Summary
Proper ovarian development requires the cell type-specific transcription factor TAF4b, a subunit of the core promoter recognition complex TFIID. We present the 35Å structure of a cell type-specific core promoter recognition complex containing TAF4b and TAF4 (4b/4-IID), which is responsible for directing transcriptional synergy between c-Jun and Sp1 at a TAF4b target promoter. As a first step towards correlating potential structure/function relationships of the prototypic TFIID versus 4b/4-IID, we have compared their 3D structures by electron microscopy and single particle reconstruction. These studies reveal that TAF4b incorporation into TFIID induces an open conformation at the lobe involved in TFIIA and putative activator interactions. Importantly, this open conformation correlates with differential activator-dependent transcription and promoter recognition by 4b/4-IID. By combining functional and structural analysis, we find that distinct localized structural changes in a mega-Dalton macromolecular assembly can significantly alter its activity and lead to a TAF4b induced re-programming of promoter specificity.
Keywords: TAF4b, transcription, c-Jun, structure
Introduction
Gene expression initiated by mammalian RNA polymerase II (Pol II) involves a highly coordinated assembly of several multi-subunit complexes to form the preinitiation complex (PIC) (Thomas and Chiang, 2006). One critical step in gene activation involves directing the core recognition complexes to specific target promoters. The multi-subunit TFIID complex, consisting of the TATA-binding protein (TBP) and at least 8-14 evolutionarily conserved TBP-associated factors (TAFs), is a principle component within the transcriptional machinery capable of recognizing and targeting specific promoter DNA. The binding of TFIID to the core promoter is followed by a sequential recruitment of other general transcription factors including TF-IIA, -IIB, -IIE, -IIH, -IIF, Mediator and Pol II that culminates with activated transcription. TFIID carries out several key activities, including promoter DNA recognition and binding, activator-dependent co-activator targeting, and chromatin binding as well as several other TAF1 associated enzymatic activities, such as histone acetylation, phosphorylation and ubiquitination (Thomas and Chiang, 2006).
The discovery of cell type- and tissue-specific TFIID complexes underscores the evolutionary diversification of this core promoter recognition co-activator complex in metazoans that likely plays an important role during development (Thomas and Chiang, 2006). For example, the first of these cell type-specific TAFs identified, TAF4b (formerly TAFII105), was found to be selectively expressed in differentiated lymphocytes, testis, and granulosa cells of the ovary (Dikstein et al., 1996; Falender et al., 2005; Freiman et al., 2001). Studies of taf4b knockout mice established that this paralogue of the ubiquitously expressed TAF4 (formerly TAFII130) is required for ovarian development by directing selective gene networks essential for proper mammalian folliculogenesis (Freiman et al., 2001).
A central step carried out by sequence-specific activators/promoter binding factors is to interact directly with various components of the PIC. Not surprisingly, at certain promoters, TAFs in the TFIID complex play a critical role in this activator-stimulated transcription initiation with TAF4 being a known target for various activators and coactivators (Thomas and Chiang, 2006). Importantly, its paralogue TAF4b is thought to direct promoter selective recognition and recruitment of activators to its target genes in a cell- or tissue-specific manner (Freiman et al., 2001; Geles et al., 2006). Surprisingly, TAF4b does not appear to require ovarian-specific activators to direct cell type-specific programs of transcription. Instead, TAF4b selectively induces the expression of c-Jun in granulosa cells to regulate a diverse set of ovarian specific gene transcription. This unexpected finding suggests that cell type-specific subunits of the core machinery such as TAF4b may serve as gene-specific co-regulators that help determine the selectivity and dynamics of activator-directed gene expression. This unusual mechanism in turn allows ubiquitously expressed activators such as c-Jun to effectively target subsets of genes during development and cellular differentiation without invoking the need for additional cell type-specific activators. Thus, a nearly ubiquitous transcriptional activator such as c-Jun can carry out temporal and spatially restricted programs of transcriptional control through an interplay with a unique “cell type-specific” TFIID complex. This underlines the importance of this complex in regulating developmental processes.
