Abstract
The effects of synchronous photo (16 h daylength) and thermo (2 °C daily fluctuation) cycles on flowering time were compared with constant light and temperature treatments using two barley mapping populations derived from the facultative cultivar ‘Dicktoo’. The ‘Dicktoo’בMorex’ (spring) population (DM) segregates for functional differences in alleles of candidate genes for VRN-H1, VRN-H3, PPD-H1, and PPD-H2. The first two loci are associated with the vernalization response and the latter two with photoperiod sensitivity. The ‘Dicktoo’בKompolti korai’ (winter) population (DK) has a known functional polymorphism only at VRN-H2, a locus associated with vernalization sensitivity. Flowering time in both populations was accelerated when there was no fluctuating factor in the environment and was delayed to the greatest extent with the application of synchronous photo and thermo cycles. Alleles at VRN-H1, VRN-H2, PPD-H1, and PPD-H2—and their interactions—were found to be significant determinants of the increase/decrease in days to flower. Under synchronous photo and thermo cycles, plants with the Dicktoo (recessive) VRN-H1 allele flowered significantly later than those with the Kompolti korai (recessive) or Morex (dominant) VRN-H1 alleles. The Dicktoo VRN-H1 allele, together with the late-flowering allele at PPD-H1 and PPD-H2, led to the greatest delay. The application of synchronous photo and thermo cycles changed the epistatic interaction between VRN-H2 and VRN-H1: plants with Dicktoo type VRN-H1 flowered late, regardless of the allele phase at VRN-H2. Our results are novel in demonstrating the large effects of minor variations in environmental signals on flowering time: for example, a 2 °C thermo cycle caused a delay in flowering time of 70 d as compared to a constant temperature.
Keywords: BM5A, Hordeum vulgare, HvFT1, HvFT3, HvPRR7, ZCCT-H
Introduction
Flowering time is one of the most important adaptive characteristics of plants. Genetic regulation of physiological processes acts to ensure that flowering occurs at seasonal optima for pollination, fertilization, and seed development. In temperate zones, vernalization sensitivity and juvenility repress flowering early in plant development, allowing the plant to avoid freezing injury (Danyluk et al., 2003) and to achieve sufficient vegetative growth for supporting the high energy demands of flowering and seed set (Boss et al., 2004). Additional regulatory pathways are involved in perceiving and transferring environmental signals to floral meristem and organ identity genes (reviewed by Boss et al., 2004; Jack, 2004; Amasino, 2005). A better understanding of the genetic components of flowering time regulation will aid in the improvement of agricultural productivity in new production zones, or in conventional zones subject to greater climatological fluctuations.
The critical genes in the complex hierarchical cascades regulating flowering are best described in Arabidopsis (Arabidopsis thaliana L.) (reviewed by Boss et al., 2004; Amasino, 2005; Imaizumi and Kay, 2006). These genes are assigned roles in one of four pathways: photoperiod (including the perception of light quality and quantity); autonomous; vernalization; and hormonal. Signals from the pathways converge at a limited number of floral integrator genes, which in turn activate meristem identity genes. Information from Arabidopsis was an indispensable platform for rapidly isolating the homologues from cereal crops. These include: PSEUDO RESPONSE REGULATOR 7 (PRR7) (Turner et al., 2005; Beales et al., 2007) and GIGANTEA (GI) (Dunford et al., 2005) in the circadian central oscillator (McClung, 2006); genes in the photoperiod regulation pathway, including photoreceptors (PhyA, PhyB, PhyC, Cry1, Cry2) (Szűcs et al., 2006) and the CONSTANS (CO) gene family (Griffiths et al., 2003); the FLOWERING LOCUS T (FT) family of integrator genes (Yan et al., 2006; Faure et al., 2007); and the meristem identity gene AP1 (BM5A in barley) (Yan et al., 2003; Danyluk et al., 2003; von Zitzewitz et al., 2005). Interestingly, for some of these genes, for example, GI, CO, and photoreceptors (Griffiths et al., 2003; Dunford et al., 2005; Szűcs et al., 2006) there is no association between phenotypic variation in flowering time and allele sequence variation.
Attention has therefore focused on Triticeae genes that are significant determinants in flowering, and these can be broadly classified as VRN (vernalization response) or PPD (photoperiod sensitivity) genes (Cockram et al., 2007; Trevaskis et al., 2007). These Triticeae genes have different designations (and often functions) than in Arabidopsis. Arabidopsis AP1 corresponds to the candidate gene of the Triticeae VRN1 locus (Danyluk et al., 2003; Yan et al., 2003), PRR7 to PPD1 (Turner et al., 2005; Beales et al., 2007) and one member of the FT gene family (FT1), to VRN3 (Yan et al., 2006; Faure et al., 2007). Another member of this family (HvFT3) is considered a candidate for the PPD-H2 locus (Faure et al., 2007). In cereals, ZCCT1 (ZCCT-H in barley), with no known Arabidopsis homologue, was identified as the candidate gene of the VRN2 locus (Yan et al., 2004; Dubcovsky et al., 2005). This gene seems to occupy an intermediate position between the vernalization and photoperiod pathways, as both low temperature vernalization and photoperiod influences its activity (Dubcovsky et al., 2006).
