet BAC library from a hybrid with diverse traits. Plant Molecular Biology Reporter. 2004;22:23–28. [Google Scholar]
- Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiology. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiology. 2003;132:629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohring S, Salamini F, Schneider K. Multiplexed, linkage group-specific SNP marker sets for rapid genetic mapping and fingerprinting of sugar beet (Beta vulgaris L.) Molecular Breeding. 2004;14:475–488. [Google Scholar]
- Munerati O. L'eredita della tendenza alla annualita nella commune barbabietola coltivata. Zeitschrift für Zuchtung, Reihe A, Pflanzenzuchtung. 1931;17:84–89. [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. The Plant Journal. 2003;36:82–93. doi: 10.1046/j.1365-313x.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Owen FV. The significance of single gene reactions in sugar beets. Proceedings of the American Society of Sugar Beet Technologists. 1954;8:392–398. [Google Scholar]
- Owen FV, Carsner E, Stout M. Photothermal induction of flowering in sugar beet. Journal of Agricultural Research. 1940;61:101–124. [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Piechulla B, Merforth N, Rudolph B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Molecular Biology. 1998;38:655–662. doi: 10.1023/a:1006094015513. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Research. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P, Heitzler P, Haenlin M, Simpson P. pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development. 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- Redei GP. Supervital mutants of Arabidopsis. Genetics. 1962;47:443–460. doi: 10.1093/genetics/47.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PA, He Y, Schmitz RJ, Amasino RM, Panella LW, Richards CM. Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris) Genetics. 2007;176:295–307. doi: 10.1534/genetics.106.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert LS, Robson F, Sharpe A, Lydiate D, Coupland G. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Molecular Biology. 1998;37:763–772. doi: 10.1023/a:1006064514311. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. The Plant Journal. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Santi L, Wang Y, Stile MR, et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. The Plant Journal. 2003;34:813–826. doi: 10.1046/j.1365-313x.2003.01767.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. The Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Kulosa D, Soerensen TR, et al. Analysis of DNA polymorphisms in sugar beet (Beta vulgaris L.) and development of an SNP-based map of expressed genes. Theoretical and Applied Genetics. 2007;115:601–615. doi: 10.1007/s00122-007-0591-4. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Watson CF, Bird CR, Ray J, Schuch W, Grierson D. Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Molecular and General Genetics. 1990;224:477–481. doi: 10.1007/BF00262443. [DOI] [PubMed] [Google Scholar]
- Song J, Yamamoto K, Shomura A, Itadani H, Zhong HS, Yano M, Sasaki T. Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Research. 1998;5:95–101. doi: 10.1093/dnares/5.2.95. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Sun CW, Chen LJ, Lin LC, Li HM. Leaf-specific upregulation of chloroplast translocon genes by a CCT motif-containing protein, CIA 2. The Plant Cell. 2001;13:2053–2061. doi: 10.1105/TPC.010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:445–474. [Google Scholar]
- Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S, Towatari M, Saito H, Enver T. Potentiation of GATA-2 activity through interactions with the promyelocytic leukemia protein (PML) and the t(15;17)-generated PML-retinoic acid receptor alpha oncoprotein. Moecular andl Cellular Biology. 2000;20:6276–6286. doi: 10.1128/mcb.20.17.6276-6286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap® 3.0, Software for the calculation of genetic linkage maps. Plant Research International. Wageningen, The Netherlands: 2001. [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, Mizuno T. A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant and Cell Physiology. 2003;44:619–629. doi: 10.1093/pcp/pcg078. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Living by the calendar: how plants know when to flower. Nature Reviews Molecular Cell Biology. 2003;4:265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- Zobell O, Coupland G, Reiss B. The family of CONSTANS-like genes in Physcomitrella patens. Plant Biology. 2005;7:266–275. doi: 10.1055/s-2005-865621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.