Abstract
The turnip crinkle virus-based vector TCV–GFPΔCP had been devised previously to study cell-to-cell and long-distance spread of virus-induced RNA silencing. TCV–GFPΔCP, which had been constructed by replacing the coat protein (CP) gene with a green fluorescent protein (GFP) coding sequence, was able to induce RNA silencing in single epidermal cells, from which RNA silencing spread from cell-to-cell. Using this unique local silencing assay together with mutagenesis analysis, two TCV genes, p8 and p9, which were involved in the intercellular spread of virus-induced RNA silencing, were identified. TCV–GFPΔCP and its p8- or p9-mutated derivatives, TCVmp8–GFPΔCP and TCVmp9–GFPΔCP, replicated efficiently but were restricted to single Nicotiana benthamiana epidermal cells. TCV–GFPΔCP, TCVmp8–GFPΔCP, or TCVmp9–GFPΔCP was able to initiate RNA silencing that targeted and degraded recombinant viral RNAs in inoculated leaves of the GFP-expressing N. benthamiana line 16c. However, cell-to-cell spread of silencing to form silencing foci was triggered only by TCV–GFPΔCP. Non-replicating TCVmp88–GFPΔCP and TCVmp28mp88–GFPΔCP with dysfunctional replicase genes, and single-stranded gfp RNA did not induce RNA silencing. Transient expression of the TCV p9 protein could effectively complement TCVmp9–GFPΔCP to facilitate intercellular spread of silencing. These data suggest that the plant cellular trafficking machinery could hijack functional viral proteins to permit cell-to-cell movement of RNA silencing.
Keywords: Cell-to-cell movement, Nicotiana benthamiana, RNA silencing, TCV, viral movement proteins
Introduction
RNA silencing involves structured or double-stranded RNA (dsRNA) that is processed into small interfering RNAs (siRNAs) of 21–24 nucleotides (nt) by Dicer, a member of the RNase III family of dsRNA-specific endonucleases (Hamilton and Baulcombe, 1999; Bernstein et al., 2001; Hamilton et al., 2002; Meister and Tuschl, 2004). One intriguing characteristic of RNA silencing is that RNA silencing is non-cell-autonomous. RNA silencing can be induced locally and spread to distal parts in plants, fungi, and nematodes (Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Winston et al., 2002; Himber et al., 2003; Mallory et al., 2003; Timmons et al., 2003). In Caenorhabditis elegans, several genes including sid-1 (systemic interference defective), rsd-2, rsd-3, and rsd-6 (RNAi spreading defective) have been identified in the signalling pathway for systemic RNA silencing (Winston et al., 2002; Feinberg and Hunter, 2003; Tijsterman et al., 2004). In particular, sid-1 encodes a transmembrane protein SID-1 that enables intercellular transport of dsRNA, which is essential for systemic but not cell-autonomous RNA silencing (Winston et al., 2002; Feinberg and Hunter, 2003). rsd-2 encodes a large protein RSD-2 which interacts with the rsd-6 gene product RSD-6 that may bind to RNA. rsd-3 encodes a protein (RSD-3) with an epsin N-terminal domain and is homologous to the human protein enthoprotin involved in vesicle trafficking. These proteins may operate at various steps of a pathway specific for systemic RNA silencing in C. elegans (Tijsterman et al., 2004).
In plants, compelling evidence indicates that local silencing signal(s) can move between cells via plasmodesmata, while the systemic silencing signal(s) spreads within the vascular system, mimicking the cell-to-cell and long-distance movement of plant viruses (Palauqui and Balzergue, 1997; Voinnet et al., 1998, 2000; Ueki and Citovsky, 2001; Himber et al., 2003; Baulcombe, 2004). Transgenes, high molecular weight RNAs and siRNAs have also been reported to induce RNA silencing that can systemically spread in plants (Braunstein et al., 2002; Klahre et al., 2002; Van Houdt et al., 2003; Garcia-Perez et al., 2004). However, the characteristics of mobile silencing signals have not been definitely reported in plants and C. elegans (Mallory et al., 2001, 2003; Parrish and Fire, 2001). It has been proposed that the 21 nt siRNAs represent the local silencing signal for limited cell-to-cell movement of RNA silencing across only 10–15 cells (Hamilton et al., 2002; Himber et al., 2003). Production of the 21 nt cell-to-cell silencing signal requires DICER-LIKE 4, and at least three silencing movement-deficient genes (SMD1, SMD2, and SMD3) are essential for intercellular spread of 21 nt siRNA (Dunoyer et al., 2005). SMD1 and SMD2 are found to be allelic to RDR2 (RNA-dependent RNA polymerase 2) and NRPD1a (RNA polymerase IVa), respectively (Dunoyer et al., 2007). Interestingly, NRPD1a, RDR2, and DCL3 (DICER-like 3), as well as ago4 (argonaute4), are required for reception of long-distance RNA silencing (Brosnan et al., 2007). On the other hand, the 24 nt siRNA is indispensable for long-range (extensive) cell-to-cell and systemic spread of silencing by acting as a phloem-specific silencing signal (Hamilton et al., 2002; Himber et al., 2003). Extensive cell-to-cell movement of RNA silencing also requires the combined activity of SDE1, a putative RNA-dependent RNA polymerase, and SDE3, a putative RNA helicase, although SDE1 and SDE3 may not directly participate in the movement of silencing (Dalmay et al., 2000, 2001; Himber et al., 2003). An RNA-dependent RNA polymerase (RDR6) was found to be required for cells to respond to the long-distance silencing signal, but not to produce or translocate the signal in N. benthamiana (Schwach et al., 2005). More recently, an SNF2 protein CLASSY1 was shown to act together with RDR2 and an RNA polymerase IVa gene product NRPRD1a in the production of siRNAs for the intercellular spread of transgene silencing (Smith et al., 2007). However, no protein has been reported to be involved directly in cell-to-cell and long-distance trafficking of RNA silencing signals in plants.