The VRN loci have been assigned roles in a general model of flowering in cereals (Yan et al., 2006; Cockram et al., 2007). Briefly, vernalization saturation represses the activity of the dominant VRN2 allele, allowing expression of the recessive alleles at VRN3 and VRN1. VRN3 enhances activity of VRN1 under a long photoperiod, resulting in earlier heading. The roles of the PPD loci within this scheme are not fully resolved. In barley, PPD-H1 appears to affect flowering under long photoperiods, and PPD-H2 under short ones (Laurie et al., 1995; Turner et al., 2005).
Genetic analyses of the flowering time phenotype in the Triticeae under greenhouse or field conditions are rendered complex by uncontrolled sources of variation. Controlled environment tests allow for trait dissection but usually a limited number of environmental cues are varied at a constrained number of levels. Vernalization treatment and photoperiod duration are the two most widely-studied cues in the Triticeae (reviewed by Cockram et al., 2007; Trevaskis et al., 2007). Usually, all other factors are held constant or their effects are ignored. The results of preliminary experiments have recently been reported, in which a daily fluctuation of 2 °C (18/16 °C day/night) versus a constant temperature (18 °C) treatment during plant growth dramatically altered flowering time in a reference panel of barley accessions (Karsai et al., 2008). Spring growth habit accessions were the least affected and facultative accessions were the most affected. These results underscore the need for rigorous characterization of all environmental cues in flowering time experiments. A deeper understanding of all environmental cues will be of assistance in developing tools to alter flowering time without manipulating vernalization or photoperiod.
In this study a ‘genetical phenomics’ approach was applied to understand the effects of fluctuating temperature on the phenotypic manifestations of five loci involved in flowering time of barley: VRN-H1, VRN-H2, VRN-H3, PPD-H1, and PPD-H2. This investigation was facilitated by the availability of (i) allele-specific primers for the candidate or putative genes of these loci, (ii) very well-characterized barley accessions representative of the three growth habit types, and (iii) two well-characterized doubled haploid (DH) mapping populations.
Materials and methods
Plant materials
The Morex (M, spring), Dicktoo (D, facultative), and Kompolti korai (K, winter) cultivars and the two DH mapping populations derived from the cross of D×M (DM) and D×K (DK) used for these experiments have been well characterized at the genotypic and phenotypic levels (Pan et al., 1994; Karsai et al., 2005, 2006, 2007, 2008; von Zitzewitz et al., 2005; Szűcs et al., 2006).
Phenotypic characterizations
Controlled environment experiments were carried out in the Phytotron facilities of the Agricultural Research Institute of HAS, Martonvásár, Hungary using Conviron PGV type growth chambers (Conviron Ltd., Winnipeg, Canada). The technical parameters of the growth chambers and control systems for temperature and light intensity, are detailed in Karsai et al. (2004). Three different growth conditions were applied for the two DH populations and the three parents for a total of 150 d: (i) continuous light and a constant temperature of 18 °C, (ii) 16/8 h light/dark per 24 h with a constant temperature of 18 °C, and (iii) 16/8 h light/dark per 24 h with a temperature cycle of 18 °C light/16 °C dark, applied synchronously with the photo cycle. The two latter growth conditions were applied in phytotron chambers, while the continuous light and constant temperature conditions were applied in a phytotron chamber and in a greenhouse for the DK and the DM populations, respectively. In the greenhouse, a small natural daily variation in light intensity and in temperature could not be excluded completely. Acronyms for the three growth conditions applied to each of the two populations are as follows: 24C, 16C, and 16T, where the number refers to the number of h of light/24 h (24 or 16), and C or T refer to the temperature treatment, constant or thermo cycle. Flowering time (FT) was recorded as the number of days elapsed between planting and flowering and referred as FT24C, FT16C, and FT16T based on the growth conditions applied. Germplasm that did not flower was assigned an FT value of 150. The same light intensity (photosynthetic photon flux density (PPFD) of 220 μmol m−2 s−1) provided by a mixture of fluorescent tubes (Sylvania cool white) and incandescent lamps was used under all three growth conditions. Relative humidity was also maintained at the same level of ∼80% under the treatments. In the case of the DM (facultative×spring) population, the DH lines and parents were grown without vernalization. In the case of the DK (facultative×winter) population, all plant material was vernalized for 6 weeks at 3 °C with a 9/15 h light/dark photoperiod regime at low light intensity (10 μmol m−2 s−1). Each genotype was replicated twice within each growth condition, giving an average plant density of approximately 60 plants m−2.
Genotyping, linkage map construction, and QTL analysis
The DM linkage map consists of 165 loci of various types (e.g. AFLP, RFLP, SSR, STS, and ASGTs (allele-specific gene tags) with a total recombination length of 1040 cM and an average marker spacing of 6.3 cM (Pan et al., 1994; Skinner et al., 2006; Szűcs et al., 2006). The DK linkage map consists of 236 loci of various types, with a total recombination length of 1107 cM and an average marker distance of 4.5 cM (Karsai et al., 2005, 2007; Szűcs et al., 2006). The VRN-H1 and VRN-H2 loci were mapped with allele-specific primers of the respective candidate genes BM5A and ZCCT-H in the DM and DK populations (von Zitzewitz et al., 2005; Karsai et al., 2005).