The three-dimensional (3D) structural visualization by electron microscopy (EM) and single-particle reconstruction has significantly advanced our understanding of structural features and protein dynamics of large macromolecular assemblies. For example, a major discovery in transcriptional activator function has been elucidated through negative stain EM of the multi-subunit Mediator/CRSP complexes (Taatjes et al., 2002). These EM studies revealed unique major structural changes in Mediator/CRSP that correlated with the binding of different activators, highlighting the potential importance of co-activator structural plasticity during transcription PIC formation.
EM studies have also revealed a common structural framework of the yeast and human TFIID complexes, which features a “horseshoe” shape consisting of three large lobes (Andel et al., 1999; Leurent et al., 2004). Importantly, EM studies also have located the regions within TFIID that interact with the general transcription factors TFIIA and TFIIB (Andel et al., 1999). Immunomapping studies further identified the relative positions of several major TAFs within yeast TFIID (Leurent et al., 2004). A recent cryo-EM study revealed more structural details of human TFIID including a striking flexibility leading to distinct open and closed conformational states (Grob et al., 2006) suggesting that TFIID, like Mediator, may adopt specific conformations related to PIC formation. However, the potential functional significance of these different conformational states of TFIID has not been determined. How these apparently dynamic structural features of this very large multi-subunit co-activator complex may be linked to potential promoter specificity and transcriptional properties remains a challenging and enigmatic question.
Here, we have begun to address aspects of structure/function relationships of a cell type-specific member of the mammalian TFIID family. To address potential links between structural features of TFIID and transcriptional activity, we have assayed the ability of the activator c-Jun to direct transcription activation in vitro using both canonical and cell type-specific TFIID complexes. In particular, we have addressed the question of whether the prototypic TAF4 subunit can functionally replace TAF4b in directing c-Jun dependent transcriptional activation of several target promoters. We have also tested the hypothesis that the presence of TAF4b within TFIID induces a distinct conformational alteration of the TFIID complex that influences the overall structure and correlates with changes in its functional specificity. In order to envision such structural changes of TFIID induced by TAF4b, we have determined the 3D structure of a cell type-specific TFIID complex containing TAF4b and compared this structure to the classical TFIID complex. Coupling these 3D EM structures with functional analysis of c-Jun dependent in vitro transcriptional activities unmasked activator and promoter specificities that correlate with the known in vivo functions of TAF4b and c-Jun in gene regulatory programs during ovarian cell development.