A virus-induced RNA silencing (VIRS) vector, based on Turnip crinkle virus (TCV), was developed to discriminate between cell-to-cell and long-distance systemic spread of RNA silencing in plants (Ryabov et al., 2004). TCV, a member of the Carmovirus genus, has a positive-sense single-stranded RNA genome (4053 nt) and encodes five proteins (Carrington et al., 1989). P28 and its read-through (p88) proteins are required for viral RNA replication. Two small overlapping genes, p8 and p9, are essential for cell-to-cell movement and systemic spread of the virus (Hacker et al., 1992; Li et al., 1998). The 3′-proximal gene codes for the 38 kDa coat protein (CP) that is indispensable for cell-to-cell movement of TCV in N. benthamiana (Cohen et al., 2000a) and for suppression of RNA silencing (Qu et al., 2003; Thomas et al., 2003). The VIRS assay vector TCV–GFPΔCP lacks the CP gene, which is replaced with a functional green fluorescent protein (GFP) coding sequence. TCV–GFPΔCP differs from other previously described plant virus-based VIRS vectors (Burch-Smith et al., 2004). First, the latter cannot distinguish between cell-to-cell and systemic spread of RNA silencing because these earlier viruses, with silencing trigger sequences, moved from cell-to-cell and long distance. Secondly, the TCV–GFPΔCP VIRS assay is different from the graft-induced RNA silencing assay, which is widely used to investigate the systemic spread of RNA silencing. Moreover, TCV–GFPΔCP infects only one type of cells (i.e. epidermal cells) after mechanical inoculation (Cohen et al., 2000a; Ryabov et al., 2004), where silencing is initiated and then the silencing signal(s) moves to other epidermal and mesophyll cells. By contrast, the types of cells in which silencing is initiated cannot be identified easily during agro-infiltration- or bombardment-induced RNA silencing. Both agroinfiltration and biolistics can deliver inducers of silencing simultaneously into various cell types, including epidermal, mesophyll, and vascular cells. The initiation and subsequent spread of silencing in different cell types may have different signalling requirements. Thus, the TCV-based VIRS assay provides a unique tool to dissect the trafficking of silencing, in particular, the cell-to-cell movement of RNA silencing in plants. Using the TCV-based VIRS assay, mutagenesis, and complementation analysis, it was shown that only replicating, but not non-replicating, TCV-based vectors could initiate VIRS, and that host cellular silencing trafficking machinery could make use of TCV gene functions for cell-to-cell spread of RNA silencing.
Materials and methods
Plasmid construction
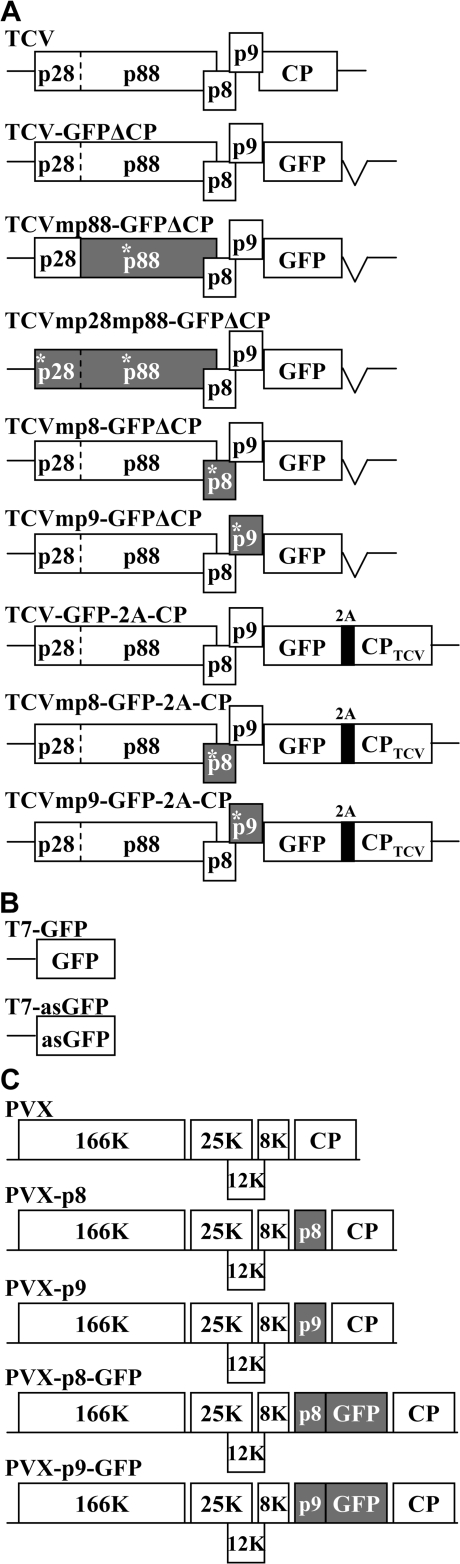
Diagrammatic representations of recombinant viruses and plasmids used in this work are shown in Fig. 1. The full-length infectious cDNA clone of the UK isolate (AY312063) of turnip crinkle virus pT7.TCV and its derivative pT7.TCV–GFPΔCP in which the CP gene was replaced with a GFP-coding sequence were described previously (Ryabov et al., 2004). Plasmid pT7.TCV–GFPΔCP was linearized at the unique AvrII or BbvCI sites at nucleotide positions 108 or 1802, respectively. Protruding 5′ termini were filled in using the Klenow fragment of DNA polymerase I and recircularized to produce pT7.TCVmp28mp88–GFPΔCP and pT7.TCVmp88–GFPΔCP (Fig. 1A). The mutation at the AvrII site reduced the coding capacity of open reading frame p28 from 251 to 16 amino acids, and no read-through (p88) protein can be produced by TCVmp28mp88–GFPΔCP. The mutation in TCVmp88–GFPΔCP at the BbvCI site introduced an Asp residue into the p88 protein sequence between Ala and Glu at amino acid positions 580 and 581.
Fig. 1.
A schematic representation of recombinant viral constructs. (A) Genomic organization of TCV and TCV-based local RNA silencing vectors. Viral genes and GFP coding regions are presented as open boxes. Mutated open reading frames have asterisks and are shaded. The self-cleaving 2A oligopeptide of Foot-and-mouth disease virus is shown as a dark box. (B) Gfp sense and anti-sense cassettes. The GFP gene was cloned into pGEM-T Easy vector in opposite orientations under the transcriptional control of the T7 RNA promoter. (C) Genomic organization of PVX and PVX-based gene expression constructs.
pT7.TCV–GFPΔCP contains a unique BglII site downstream of the stop codon for the p9 movement protein and a single PacI site downstream of the 3′ end of the TCV genome. A fragment containing the gfp-2a-cp gene coding for GFP–2A–CP and the 3′-terminal untranslated region of TCV was generated by overlap extension PCR (Higuchi et al., 1988). The self-cleaving 2A oligopeptide coding sequence was originated from the Foot-and-mouth disease virus (Santa Cruz et al., 1996). PCR amplifications were performed using pTXS.GFP–CP, a derivative of potato virus X (PVX) vector containing the gfp-2a fusion gene (Santa Cruz et al., 1996) and pT7.TCV as templates, and a gfp-specific primer 5′-GGTTagatctATGAGTAAAGGAGAAG-3′ with a BglII site (shown in lower case, preceding the 5′-terminal sequence of gfp-2a), a pair of self-complementary mutagenic primers 5′-CGAGTCCAACCCTGGGCCCGCGATGGAAAATGATCCTAG-3′ and 5′-CTAGGATCATTTTCCATCGCGGGCCCAGGGTTGGACTCG-3′ (regions corresponding to 5′-terminus of TCV cp gene are shown in bold), and the M13 reverse primer as the flanking primer. The amplified fragment was cloned into the BglII and PacI sites of pTCV–GFPΔCP to produce pTCV–GFP–2A–CP (Fig. 1A).