For this experiment, the VRN-H3 and the PPD-H1 loci were mapped in the DM population as codominant CAPS markers designed for HvFT1 (Yan et al., 2006) and for HvPRR7 (Turner et al., 2005), respectively. The PPD-H2 locus was mapped as a dominant marker based on the presence/absence of the HvFT3 gene as described by Faure et al. (2007). In the DK population two SNPs were identified in the aligned HvFT1 (VRN-H3) sequences of Dicktoo (EU007827) and Kompolti korai (EU007828). The exon 3 SNP was utilized as the basis for a CAPS marker employing the primer pairs of HvFT1.03F (5′-CTT GCT CCC TCA TAC CCT AG-3′) and HvFT1.04R (5′-GCT TAA TTC GTG GCT GGC TTC-3′) and product digestion with BsrDI. The PPD-H1 and PPD-H2 loci were not mapped in DK because Dicktoo and Kompolti korai are monomorphic for the tested CCT domain SNP [SNP22 based on Turner et al. (2005)] in HvPRR7 and both accessions have the deletion of HvFT3. Linkage maps were constructed using JoinMap 4.0 (van Ooijen, 2006). QTL analyses were performed using composite interval mapping (CIM) Model 6, with forward regression and backward elimination as implemented in WinQTL Cartographer v. 2.5 (Wang et al., 2007). Threshold levels were set using 500 permutations.
Results
Phenotypic and genotypic characterization of the parental lines
Flowering was most accelerated in Morex, Dicktoo, and Kompolti korai when there were no thermo or photo fluctuations (24C) (Table 1). Flowering was delayed by the application of a daily photo cycle at a constant temperature (16C) but simultaneous photo and thermo cycling (16T) had the greatest effect on delaying flowering (Table 1). Morex was the least sensitive, while Dicktoo the most sensitive, to the fluctuating factors.
Table 1.
Flowering characteristics of the Dicktoo (facultative)×Morex (spring) (DM) and the Dicktoo (facultative)×Kompolti korai (winter) (DK) barley doubled haploid mapping populations under various environmental conditions
| Growth conditiona | Dicktoo | Morex | DM population | Dicktoo | Kompolti | DK population | ||||
| Meanb | Interval | LSD | Meanb | Interval | LSD | |||||
| 24Cc | 42 | 38 | 42 a | 25–78 | 3.0 | 28 | 53 | 42 a | 26–80 | 3.5 |
| 16C | 37 | 40 | 44 a | 27–95 | 4.0 | 39 | 61 | 52 b | 31–94 | 5.1 |
| 16T | 128 | 54 | 74 c | 39–150 | 4.8 | 109 | 74 | 77 c | 43–104 | 6.6 |
24C, continuous light and constant temperature; 16C, 16 h photoperiod and constant temperature; 16T, 16 h photoperiod and thermo cycle.
Mean values followed by the same letter within column are not significantly different from each other at the P=0.05 level.
Values for Dicktoo, Morex, and the DM population are from Pan et al., 1994. See Materials and methods for details.
Allele-specific primers revealed diversity at VRN and PPD loci in the parental lines (Table 2). In the DM population, the effect of the VRN-H1, VRN-H3, PPD-H1, and PPD-H2 loci could be evaluated based on the functional allele differences between the two parental lines in the respective candidate genes. In the DK population, there was only functional segregation in the candidate gene of the VRN-H2 locus. The minor polymorphisms in the candidate genes for the VRN-H1 and VRN-H3 loci are not known to affect phenotype, but these variations allowed us to monitor allelic segregation at these loci. In the remainder of this report, the first letter of the parental cultivar's name will be used to refer to the allele phases in each individual VRN-H and PPD-H locus.
Table 2.
Allele compositions of three barley parental lines at vernalization response (VRN) and photoperiod sensitivity (PPD) loci based on polymorphisms associated with the candidate or putative gene functions of each locus
| Locus/gene | Chromosome | Site of functional polymorphism | Allele types of the varietiesa | ||
| Dicktoo (facultative) | Morex (spring) | Kompolti (winter) | |||
| VRN-H1/BM5A | 5H | Intron 1 deletion (1) | Winter | Spring | Winter |
| VRN-H2/ZCCT-H | 4H | Presence/absence (2) | Spring | Spring | Winter |
| VRN-H3/HvFT1 | 7H | SNP haplotype in intron 1 (3) | Winter | Spring | Winter |
| PPD-H1/HvPRR7 | 2H | SNP in the CCT domain (4) | Early flowering under long days | Late flowering under long days | Early flowering under long days |
| PPD-H2/HvFT3 | 1H | Presence/absence (5) | Late flowering under short days | Early flowering under short days | Late flowering under short days |
Primers used for detection and the allele nomenclatures are based on the relevant references: (1) von Zitzewitz et al. (2005); (2) Karsai et al. (2005); (3) Yan et al. (2006); (4) Turner et al. (2005); (5) Faure et al. (2007).