Results
Isolation of cell type-specific TAF4b-TFIIDs
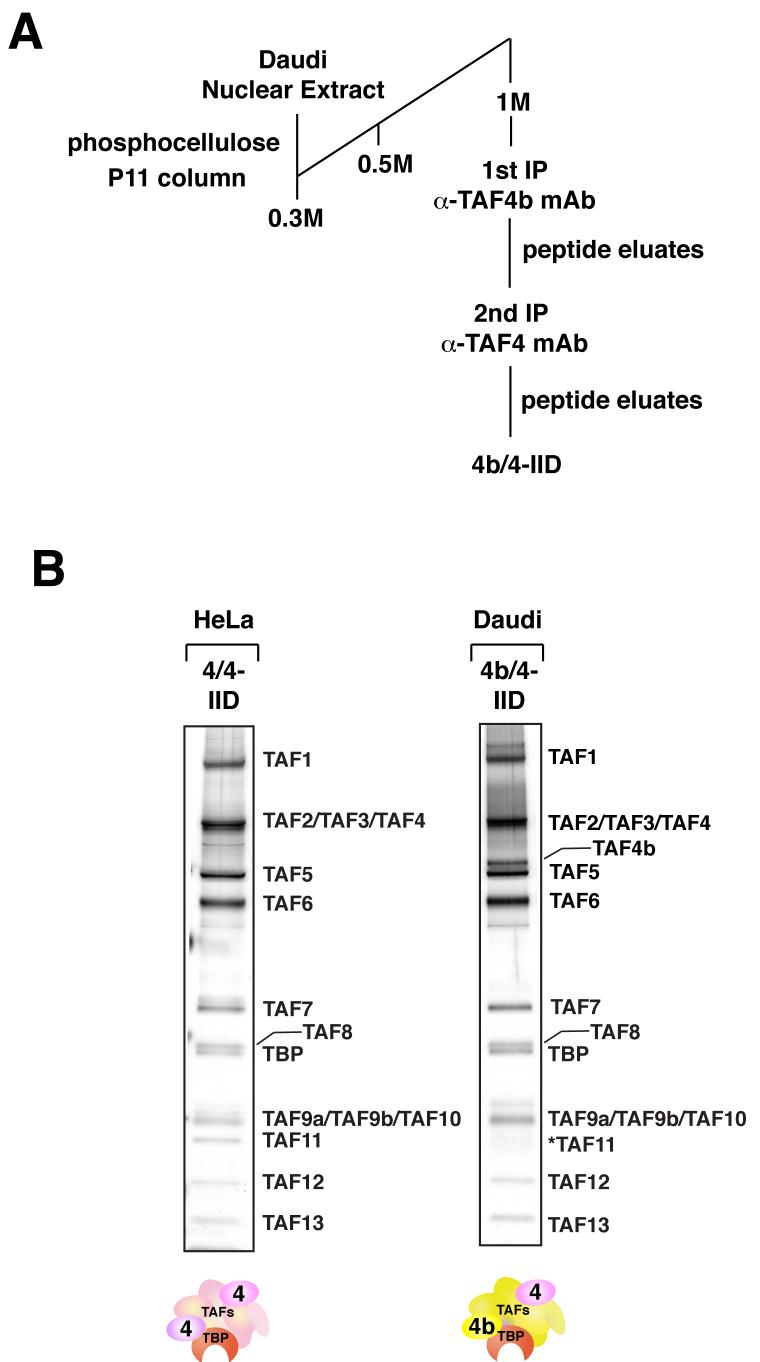
Although in vivo the cell type-specific function of TAF4b-containing TFIID complex (4b-IID) has largely been documented in the reproductive system, additional studies found that TAF4b is also expressed in relatively high levels in differentiated human B cells (Daudi) (Dikstein et al., 1996). This rapidly growing cell culture system afforded us a convenient and unique source to isolate a native TFIID complex containing TAF4b. In order to obtain highly purified 4b-IID, we generated a panel of monoclonal antibodies against the amino terminus of TAF4b that bears little sequence identity with its homologue TAF4. We selected a monoclonal antibody that specifically binds TAF4b (amino acids 178 to 204) to carry out antibody affinity chromatography of 4b-IID. Nearly homogenous native 4b-IID can be eluted from immunoprecipitations of Daudi nuclear extracts using a synthetic peptide corresponding to the cognate epitope.
Our earlier studies found that TAF4 is always present in the 4b-IID preparations even when using three independent immuoprecipitation procedures, with antibodies targeting either TBP, TAF4, or TAF4b (Dikstein et al., 1996; Geles et al., 2006). These studies indicate that TAF4 and TAF4b can co-exist in at least a portion of the TFIID complexes present in TAF4b-containing cell types (i.e. Daudi cells and ovarian granulosa cells as opposed to HeLa cells). This was not entirely unexpected as it has been shown by various assays that holo-TFIID derived from HeLa cells and yeast likely contains two copies of TAF4 (Guermah et al., 2001; Leurent et al., 2004). To isolate homogenous preparations of TFIID containing at least one subunit of TAF4b per complex, we carried out tandem immunoprecipitations from Daudi cells using peptide-elutable monoclonal antibodies against both TAF4b and TAF4, sequentially (Figure 1A). After this double affinity purification step, we obtained a TFIID complex containing near stoichiometric amounts of TAF4b and TAF4, designated as 4b/4-IID (Figure 1B). In addition, we found that we can isolate TAF4-containing TFIID that lacks TAF4b from Daudi cells by immunopurifying TBP and TAF4 from fractions that were previously immunodepleted of TAF4b (data not shown). This TFIID complex lacking TAF4b appears to be identical to the canonical TAF4 affinity purified TFIID from HeLa cells by SDS-PAGE and silver stain analysis (data not shown).
Figure 1. Isolation of cell type-specific 4b-IIDs.
(A) Schematic representation of the purification procedure of 4b-IID from Daudi cells. 4b/4-IID was obtained by tandem immunoprecipitations from the fractions eluted from the phosphocellulose 1M KCl step using anti-TAF4b and anti-TAF4 antibodies. (B) Distinct TFIID complexes (i.e. HeLa 4/4-IID and Daudi 4b/4-IID) were analyzed by 4-12% SDS-PAGE (Invitrogen) and visualized by silver staining. The star indicates a weak band corresponding to TAF11.