To construct p8-mutated TCV clones, DNA fragments were generated by PCR using the mutagenic primer 5′-GATAGATGGATCCTtAAtGAATTCCCTACAACTCTC-3′ (corresponding to nucleotides 2352–2387 of TCV), the M13 reverse primer, and pT7.TCV–GFP–ΔCP or pT7.TCV–GFP–2A–CP as templates. Modified nucleotides to substitute the fourth and fifth codons of the p8 gene with the stop codons TAA and TGA are shown in lower case. The resulting PCR products were digested with BamHI and cloned to replace a BamHI fragment of pT7.TCV–GFP–ΔCP to produce pT7.TCV–mp8GFP–ΔCP and pT7.TCV–mp8GFP–2A–CP (Fig. 1A). To construct p9-mutated TCV clones, DNA fragments were generated by overlap extension PCR using the primer 5′- CCAAGATGCTAGGTTCAG-3′ (corresponding to nucleotides 2219–2236 of TCV), the mutagenic primers 5′-AGAGAcGAAGGTTCTGtaAGCCACGGGGGTACTTG-3′ and 5′- CCGTGGCTtaCAGAACCTTCgTCTCTTTTCTTGT-3′, the M13 reverse primer as the flanking primer, and pT7.TCV–GFPΔCP or pT7.TCV–GFP–2A–CP as templates. Modified nucleotides to mutate the ATG start codon of the p9 gene to ACG and to substitute the fifth codon with a stop codon (TAA) are shown in lower case. The resulting PCR products were then digested with BamHI and cloned to replace a BamHI fragment of pT7.TCV–GFPΔCP to produce pT7.TCV–mp9GFP–ΔCP and pT7.TCV–mp9GFP–2A–CP (Fig. 1A).
The GFP-coding sequence was PCR amplified using a pair of primers 5′-ATGAGTAAAGGAGAAGAA-3′ and 5′-TTTGTATAGTTCATCCATGCCA-3′ with pTXS.GFP–CP (Santa Cruz et al., 1996) as template. The gfp gene was cloned directly into pGEM-T Easy vector (Promega) to produce pT7–GFP and pT7–antisenseGFP (Fig. 1B). The TCV p8 and p9 genes were PCR amplified using pfu DNA polymerase (Promega), pTCV DNA template, and a set of gene-specific primers 5′-TTAGTGATcGATGGATCCTGAACGAATTCCC-3′ and 5′-CATGTCcGgccgGAAGTTGAAGTTGATTGAGAC-3′ for p8, and 5′-AAGAAAAtcgATGAAGGTTCTGCTAGTCACGGGGG-3′ and 5′-ACTCTAcGgcCgTTTTCCGTTTCCAGTGTTGATG-3′ for p9. Modified nucleotides are shown in lower case and introduced ClaI and EagI sites are underlined. PCR products corresponding to the two genes were digested with ClaI and EagI, and cloned into ClaI/EagI-digested PVX-based vectors p45P46 and p45P46–GFP (Van Wezel et al., 2001) to produce PVX-p8, PVX-p9, PVX-p8–GFP, and PVX-p9–GFP (Fig. 1C). The integrity of all recombinant clones and the presence of gene mutations were confirmed by sequencing.
Virus infection and replication assays
RNA transcripts were produced by in vitro transcription from each construct and mechanically inoculated onto N. benthamiana plants as described (Van Wezel et al., 2001). Plants were maintained in an insect-free glasshouse at 25 °C. To measure the accumulation of viral RNA and the level of gfp RNA, total RNA was extracted from leaf tissues using the RNeasy plant mini kit (Qiagen) and assayed by reverse transcriptase-polymerase chain reaction (RT-PCR) as described (Ryabov et al., 2004).
Epifluorescence microscopy
Inoculated N. benthamiana leaves were collected, examined with a Zeiss Axiophot microscope through a green filter and photographed with a Nikon Coolpix990 digital camera as previously described (Dong et al., 2003).
Local RNA silencing assays
Seedlings of transgenic N. benthamiana line 16c that constitutively express GFP (Brigneti et al., 1998) were mechanically inoculated with RNA transcripts produced by in vitro transcription from Pac I-linearized TCV- or SpeI-linearized PVX-based constructs. Induction and cell-to-cell spread of RNA silencing of GFP expression were routinely examined under long-wavelength UV light and recorded photographically using a Nikon Digital Camera Coolpix990. Regions of the leaf lamina where silencing of gfp RNA occurred showed red chlorophyll fluorescence (red foci) while tissues expressing GFP showed green fluorescence. Leaf samples were taken for RNA extraction and RT-PCR assays as described (Ryabov et al., 2004).
Results
VIGS constructs
The movement-deficient TCV–GFPΔCP is able to trigger efficient intercellular spread of RNA silencing (Ryabov et al., 2004), suggesting that viral factors may be exploited by plants to spread the RNA silencing signal between cells. To test the potential involvement of TCV genes in the cell-to-cell movement of RNA silencing, a series of TCV–GFPΔCP-based VIRS vectors was constructed by introducing mutations into individual viral genes (Fig. 1A), as well as recombinant plasmids for the production of sense and anti-sense strands of gfp RNA (Fig. 1B), and PVX-based vectors for expressing TCV p8 and p9, and p8– and p9–GFP fusion proteins (Fig. 1C). TCVmp88–GFPΔCP had the capacity to produce a mutated p88, while TCVmp28mp88–GFPΔCP could only express a truncated p28 but no read-through p88. A stop codon (TAA) was introduced into the 5′ end of the p8 and p9 genes in TCVmp8–GFPΔCP and TCVmp9–GFPΔCP, respectively, in addition to separate mutations which disrupted the initiation codons in the p8 and p9 genes.