Effects of photo and thermo fluctuations on flowering in DM
There were similar patterns of flowering time amongst parents and DH lines under the three growth conditions (correlations 0.79–0.88, see Supplementary Fig. S1 at JXB online). As shown in Table 1, the population as a whole was earliest to flower under 24C, but this mean value was not significantly different from that for 16C. Flowering was significantly delayed under 16T and eight DH lines did not flower in 150 d. QTL analyses revealed that PPD-H1 and VRN-H1 were the most significant determinants of the observed phenotypic variation in flowering time under the three growth conditions (Table 3; see Supplementary Fig. S2 at JXB online). The M allele at PPD-H1 and the D allele at VRN-H1 delayed flowering. The additive effect of the M allele at PPD-H1 was to delay flowering for 12 d and 13 d under 16C and 16T, respectively. The difference in additive effect of the D allele at VRN-H1 was much greater under the 16T condition (9 d and 24 d under 16C and 16T, respectively). Of the two genes, VRN-H1 determined a greater proportion of the phenotypic variance at 24C and 16T and PPD-H1 at 16C.
Table 3.
Effects of the vernalization response (VRN) and photoperiod sensitivity (PPD) loci on flowering time in two barley mapping populations under various environmental conditions
| Growth conditiona | VRN-H1 | VRN-H2 | VRN-H3 | PPD-H1 | PPD-H2 | ||||||||||
| LOD | R2 (%) | Add. eff. | LOD | R2 (%) | Add. eff. | LOD | R2 (%) | Add. eff. | LOD | R2 (%) | Add. eff. | LOD | R2 (%) | Add. eff. | |
| Dicktoo×Morex | |||||||||||||||
| 24C | 32.1 | 49.0 | 11 | – | ns | 23.5 | 40.0 | –9 | ns | ||||||
| 16C | 22.7 | 30.3 | 9 | – | ns | 31.2 | 55.0 | –12 | ns | ||||||
| 16T | 22.3 | 41.6 | 24 | – | ns | 12.0 | 17.2 | –13 | 7 | 7.8 | 9 | ||||
| Dicktoo×Kompolti korai | |||||||||||||||
| 24C | 4.6 | 2.1 | 2 | 50.1 | 77.8 | –12 | ns | – | – | ||||||
| 16C | 3.7 | 3.2 | 3 | 34.5 | 63.8 | –12 | ns | – | – | ||||||
| 16T | 24.6 | 49.5 | 12 | 11.0 | 15.5 | –7 | ns | – | – | ||||||
24C, continuous light and constant temperature; 16C, 16 h photoperiod and constant temperature; 16T, 16 h photoperiod and thermo cycle.
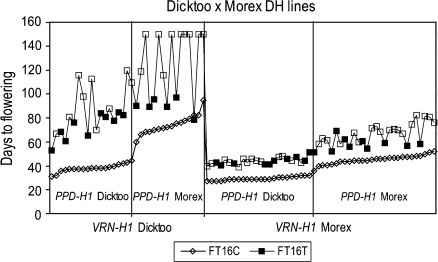
The combined effects of VRN-H1 and PPD-H1 explained most of the phenotypic variation in the experiment (two-locus R2 values were 81.4%, 83.5%, and 74.0% for FT24C, FT16C, and FT16T, respectively). As a result, the mean flowering times of lines with the parental allele combinations at the two loci were statistically the same as the respective parents under 24C and 16C. At 16T, however, the DD (VRN-H1/PPD-H1) lines headed significantly earlier than Dicktoo (84 d versus 128 d, respectively), while the MM lines were significantly later than Morex (66 d versus 54 d, respectively). The non-parental allele combinations were responsible for the significant phenotypic transgressive segregation shown in Fig. S1 in Supplementary data at JXB online. Lines with MD alleles at VRN-H1/PPD-H1 flowered significantly earlier, while lines with DM alleles at VRN-H1/PPD-H1 flowered significantly later than the parents and parental allele combinations. In addition, flowering of the non-parental combinations were significantly influenced by the application of a thermo cycle. The MD (VRN-H1/PPD-H1) was the only subclass with a relatively uniform reaction to the thermo cycle. Conversely, the largest scattering was observed in the DM (VRN-H1/PPD-H1) subclass (Fig. 1).
Fig. 1.
Effect of synchronous photo and thermo cycles on the major developmental genes in the Dicktoo (facultative)×Morex (spring) barley population. Within the graph of FT16T, the empty square represents the Dicktoo null allele in PPD-H2, while the full square stands for the Morex allele.