For comparison purposes and to avoid potential TAF4b contamination, we purified the canonical TAF4-containing TFIID complex, designated as 4/4-IID, from HeLa nuclear extracts that lack TAF4b using a single TAF4 immunoprecipitation step. We analyzed the subunit composition of these two complexes by SDS-PAGE/silver staining analysis (Figure 1B) and also confirmed the presence of the TAF subunits by MudPIT mass spectrometry (Supplementary data S1). In short, our sequential antibody affinity purification strategy allowed the isolation of a highly homogenous 4b/4-IID complex that resolved by a size exclusion chromatography as predominantly one native species greater than 1MDa (Supplemental data S2). Thus we confirmed the presence of at least two TFIID-like complexes represented by 4/4-IID and 4b/4-IID that apparently co-exist in Daudi cells (data not shown). A similar assortment of TFIID complexes were also found in ovarian granulosa cells (data not shown).
Differential core promoter selectivity directed by distinct TFIID complexes
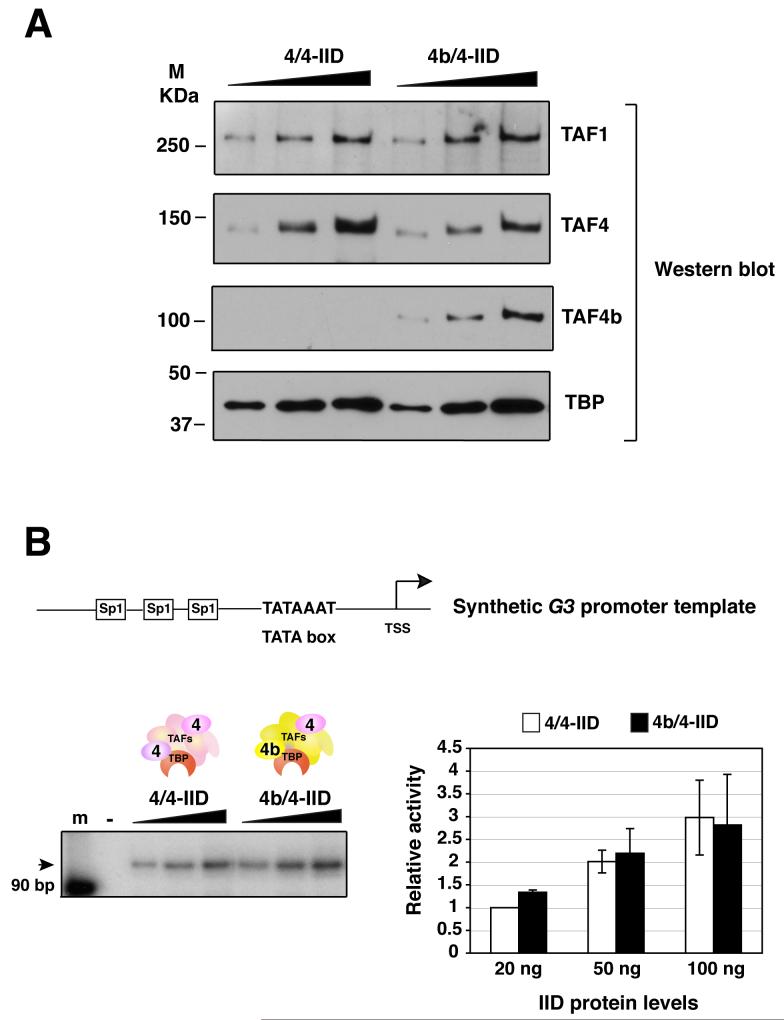
One of the well established functions of TFIID is to recognize and bind specific core promoter sequences, thereby recruiting and stabilizing other basal factors during PIC formation prior to transcription initiation. We therefore first set out to examine whether these two distinct TFIID complexes (i.e. 4/4- and 4b/4-IID) are equally efficient at directing basal transcription in the absence of activators. We utilized highly purified basal factors (TF-IIA, -IIB, -IIE, -IIF, -IIH and RNA Pol II) to reconstitute transcription in vitro to test activity of these two distinct TFIID complexes. For our initial studies, we used the synthetic G3 DNA template that contains a canonical TATA box, which is optimal for TFIID recognition and binding. Equivalent amounts of these two different purified TFIIDs at various concentrations were tested in this core promoter dependent assay. The relative protein levels of the TFIIDs were assessed by immunoblotting using antibodies against TBP, TAF4b, TAF4, and TAF1 (Figure 2A). These two TFIID complexes were essentially equivalent in directing accurate transcription initiation from a synthetic core promoter/TATA box containing template as determined by primer extension analysis in our in vitro reconstituted transcription assay and most importantly their relative specific activities for basal transcription were comparable (Figure 2B).