Effects of viral gene mutations on the cell-to-cell movement and replication of TCV
Total RNA was extracted from non-transgenic N. benthamiana leaves inoculated with equal amounts of RNA transcripts of TCV–GFPΔCP or its mutant derivatives at 3, 12, and 24 d post-inoculation (dpi) and analysed by RT-PCR (Fig. 2). TCV-specific RT-PCR products were readily detected in TCV–GFPΔCP, TCVmp8–GFPΔCP, or TCVmp9–GFPΔCP infections (Fig. 2A). All three recombinant viruses maintained their readily detectable gfp sequence (Fig. 2B). However, no virus- or gfp-specific sequences were detected in mock-inoculated leaves or in leaves inoculated with TCVmp88–GFPΔCP, TCVmp28mp88–GFPΔCP, or the sense or anti-sense strand of gfp RNA, although an 180 bp 18S rRNA-specific RT-PCR product was present in all samples (Fig. 2C). RT-PCR reactions without AMV RT (negative controls) gave no positive signal for any target sequences.
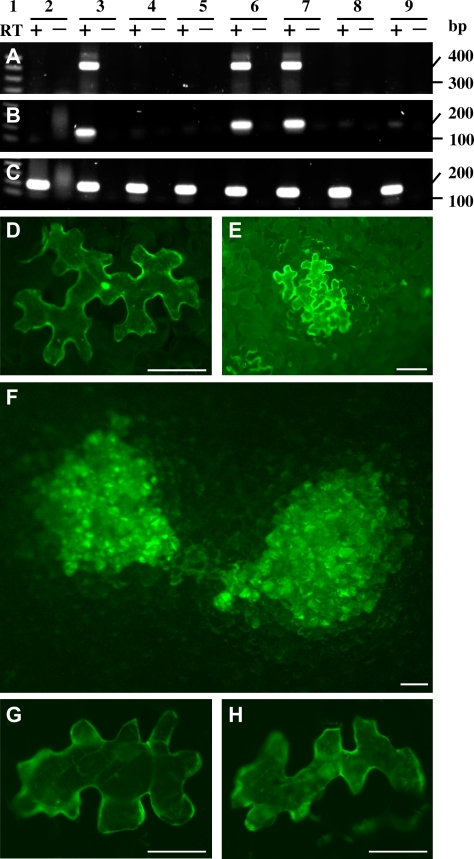
Fig. 2.
Requirement of functional p8 and p9 for cell-to-cell movement, but not for replication of TCV in single Nicotiana benthamiana epidermal cells. (A–C) Effect of various gene mutations on TCV replication. Detection of TCV (A), gfp RNA (B), and 18S rRNA (C) were performed by RT-PCR (+) using DNase I-pretreated RNA samples (10 ng). RT was excluded in RT-PCR control reactions (–) for each detection. Total RNA samples were extracted from N. benthamiana leaves mock-inoculated (2) or inoculated with TCV–GFPΔCP (3), TCVmp88–GFPΔCP (4), TCVmp28mp88–GFPΔCP (5), TCVmp8–GFPΔCP (6), TCVmp9–GFPΔCP (7), sense (8) or anti-sense (9) gfp ssRNA RNAs at 24 d post-inoculation (dpi). Similar results were obtained using RNAs extracted from leaves collected at 3 and 12 dpi (data not shown). The sizes (in base pairs, bp) and positions of a 1 kb DNA ladder (Sigma) (lane 1) are indicated. (D–F) Fluorescent microscopy of GFP expression in N. benthamiana leaves. TCV–GFPΔCP restricted to single epidermal cells showed green GFP fluorescence (D). TCV–GFP–2A–CP trafficked from cell-to-cell in a group of four or five epidermal cells started to show GFP expression at 2 dpi (E) and multi-cellular GFP-fluorescent lesions appeared at 6 dpi (F). Both TCVmp8–GFP–2A–CP (G) and TCVmp9–GFP–2A–CP (H) were limited to single epidermal cells. Photographs were taken using a Zeiss Axiophot fluorescent microscope through a green filter. Scale bar=100 μm.
Next, the type of cells that could be infected by TCV–GFPΔCP and its mutant derivatives after mechanical inoculation was investigated. Consistent with the inability of TCVmp88–GFPΔCP, TCVmp28mp88–GFPΔCP, or gfp sense and anti-sense strand RNA to replicate, no GFP fluorescent cells were observed after intensive microscopic scrutiny of N. benthamiana leaves mechanically inoculated with those constructs. TCV–GFPΔCP was restricted to single epidermal cells showing the green florescence of viral GFP transient expression (Fig. 2D). To correlate effects of mutations in p8 or p9 with intercellular movement of TCV, the mobility of TCV–GFP–2A–CP, TCVmp8–GFP–2A–CP, and TCVmp9–GFP–2A–CP in plants was assayed (Fig. 2). A group of four to five epidermal cells appeared to show GFP fluorescence on leaves inoculated with TCV–GFP–2A–CP at 2 dpi (Fig. 2E). Up to 100 different cells were infected and formed multi-cellular GFP-fluorescent spots at 6 dpi (Fig. 2F). By contrast, p8- or p9-mutated TCVmp8–GFP–2A–CP or TCVmp9–GFP–2A–CP was limited in single epidermal cells. Not surprisingly, double-mutant viruses TCVmp8–GFPΔCP and TCVmp9–GFPΔCP, similar to TCVmp8–GFP–2A–CP and TCVmp9–GFP–2A–CP, did not move and only unicellular GFP fluorescence was observed in single epidermal cells (Fig. 2G, H). The p8 or p9 mutant virus produced unicellular GFP fluorescence that was visible at 2 dpi, and remained within single cells at 24 dpi.
TCV p8 and p9 are associated with the intercellular spread of RNA silencing
Mechanical inoculation of TCV–GFPΔCP onto GFP transgenic N. benthamiana line 16c plants initiated gfp RNA silencing, which then spread effectively from cell-to-cell to form gfp-silenced foci, which appeared as red spots due to the chlorophyll fluorescence. Small red silencing foci of gfp-silenced cells were seen at 3 dpi under epifluorescence microscope (Fig. 3B). The gfp-RNA silenced foci increased in size progressively (Fig. 3C). Such ‘silencing foci’ eventually became visible to the naked eye under long-wavelength UV light and were scattered throughout the whole lamina of each inoculated line 16c plant leaf (Fig. 3D), indicating efficient cell-to-cell movement of RNA silencing. As in mock-inoculated leaves, non-replicating TCVmp88–GFPΔCP, TCVmp28mp88–GFPΔCP, and sense or anti-sense gfp mRNA failed to induce any visible phenotype of gfp RNA silencing. No red silencing cells were observed on inoculated leaves despite intensive scrutiny under an epifluorescence microscope (Fig. 3A, E, F, I, J). In three independent experiments, both TCVmp8–GFPΔCP and TCVmp9–GFPΔCP rarely triggered local gfp-silencing, observed as red foci, hallmark of the cell-to-cell spread of RNA silencing (Figs 3G, H, 4A), although both recombinant viruses, like their parental virus TCV–GFPΔCP, infected and replicated efficiently in single epidermal cell (Figs 2, 4B, C). This was in contrast to the significantly higher number of silencing foci triggered by TCV–GFPΔCP (Fig. 4A).