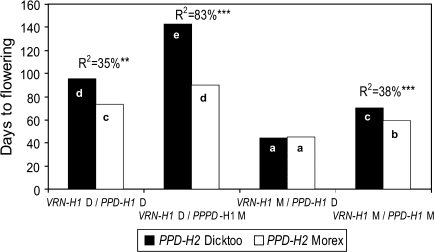
PPD-H2 had a significant main effect only under 16T, where it explained 8% of the phenotypic variance (Table 3; see Supplementary Fig. S2 at JXB online). This locus had a greater effect on flowering time, however, in association with certain alleles at other loci (Fig. 2). Considering the four possible VRN-H1 and PPD-H1 allele combinations, the allele phase at PPD-H2 had the most pronounced effect when the late flowering (M) allele at PPD-H1 was juxtaposed with the winter (D) allele at VRN-H1 under 16T. In this case, the allele phase at PPD-H2 explained 83% of the phenotypic variance and the Dicktoo allele (lack of the HvFT3 gene) resulted in extremely delayed flowering; eight DH lines of the 10 carrying this combination did not flower (Fig. 1). There was only one combination of PPD-H1 and VRN-H1 alleles where the allele at PPD-H2 had no effect at all on flowering and that was the early flowering (D) allele at PPD-H1 configured with the spring (M) allele at VRN-H1.
Fig. 2.
Effects of three-way locus interactions between VRN-H1, PPD-H1, and PPD-H2 on flowering time under synchronous photo and thermo cycles in the Dicktoo (facultative)×Morex (spring) barley population. Group averages with the same letter are not significantly different from each other at the P=0.05 level.
No QTL main effect was detected at VRN-H3 (Table 3). The effect of VRN-H3 and the three-way loci interactions with the VRN-H1 and PPD-H1 loci could not be discerned due to segregation distortion in two of the four allele classes at VRN-H1 and PPD-H1.
Effects of photo and thermo fluctuations on flowering in DK
In DK, the average flowering time of the population was also the earliest under 24C (Table 1). Varying photoperiod duration and holding temperature constant (16C) significantly delayed the average flowering of the population (10 d). The delay was relatively uniform: the correlation was 0.92 between FT24C and FT16C (see Supplementary Fig. S1 at JXB online). The combined effects of light and thermo cycles further delayed the mean flowering time of the population by 25 d. Individual line responses were not as consistent: the correlations between FT16T and the other two growth conditions were both 0.52 (see Supplementary Fig. S1 at JXB online). Even under 16T all lines flowered within 110 d.
VRN-H2 had a very large effect on flowering time at 24C and 16C, accounting for 78% and 64% of the phenotypic variance, respectively (Table 3, see Supplementary Fig. S2 at JXB online). The winter (K) allele main additive effect was a 12 d delay in flowering. The VRN-H1 locus had a significant effect under these conditions, but it explained a very low portion of the phenotypic variance. The application of synchronous light and thermo cycles resulted in a shift in the significance of the effects of these two loci: at 16T VRN-H1 explained close to 50% of the phenotypic variance and VRN-H2 only 16%.
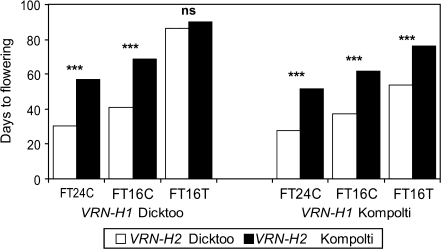
VRN-H2 and VRN-H1 jointly accounted for most of the phenotypic variation, irrespective of growth condition: the two-locus R2 values were 0.91 for FT24C, 0.83 for FT16C, and 0.69 for FT16T. The average flowering times of lines with parental allele combinations at these two loci were statistically the same as the respective parents under all three conditions, with one exception. At 16T, the average flowering of the DD lines was again significantly earlier than that of Dicktoo (86 d versus 109 d). As shown in Fig. 3, there is a pattern of growth condition-dependent epistasis between these two loci. Two features are noteworthy. First, the K allele at VRN-H1 always resulted in significantly earlier flowering than the D allele, regardless of growth condition or allele phase of VRN-H2. Second, the winter allele (K) at VRN-H2 delayed flowering, with one exception: under 16T, the D allele at the VRN-H1 locus resulted in extremely delayed flowering, irrespective of the allele phase at VRN-H2.
Fig. 3.
Effects of daily fluctuating environmental factors on the association between the allele phases of VRN-H2 and VRN-H1 genes in the Dicktoo (facultative)×Kompolti korai (winter) barley mapping population.
The VRN-H3 locus had no significant effect on flowering time in the DK population, either alone, or in association with the allele phases of VRN-H2 and VRN-H1.
Effects of Dicktoo alleles at the major flowering loci
The unique effects of D alleles at the flowering time loci monitored in these experiments show excellent correspondence across genetic backgrounds and growth conditions. For example, the flowering times of the subsets of lines with D alleles at all five VRN and PPD loci (6 lines in the case of DM and 10 lines in the case of DK populations) were not significantly different at 16C and 16T, in the two conditions which were identical for the two populations. The corresponding FT16C and FT16T means and P values for the 6 DM and the 10 DK lines were 40 d versus 42 d (P=0.33) and 99 versus 92 d (P=0.44), respectively.
Discussion
The primary environmental factors controlling the major developmental loci in cereals are low temperature on VRN loci (Yan et al., 2003, 2004, 2006) and photoperiod duration on PPD loci (Laurie et al., 2004; Turner et al., 2005; Beales et al., 2007). The associations between these environmental cues and corresponding genes were identified via QTL analyses and quantitative genetics approaches (Laurie et al., 1994, 1995; Karsai et al., 2005; Kóti et al., 2006; Szűcs et al., 2007) and by monitoring changes in gene activities (von Zitzewitz et al., 2005; Turner et al., 2005; Dubcovsky et al., 2006; Trevaskis et al., 2006; Beales et al., 2007). There is evidence, however, that secondary and interaction effects may also regulate these loci, i.e. photoperiod on VRN loci (von Zitzewitz et al., 2005; Dubcovsky et al., 2006; Karsai et al., 2006; Szűcs et al., 2006; Trevaskis et al., 2006).