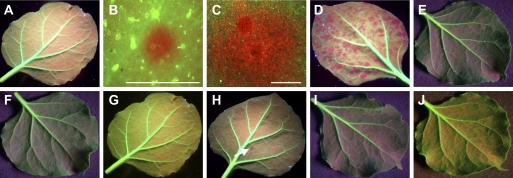
Fig. 3.
Local induction and cell-to-cell spread of RNA silencing in GFP transgenic Nicotiana benthamiana line 16c plants. Plants were mock-inoculated (A) or mechanically inoculated with RNA transcripts of TCV–GFPΔCP (B–D), TCVmp88–GFPΔCP (E), TCVmp28mp88–GFPΔCP (F), TCVmp8–GFPΔCP (G), TCVmp9–GFPΔCP (H), sense (I), or anti-sense (J) gfp ssRNA RNAs. Photographs of (A) and (C–J) were taken at 9 dpi and that of (B) at 3 dpi with a Nikon Coolpix990 digital camera under long-wavelength UV illumination through a yellow filter. TCV–GFPΔCP-inoculated leaves (B, C) were examined by fluorescent microscopy through a red filter to show progressive enlargement of individual gfp RNA silencing foci. Regions of gfp RNA silencing appeared red due to chlorophyll fluorescence, while GFP-expressing tissues showed green fluorescence. Only one red focus of gfp RNA silencing (arrow) is seen on the leaf inoculated with TCV–mp9GFPΔCP (H). Scale bar=1 mm.
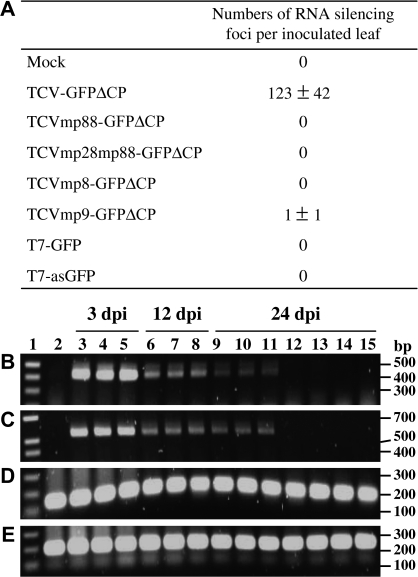
Fig. 4.
Effects of mutations in p8 and p9 on the ability of TCV–GFPΔCP to induce local RNA silencing. (A) Numbers of silencing foci per inoculated leaf. Numbers were counted at 9 dpi in mock-inoculated line 16c plants, and in plants inoculated with RNA transcripts of TCV–GFPΔCP and its derivatives. Data are represented as means ±standard deviations per inoculated leaf by counting the numbers of silencing foci on individual leaves on six plants in three independent experiments. (B–E) Effect of local RNA silencing on viral RNA accumulation. RT-PCR was performed to detect TCV- (B), TCV-gfp- (C), gfp- (D), or 18S rRNA- (E) specific RNAs using 10 ng total RNA samples extracted from line 16c plant leaves following mock inoculation (lane 2), or inoculated with TCV–GFPΔCP (lanes 3, 6, 9), TCVmp8–GFPΔCP (lanes 4, 7, 10), TCVmp9–GFPΔCP (lanes 5, 8, 11), TCVmp88–GFPΔCP (lane 12), TCVmp28mp88–GFPΔCP (lane 13), sense (lane 14), or antisense (lane 15) gfp RNA. Leaves were harvested at 3, 12, and 24 dpi. The sizes of amplicons (in base pairs, bp) and the positions of the 1 kb DNA ladder (Sigma) (lane 1) are indicated.
Levels of gfp and viral RNAs in inoculated line 16c leaves were further analysed by RT-PCR (Fig. 4). Consistent with limited induction of local RNA silencing, transgenically expressed and transient viral gfp RNAs were readily detectable as a 120 bp RT-PCR product in all samples taken at various stages (Fig. 4D). Using two TCV-specific primers, a 410 bp RT-PCR fragment of TCV RNA was detected at 3 dpi, and the level declined at 12 dpi and 24 dpi, but no TCV-specific RNA was found in the mock-inoculated controls (Fig. 4B). Similar results were obtained by RT-PCR using a 5′ primer specific to TCV and a 3′ primer specific to the inserted gfp sequence to detect a 530 bp RNA spanning both the 410 bp TCV- and 120 bp gfp-specific sequences (Fig. 4C). Due to their inability to replicate, no TCVmp88–GFPΔCP-, TCVmp28mp88–GFPΔCP-, or gfp-specific RNA was detected, although an 18S rRNA-specific RT-PCR product was seen in all samples (Fig. 4E).
Transient expression of the TCV p9 protein complements TCVmp9–GFPΔCP to facilitate the intercellular trafficking of RNA silencing
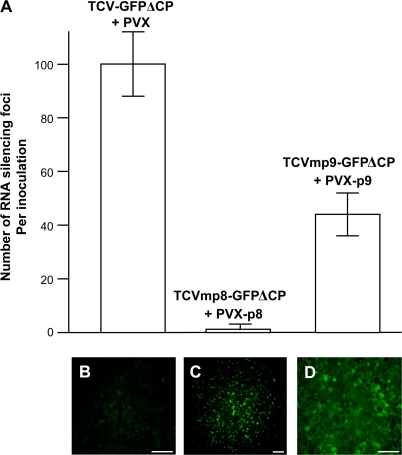
To find out if transient expression of the TCV p8 or p9 protein could complement TCVmp8GFPΔCP or TCVmp9–GFPΔCP, respectively, to facilitate the cell-to-cell spread of RNA silencing, the TCV p8 or p9 genes were cloned into a modified PVX-based vector in which transient expression of p8 or p9 was under the control of a duplicated PVX CP sub-genomic promoter (Fig. 1C). Co-inoculation of line 16c plants with wild-type PVX had no effect on TCV–GFPΔCP to induce and spread gfp RNA silencing and ∼100 of the characteristic silencing red foci per co-inoculated leaf were observed (Fig. 5A). Then line 16c plants were inoculated with mixtures of RNA transcripts of TCVmp8–GFPΔCP and PVX–p8, or TCVmp9–GFPΔCP and PVX–p9 in two separate experiments. PVX–p8 did not complement TCVmp8–GFPΔCP to promote cell-to-cell movement of gfp RNA silencing. By contrast, the deficiency of TCVmp9–GFPΔCP for intercellular spread of RNA silencing was effectively recovered by PVX–p9. Here, many typical gfp-silenced red foci became visible on 16c plant leaves co-inoculated with TCVmp9–GFPΔCP and PVX–p9 (Fig. 5A). However, these silencing foci took longer to develop and were visible under the fluorescent microscope only at 8–10 dpi and later under long-wavelength UV light. Importantly, no cell-to-cell movement of TCVmp8–GFPΔCP or TCVmp9–GFPΔCP in plants co-inoculated with PVX, PVX–p8, or PVX–p9 was observed (data not shown).
Fig. 5.
Transient expression of TCV p9 protein complements TCVmp9–GFPΔCP to promote cell-to-cell spread of RNA silencing. (A) Average numbers of silencing foci per inoculation are shown. Numbers were counted at 18 dpi in line 16c plants co-inoculated with TCV–GFPΔCP+PVX, TCVmp8–GFPΔCP+PVX-p8, or TCVmp9–GFPΔCP+PVX-p9. (B–D) Non-transformed N. benthamiana plants were infected with PVX (B), PVX-p8–GFP (C), or PVX-p9–GFP (D). Photographs of lesions which resulted from virus infection were taken at 5 dpi using a Zeiss Axiophot fluorescent microscope through a green filter. GFP-expressing cells showed green fluorescence predominately in their nuclei (C) or throughout their cytoplasm (D). Scale bar=100 μm.
Expression of the TCV p8 and p9 proteins was confirmed indirectly by examining the production of p8–GFP and p9–GFP fusion proteins from PVX–p8–GFP and PVX–p9–GFP, the gene expression vectors corresponding to PVX–p8 and PVX–p9, respectively (Fig. 1C). Fluorescent microscopy showed that infection of N. benthamiana with PVX–p8–GFP or PVX–p9–GFP resulted in multiple cells with strong green fluorescence generated from the p8–GFP and p9–GFP fusion proteins (Fig. 5C, D) in contrast to the background fluorescence associated with PVX control infection (Fig. 5B).
Discussion
In this paper, it is reported that TCV–GFPΔCP, p8- or p9-mutated TCV–GFPΔCP possessed replication capacities and was confined to single epidermal cells after mechanical inoculation of N. benthamiana. Nonsense mutations in p8 and p9 had no effect on viral RNA accumulation. However, truncations of replicase proteins p28 or p88 inhibited replication of TCV–GFPΔCP. The present data also indicate that TCV–GFP–2A–CP expressed a functional CP either in the fusion form or free CP via cleavage of GFP–2A–CP fusion protein by the foot-and-mouth disease virus 2A peptide (Santa Cruz et al., 1996), and that this was required for cell-to-cell movement of TCV in N. benthamiana. Moreover, due to their inability to spread from cell-to-cell, the levels of TCV–GFPΔCP, TCVmp8–GFPΔCP, and TCVmp9–GFPΔCP RNAs detected in inoculated leaves (Fig. 2) probably reflected equivalent accumulation of recombinant viral RNAs in single epidermal cells.
Mutations in individual genes of the VIRS vector TCV–GFPΔCP could generate mutant viruses (Fig. 1A) with altered abilities to replicate. For example, mutant viruses that carried the silencing inducer gfp sequences could accumulate to different levels in individual epidermal cells, and the dose of the inducer might be inappropriate to initiate gfp RNA silencing for silencing to spread. Consistent with this hypothesis, it was demonstrated that viral RNA replication was a prerequisite for TCV–GFPΔCP to trigger local VIRS. Mutations in p28 and p88 were lethal in TCVmp88–GFPΔCP and TCVmp28mp88–GFPΔCP. These two mutant viruses did not replicate and their lack of dsRNA may have contributed to their failure to induce RNA silencing (Figs 3, 4). This phenomenon implies that endogenous RNA-dependent RNA polymerases identified as being necessary to convert ssRNA into the dsRNA initiator of silencing in N. benthamiana and Arabidopsis (Dalmay et al., 2000; Mourrain et al., 2000; Schwach et al., 2005) may not be active in epidermal cells. This idea is supported by the fact that direct inoculation of 16c leaves with gfp sense or anti-sense ssRNA failed to trigger RNA silencing (Fig. 3).
On the other hand, TCV CP is a potent suppressor of RNA silencing and p8 or p9 is not involved in silencing suppression (Qu et al., 2003; Thomas et al., 2003). Expression of CP during TCV co-infection is able to arrest TCV–GFPΔCP-mediated local induction of RNA silencing (Ryabov et al., 2004) and the effectiveness to trigger RNA silencing by TCV–GFP–2A–CP was dramatically reduced (data not shown). Deletion of the CP gene from the viral genome enabled the three replicating but intercellular movement-deficient viruses TCV–GFPΔCP, TCVmp8–GFPΔCP, and TCVmp9–GFPΔCP to initiate and maintain silencing of both transgenic and transient gfp RNAs in individual epidermal cells, which effectively targeted and degraded recombinant viral RNAs during the course of infection (Figs 2, 4B, C). However, the cell-to-cell movement of silencing occurred only in association with TCV–GFPΔCP, and not with TCVmp8–GFPΔCP or TCVmp9–GFPΔCP. Therefore, knocking-out the p8 or p9 gene resulted in a loss-of-function of TCV–GFPΔCP for promoting cell-to-cell movement of RNA silencing. This is further evident by the fact that transient expression of the p9 protein from PVX/p9 led to a gain-of-function of TCVmp9–GFPΔCP by facilitating the cell-to-cell spread of silencing, although the p8 protein expressed in a similar manner failed to complement the deficiency of TCVmp8–GFPΔCP (Fig. 5).
The precise mechanism for the different silencing phenotypes, which were triggered by TCV–GFPΔCP, TCVmp8–GFPΔCP, or TCVmp9–GFPΔCP, remains to be elucidated. One possibility is that the silencing foci could be caused by limited cell-to-cell spread of TCV–GFPΔCP due to the activity of viral movement protein p9 and/or p8, which in turn caused gfp RNA silencing in the adjacent cells. However, fluorescent microscopic examination of leaves inoculated with the three CP-less TCV mutants clearly showed that they were restricted to single epidermal cells. There was also no difference in the florescence intensity in surrounding cells, indicating that TCV–GFPΔCP and its p8- or p9-mutated derivatives were indeed incapable of cell-to-cell movement (Fig. 2). Furthermore, it is unlikely that TCV–GFPΔCP could move across so many cells in which VIRS occurred in the absence of silencing suppressor. In this scenario, any limitedly spread viral RNA would be immediately silenced because the silencing suppressor was not expressed and there would be no further spread of TCV–GFPΔCP. Thus, induction of local RNA silencing foci by TCV–GFPΔCP was unlikely to be caused by limited cell-to-cell diffusion of this movement-deficient virus.