Our results are novel in demonstrating the large effects of minor variations in environmental signals on flowering time. A 2 °C thermo cycle caused a delay in flowering time of 70 d as compared to a constant temperature. The phenotypic reactions of the two mapping populations were in accordance with the results observed for the parental lines in previous experiments (Karsai et al., 2008). Flowering of both populations was the earliest under constant environmental conditions and was the most delayed when long photoperiod cycles and thermo cycles of 2 °C were applied in synchronous fashion. It is shown that this variation in timing is due to allelic variation at four flowering time-related loci: VRN-H1, VRN-H2, PPD-H1, and PPD-H2, and certain inter-locus allelic interactions.
The application of synchronous photo and thermo cycles significantly delayed the flowering time of plants with Dicktoo type VRN-H1 in both populations. The vernalization-critical regulatory regions of Dicktoo and Morex alleles at BM5A, the candidate gene at the VRN-H1 locus, are polymorphic (Fu et al., 2005; von Zitzewitz et al., 2005), but identical between Dicktoo and Kompolti korai (Szűcs et al., 2007). The significant differences found between the effects of VRN-H1 alleles on the flowering times of the two latter varieties may be due to as yet uncharacterized functional polymorphisms in other regions of the 17 kb gene or to the effects of a tightly linked gene, for example, HvPhyC (Szűcs et al., 2006). Support for the former possibility is that BM5A has been shown to respond to other environmental signals in addition to temperature: there is no transcription of the winter growth habit type Dicktoo BM5A allele under short photoperiod (Danyluk et al., 2003; von Zitzewitz et al., 2005). Further gene expression studies under the specific environmental conditions of fluctuating factors are necessary for determining which gene(s) may be responsible for the significantly delayed flowering associated with the Dicktoo VRN-H1 locus. In addition to influencing the significance of alleles at the VRN-H1 locus, the synchronously applied photo and thermo cycles also modified the interactions between VRN-H1 and the other flowering time loci in both populations. In the Dicktoo×Morex population, environment-specific interactions between VRN-H1, PPD-H1, and PPD-H2 loci could be detected.
HvPRR7, the candidate gene for the PPD-H1 locus, has an Arabidopsis homologue (AtPRR7) that is directly involved in the central oscillator cycles of the circadian clock (Salomé and McClung, 2005; Turner et al., 2005). The activity of HvPRR7 shows daily fluctuation, suggesting a connection with circadian rhythm in barley. A single nucleotide change in the CCT domain of HvPRR7 results in a loss of function mutation of tremendous adaptive importance: mutant genotypes are insensitive to long photoperiods, leading to later flowering under higher latitude summer conditions. This mutation does not affect the expression of HvPRR7 and its daily fluctuations in activity (Turner et al., 2005). Our results indicate that the effects of the synchronous photo and thermo cycling on the PPD-H1 locus were independent of allele phase in the CCT domain (Fig. 1). Therefore, the observed phenotypic variation must be due to polymorphisms in other regions of the gene besides the CCT domain, or to another linked, but unknown, gene. Support for the former comes from the report that, in wheat, a large deletion in the promoter region of PPD-D1 is responsible for the shift in peak activity within the daily circadian rhythm, resulting in altered circadian periodicity and, consequently, photoperiod insensitivity (Beales et al., 2007).
The epistatic interactions between VRN-H1 and PPD-H1 were previously identified in the Dicktoo×Morex population under long photoperiod treatments, both in greenhouse and in growth chamber tests (Pan et al., 1994; Karsai et al., 1997). In the current experiments, it is confirmed that the combination of the recessive BM5A allele at VRN-H1 and the insensitive HvPRR7 allele at PPD-H1 resulted in the latest flowering genotypes in each treatment. In plants with this allele combination, the allele phase at PPD-H2 became a significant determinant of flowering time under synchronous photo and thermo cycles.
Allelic variation at the PPD-H2 locus is of great agronomic importance: at the QTL level, this locus has significant effects on flowering time under short photoperiod conditions and in autumn field-sown experiments (Pan et al., 1994; Laurie et al., 1995; Cuesta-Marcos et al., 2008). HvFT3 is the candidate gene for the PPD-H2 locus (Faure et al., 2007). Under short-day conditions, the presence of this gene (characteristic of most spring barleys) is associated with early flowering, while the deletion of the gene (characteristic of most winter barleys) is associated with late flowering. The association between HvFT3 gene and the PPD-H2 locus is based on phenotype and allele associations, and gene expression patterns: HvFT3 is expressed only under short photoperiods and is not detectable under long light regimes (Faure et al., 2007). Our results show that the PPD-H2 locus is a significant determinant of flowering time under long photoperiods as well, but only in the case of applying synchronous photo and thermo cycles, and when specific allelic configurations are present at the PPD-H1 and VRN-H1 loci. The difference between our results and those of Faure et al. (2007) may be due to their use of a genotype with the ‘early’ (sensitive) allele at PPD-H1 and a spring allele at VRN-H1. The largest delay of flowering due to the allele phase of the PPD-H2 locus was observed in plants with the late flowering (insensitive) allele at PPD-H1 and the repressible winter allele at VRN-H1. In this case, the null/deleted (sensitive) HvFT3 allele at PPD-H2 resulted in complete repression of flowering under a long photoperiod with synchronous photo and thermo cycles, while the presence of the HvFT3 gene in PPD-H2 resulted in significantly earlier flowering.
In the Dicktoo×Kompolti korai population, the epistatic interactions between VRN-H2 and VRN-H1 loci could be examined in detail. ZCCT-H, the candidate gene at the VRN-H2 locus, is hypothesized to encode the transcriptional repressor of the winter allele at VRN-H1 (Yan et al., 2004; Dubcovsky et al., 2006; Kóti et al., 2006; Szűcs et al., 2007). This gene is present in Kompolti korai and absent in Dicktoo. It was identified in the DK population as the only significant QTL for vernalization response (Karsai et al., 2005). ZCCT expression is highest prior to vernalization and decreases rapidly during vernalization (Yan et al., 2004). Photoperiod duration also affects ZCCT activity: a level of expression occurs under long, but not short photoperiods (Dubcovsky et al., 2006; Trevaskis et al., 2006). The connection between photoperiod sensitivity and the allele phase of the VRN-H2 locus was also identified via functional QTL analyses (Karsai et al., 2006). Trevaskis et al. (2006) demonstrated that ZCCT-H gene expression levels fluctuate on a daily basis, suggesting that this gene is under circadian clock control. Under a long photoperiod and constant temperature it was found that the presence of ZCCT-H delayed flowering to the same extent (20–30 d) in plants with either the Dicktoo or the Kompolti VRN-H1 alleles (Fig. 2). The application of synchronous photo and thermo cycles dramatically changed the epistatic interaction between VRN-H2 and VRN-H1. Plants with Dicktoo type VRN-H1 flowered late, independent of the presence/absence of the ZCCT-H gene (Fig. 2).
Fluctuations in light and temperature have more modest delaying effects on flowering time of barley under greenhouse conditions, where there are gradual and small changes in light and temperature (Szűcs et al., 2007), than under the conditions reported here. The synchrony, as well as magnitude, of cycling can be important: asynchronous cycling of temperature and photoperiod was reported to accelerate flowering in Sorghum, as compared to synchronous cycling (Morgan et al., 1987; Ellis et al., 1997). Morgan et al. (1987) suggested that the daily temperature cycles and the asynchrony between photo and thermo cycles could be an additional environmental signal to supplement photoperiodic control of development, and thus play an important role in entraining circadian rhythms.
In summary, the application of synchronous photo and thermo cycles under controlled environment conditions led to dramatic changes in the flowering time phenotype. A genetical dissection of these changes via QTL analysis revealed novel effects and interactions of barley VRN and PPD loci. It was found that the facultative growth habit and short-day sensitivity of Dicktoo is due to the VRN-H2 deletion coupled with the recessive allele at VRN-H1, as previously described (von Zitzewitz et al., 2005), as well as the null (late-flowering) allele at the PPD-H2 locus. This particular allele combination might render Dicktoo sensitive to fluctuations in photoperiod and temperature. We hypothesize that this cycling in photoperiod and temperature led to changes in the entrainment of the circadian rhythm, which, in turn, altered activity of PPD-H1 and VRN-H1. Our results suggest that the VRN-H2 and PPD-H2 loci are also involved in the regulation of and/or are regulated by the circadian rhythm.
Supplementary data
Supplementary data can be found at JXB online.
Fig. S1. Phenotypic frequency distributions of flowering time in the Dicktoo×Morex (a–c) and Dicktoo×Kompolti korai (d–f) barley mapping populations under various combinations of daily fluctuating factors; (a, d) show continuous light and a constant temperature of 18 °C; (b, e) show a 16 h photo cycle and a constant temperature of 18 °C; (c, f) show a 16 h photo cycle and a 18/16 °C thermo cycle applied synchronously.
Fig. S2. QTL analysis results of flowering time in two barley mapping populations under the three environmental conditions (24C, continuous light and constant temperature of 18 °C; 16C, 16 h photo cycle and constant temperature of 18 °C; 16T, 16 h photo cycle and 18/16 °C thermo cycle applied synchronously).
Supplementary Material
Acknowledgments
The publication of the experimental results was funded by a Bolyai János Research Grant from the Hungarian Academy of Sciences and by the research grant (NK72913) of the National Scientific Research Fund (OTKA).