Alternatively, cell-to-cell spread of RNA silencing may involve the transport of mobile silencing signals through plasmodesmata. This process mimics the intercellular spread of plant RNA viruses, which is facilitated by virus-coded cell-to-cell movement proteins that frequently interact with RNA in a non-sequence-specific manner. Indeed, TCV proteins p8 and p9 are viral RNA cell-to-cell movement proteins (Hacker et al., 1992; Li et al., 1998; Cohen et al., 2000a). It is possible that p8 and p9 could also promote the spread of RNA silencing signals between cells. Interestingly, p8 is a non-specific RNA-binding protein that is localized in plant cell nuclei and also interacts specifically with the Arabidopsis transmembrane Atp8 protein that possesses two RGD motifs (Wobbe et al., 1998; Cohen et al., 2000b; Lin and Heaton, 2001). RGD-containing proteins function in numerous cell-signalling pathways through RGD-integrin recognition and interactions with the actin-based cytoskeleton (Ruoslahti, 1996). However, p9 is not confined to the cell nucleus, but is found throughout the cytoplasma, and does not interact with the Atp8 protein. Bioinformatics analysis (Hirokawa et al., 1998) indicated that p9 consists of 85 amino acid residues and has two separate transmembrane helices of 17 and 23 residues that could direct this protein into cell membranes (data not shown). Thus, the biochemical, molecular, and cellular requirements for p8- and p9-mediated transport of RNA silencing signals could differ. On the other hand, the expression studies with the GFP fusion proteins (Fig. 5C, D) suggest that far more p9–GFP than p8–GFP accumulates in plants. These factors may collectively account for the disparity that transient expression of p9 restored the ability of TCVmp9–GFPΔCP to promote the cell-to-cell movement of RNA silencing while p8 was unable to complement the deficiency for intercellular spread of RNA silencing in TCVmp8–GFPΔCP.
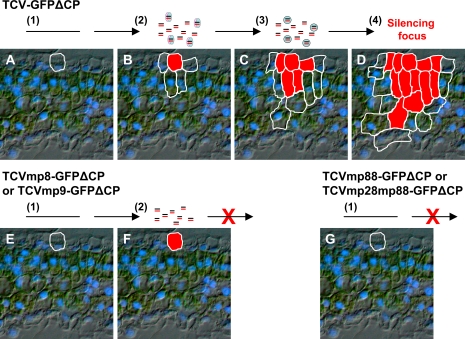
As a general mechanism against silencing, virus infections may have a detrimental effect on machinery for spreading the host silencing, resulting in a shutdown of intercellular communication for cellular RNA silencing defence between the initially infected and neighbouring cells. In this scenario, host cells would have to exploit virus-encoded protein(s) that could facilitate the intercellular trafficking of silencing signal to avoid further infection. Based on this assumption, we propose a model to explain why TCV–GFPΔCP, TCVmp8–GFPΔCP, or TCVmp9–GFPΔCP could induce RNA silencing, and why only TCV–GFPΔCP promoted the spread of silencing from single epidermal cells to adjacent cells to form visible silencing foci (Fig. 6). TCV–GFPΔCP, TCVmp8–GFPΔCP, or TCVmp9–GFPΔCP infects but cannot spread from initially invaded single epidermal cells (Fig. 6A, E). There they replicate to produce positive and negative viral ssRNA strands. dsRNA structures could be formed within or/and between these ssRNAs that trigger unicellular RNA silencing (Fig. 6B, F). At this stage, the primary mobile RNA silencing signal, presumably the 21 nt siRNA, is produced by a DICER-LIKE endonuclease (Dunoyer et al., 2005). However, the primary mobile silencing signal is unable to move out from the virus-infected epidermal cell because the host cell transport mechanism has been shut down by viral infection. Instead, the host cell could hijack the p8 and p9 proteins expressed by TCV–GFPΔCP to promote the trafficking of the primary silencing signal to adjacent cells. The primary silencing signal could then induce secondary RNA silencing in these uninfected cells, which leads to subsequent production of a secondary mobile silencing signal (Fig. 6C). Due to the absence of virus infection, trafficking of the secondary silencing signal to further cells is probably mediated by a host cell mechanism as has been proposed by others (Himber et al., 2003; Dunoyer et al., 2005), resulting in the progressive development of an RNA silencing focus (Fig. 6D). However, in the absence of p8 or p9, trafficking of the primary silencing signal from the initially virus-infected cell to neighbouring ‘recipient’ cells could not take place. In this case, the cascade of the induction and movement of RNA silencing is unlikely to continue. Thus, RNA silencing triggered by TCVmp8–GFPΔCP or TCVmp9–GFPΔCP stops spreading beyond the primary virus-infected cell to form a visible red focus of gfp RNA silencing (Fig. 6F). On the other hand, TCVmp88–GFPΔCP, TCVmp28mp88–GFPΔCP, sense or anti-sense gfp ssRNA induces no RNA silencing because these ssRNAs could not be converted into dsRNA silencing triggers in epidermal cells (Fig. 6G).
Fig. 6.
Schematic model for p8- or p9-mediated cell-to-cell spread of RNA silencing. (A–D) Induction and intercellular spread of RNA silencing induced by TCV–GFPΔCP. (E, F) RNA silencing in single-epidermal cells infected by TCVmp8–GFPΔCP or TCVmp9–GFPΔCP. (G) No RNA silencing induction by TCVmp88–GFPΔCP or TCVmp28mp88–GFPΔCP. The sequences of events are as follows: (1) viral RNA transcript enters an epidermal cell following mechanical inoculation (A, E, G); (2) viral RNA then replicates and triggers RNA silencing in the first invaded epidermal cell (B, F). At this stage, the primary silencing signals are produced. Without replication, these viruses cannot trigger RNA silencing (G). ‘=’ stands for mobile RNA silencing signal, ellipses for p8 or p9, and circles for host factors required for silencing cell-to-cell movement.