References
- Amasino RM. Vernalization and flowering time. Current Opinion in Biotechnology. 2005;16:154–158. doi: 10.1016/j.copbio.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting and resetting. The Plant Cell. 2004;16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O'Sullivan D, Powell W, Laurie DA, Greenland AJ. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. Journal of Experimental Botany. 2007;58:1231–1244. doi: 10.1093/jxb/erm042. [DOI] [PubMed] [Google Scholar]
- Cuesta-Marcos A, Igartua E, Ciudad FJ, et al. Heading date QTL in a spring×winter barley cross evaluated in Mediterranean environments. Molecular Breeding. 2008;21:455–471. [Google Scholar]
- Danyluk J, Kane ND, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiology. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen CL, Yan L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Molecular Breeding. 2005;15:395–407. [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Molecular Biology. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunford RP, Griffith S, Christodoulou V, Laurie DA. Characterisation of barley (Hordeum vulgare L.) homologue of the Arabidopsis flowering time regulator GIGANTEA. Theoretical and Applied Genetics. 2005;110:925–931. doi: 10.1007/s00122-004-1912-5. [DOI] [PubMed] [Google Scholar]
- Ellis RH, Qi A, Craufurd Q, Summerfield RJ, Roberts EH. Effect of photoperiod, temperature and asynchrony between thermoperiod and photoperiod on development to panicle initiation in Sorghum. Annals of Botany. 1997;79:169–178. [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare L) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szűcs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley (Hordeum vulgare L.), rice (Oryza sativa) and Arabidopsis thaliana. Plant Physiology. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Jack T. Molecular and genetic mechanisms of floral control. The Plant Cell. 2004;16:S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai I, Mészáros K, Hayes PM, Bedő Z. Effects of loci on chromosomes 2(2H) and 7(5H) on developmental patterns in barley (Hordeum vulgare L.) under different photoperiod regimes. Theoretical and Applied Genetics. 1997;94:612–618. [Google Scholar]
- Karsai I, Hayes PM, Kling J, Matus IA, Mészáros K, Láng L, Bedő Z, Sato K. Genetic variation in component traits of heading date in Hordeum vulgare subsp. spontaneum accessions characterized in controlled environments. Crop Science. 2004;44:1622–1632. [Google Scholar]
- Karsai I, Szűcs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedő Z. The Vrn-H2 locus is a major determinant of flowering time in a facultative×winter growth habit barley (Hordeum vulgare L.) mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Karsai I, Mészáros K, Szűcs P, Hayes PM, Láng L, Bedő Z. The influence of photoperiod on the Vrn-H2 locus (4H) which is a major determinant of plant development and reproductive fitness traits in a facultative×winter barley (Hordeum vulgare L.) mapping population. Plant Breeding. 2006;125:468–472. [Google Scholar]
- Karsai I, Szűcs P, Mészáros K, Puskás K, Bedő Z, Veisz O. Barley (Hordeum vulgare L.) marker linkage map: a case study of various marker types and of mapping population structure. Cereal Research Communications. 2007;35:1551–1562. [Google Scholar]
- Karsai I, Kőszegi B, Kovács G, Szűcs P, Mészáros K, Bedő Z, Veisz O. Effects of temperature and light intensity on flowering of barley (Hordeum vulgare L.) Acta Biologica Hungarica. 2008;59:205–212. doi: 10.1556/ABiol.59.2008.2.7. [DOI] [PubMed] [Google Scholar]
- Kóti K, Karsai I, Szűcs P, Horváth Cs, Mészáros K, Kiss GB, Bedő Z, Hayes PM. Validation of the two-gene epistatic model for vernalization response in a winter×spring barley cross. Euphytica. 2006;152:17–24. [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. Genetic analysis of a photoperiod response gene on the short arm of chromosome 2(2H) of Hordeum vulgare (barley) Heredity. 1994;72:619–627. [Google Scholar]
- Laurie DA, Pratchett N, Snape JW, Bezant JH. RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter×spring barley (Hordeum vulgare L.) cross. Genome. 1995;38:575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Griffiths S, Dunford RP, Christodoulou V, Taylor SA, Cockram J, Beales J, Turner A. Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crops Research. 2004;90:87–99. [Google Scholar]
- McClung CR. Plant circadian rhythms. The Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PW, Guy LW, Pao CI. Genetic regulation of development in Sorghum bicolor. Plant Physiology. 1987;83:448–450. doi: 10.1104/pp.83.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedő Z. Genetic analysis of the components of winterhardiness in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 1994;89:900–910. doi: 10.1007/BF00224516. [DOI] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. The Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JS, Szűcs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM. Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theoretical and Applied Genetics. 2006;112:832–842. doi: 10.1007/s00122-005-0185-y. [DOI] [PubMed] [Google Scholar]
- Szűcs P, Karsai I, von Zitzewitz J, Mészáros K, Cooper LLD, Gu YQ, Chen THH, Hayes PM, Skinner JS. Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theoretical and Applied Genetics. 2006;112:1277–1285. doi: 10.1007/s00122-006-0229-y. [DOI] [PubMed] [Google Scholar]
- Szűcs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Molecular Genetics and Genomics. 2007;277:249–261. doi: 10.1007/s00438-006-0195-8. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science. 2007;12:352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The Pseudo-Response Regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW. JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen, The Netherlands: Kyazma BV; 2006. [Google Scholar]
- von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TT, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. North Carolina State University, Raleigh, NC: Department of Statistics; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.