This model may also be applied to the intercellular spread of RNA silencing that occurs in the absence of virus infection. A mobile silencing signal generated in individual cells, for example by agroinfiltration-, bombardment-, or microinjection-induced RNA silencing, may use host factors to promote its own cell-to-cell movement, or diffuse passively between cells via plasmodesmata. However, the latter process is unlikely to occur in plants. Microinjection of 21 nt or 25 nt ssRNAs and dsRNAs into single cells has shown that none of these small RNA molecules is able to move out of the injected cell, providing direct evidence that plasmodesmata do not permit cell-to-cell diffusion of small RNA species (Yoo et al., 2004). These data are also consistent with the idea that intercellular trafficking of siRNA silencing signals requires protein components. Indeed, a small RNA binding protein (PSRP1) characterized from pumpkin (Cucurbita maxima) phloem sap selectively binds 25 nt ssRNA and mediates cell-to-cell trafficking of 25 nt ssRNA, but not dsRNA molecules, although the potential role of PSRP1 in signalling silencing remains to be elucidated (Yoo et al., 2004).
The present findings raise an interesting evolutionary question about why a plant virus encodes proteins that may promote the spread of silencing signal of an innate antiviral defence. It is speculated that during the co-evolution of plants and viruses, host plants may have adapted to viruses that often take advantage of cellular machinery for viral gene expression, replication, and movement. In return, plants could also have evolved the ability to use viral proteins to fend off viral pathogens. It is not surprising, for example, that many plant viruses encode elicitor proteins that are recognized by host plants to trigger a hypersensitive response, another form of innate defence mechanism that counteracts virus infection. Nevertheless, the specific viral proteins together with the TCV–GFPΔCP-based VIRS assay could provide a novel route to dissect the biochemical and molecular components directly involved in the intercellular movement of RNA silencing in plants.
Acknowledgments
We thank DC Baulcombe for providing seeds of GFP transgenic N. benthamiana line 16c and TMA Wilson for critical reading of the manuscript. This project was supported by BBSRC funding to Warwick HRI. XZ was funded by the Royal Society–China Association for Science and Technology Visiting Fellowship, and YZ was supported by the University of Warwick Research Development Fund.
References
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Braunstein TH, Moury B, Johannessen M, Albrechtsen M. Specific degradation of 3′ region of GUS mRNA in posttranscriptional silenced tobacco lines may be related to 5′-3′ spreading of silencing. RNA. 2002;8:1034–1044. doi: 10.1017/s1355838202026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li W-X, Ji L-H, Ding S-W, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO Journal. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proceedings of National Academy Sciences, USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. The Plant Journal. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Heaton LA, Zudema D, Hillman BI, Morris TJ. The genome structure of turnip crinkle virus. Virology. 1989;281:219–226. doi: 10.1016/0042-6822(89)90369-3. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Gisel A, Zambryski PG. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged Turnip crinkle viruses. Virology. 2000a;273:258–266. doi: 10.1006/viro.2000.0441. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Qu F, Gisel A, Morris TJ, Zambryski PG. Nuclear localization of Turnip crinkle virus movement protein p8. Virology. 2000b;273:276–285. doi: 10.1006/viro.2000.0440. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for posttranscriptional gene silencing in Arabidopsis. EMBO Journal. 2001;20:2069–2077. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Van Wezel R, Stanley J, Hong Y. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. Journal of Virology. 2003;77:7026–7033. doi: 10.1128/JVI.77.12.7026-7033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nature Genetics. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nature Genetics. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez RD, Houdt HV, Depicker A. Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. The Plant Journal. 2004;38:594–602. doi: 10.1111/j.1365-313X.2004.02067.x. [DOI] [PubMed] [Google Scholar]
- Hacker DL, Petty IT, Wei N, Morris TJ. Turnip crinkle virus genes required for RNA replication and virus movement. Virology. 1992;186:1–8. doi: 10.1016/0042-6822(92)90055-t. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A novel species of small antisense RNA in post-transcriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Voinnet O, Chappell L, Baulcombe DC. Two classes of short interfering RNA in RNA silencing. EMBO Journal. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saaki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Research. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and independent cell-to-cell movement of RNA silencing. EMBO Journal. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Seah BC, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F., Jr High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proceedings of National Academy Science, USA. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-Z, Qu F, Morris TJ. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology. 1998;244:405–416. doi: 10.1006/viro.1998.9125. [DOI] [PubMed] [Google Scholar]
- Lin B, Heaton LA. An Arabidopsis thaliana protein interacts with a movement protein of Turnip crinkle virus in yeast cells and in vitro. Journal of General Virology. 2001;82:1245–1251. doi: 10.1099/0022-1317-82-5-1245. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance VB. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. The Plant Cell. 2001;13:571–583. doi: 10.1105/tpc.13.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Mlotshwa S, Bowman LH, Vance VB. The capacity of transgenic tobacco to send a systemic RNA signal depends on the nature of the inducing transgene locus. The Plant Journal. 2003;35:82–92. doi: 10.1046/j.1365-313x.2003.01785.x. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Balzergue S. Activation of systemic acquired silencing by localised introduction of DNA. Current Biology. 1997;9:59–66. doi: 10.1016/s0960-9822(99)80016-5. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE1 and RDE4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. Journal of Virology. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E. RGD and other recognition sequences for integrins. Annual Review of Cell and Developmental Biology. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Ryabov EV, Van Wezel R, Walsh J, Hong Y. Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. Journal of Virology. 2004;78:3149–3154. doi: 10.1128/JVI.78.6.3149-3154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S, Chapman S, Roberts AG, Roberts IM, Prior DAM, Oparka K. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proceedings of National Academy Science, USA. 1996;93:6286–6290. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. The Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology. 2003;306:33–41. doi: 10.1016/s0042-6822(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk HA. Genes required for systemic RNA interference in Caenorhabditis elegans. Current Biology. 2004;14:111–116. doi: 10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- Timmons L, Tabara H, Mello CC, Fire AZ. Inducible systemic RNA silencing in Caenorhabditis elegans. Molecular Biology of the Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S, Citovsky V. Inhibition of systemic onset of post-transcriptional gene silencing by non-toxic concentration of cadmium. The Plant Journal. 2001;28:283–291. doi: 10.1046/j.1365-313x.2001.01145.x. [DOI] [PubMed] [Google Scholar]
- Van Houdt H, Bleys A, Depicker A. RNA target sequences promote spreading of RNA silencing. Plant Physiology. 2003;131:245–253. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wezel R, Liu H, Tien P, Stanley J, Hong Y. Gene C2 of the monopartite geminivirus Tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Molecular Plant-Microbe Interaction. 2001;14:1125–1128. doi: 10.1094/MPMI.2001.14.9.1125. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localised induction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Wobbe KK, Akgoz M, Dempsey DA, Klessig DF. A single amino acid change in Turnip crinkle virus movement protein p8 affects RNA binding and virulence on Arabidopsis thaliana. Journal of Virology. 1998;72:6247–6250. doi: 10.1128/jvi.72.7.6247-6250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B-C, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee Y-M, Lough TJ, Lucas WJ. A systemic small RNA signalling system in plants. The Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]