Figure 2. The basal transcriptional activities of distinct TFIID complexes.
(A) The protein levels of 4/4- and 4b/4-IID complexes were examined by immunoblotting analysis using monoclonal antibodies against TAF1, TAF4, TAF4b and TBP.
(B) Equivalent amounts (0, 20, 50, and 100 ng) of 4/4-IID and 4b/4-IID were added respectively into each reaction containing synthetic G3 promoter DNA template. The activities were quantified using a Typhoon scanner (Amersham Biosciences). Relative activities were calculated in comparison with the activity obtained from the lowest amount (20 ng) of 4/4-IID used. Each reaction was repeated several times and the representative data are shown (left panel). The arrows indicate the target transcripts. Error bars represent the standard deviations from at least three independent experiments (right panel).
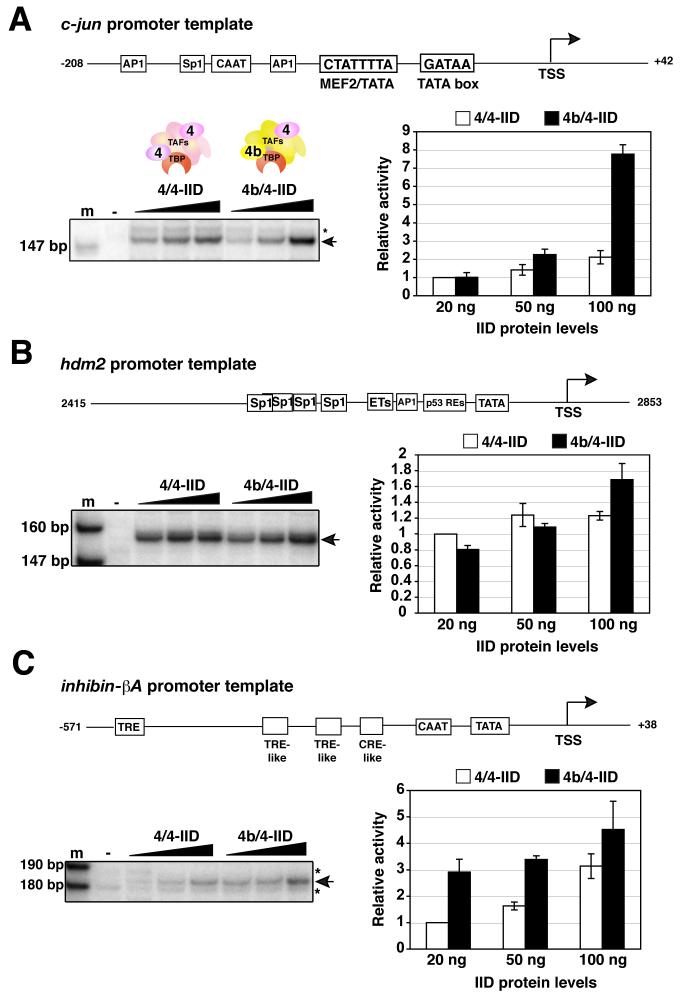
Our previous studies had found that TAF4b is also able to activate a select set of specific gene promoters (Geles et al., 2006). Included in this class of in vivo responsive genes was the promoter for the transcriptional activator, c-Jun. Interestingly, the induction of c-Jun by TAF4b in mammalian ovaries appears to be important for proliferation of granulosa cells and necessary for proper progression of folliculogenesis in vivo (Voronina et al., 2006). Armed with the well characterized c-jun promoter that contains a non-canonical TATA box, we next asked whether the two distinct TFIID complexes might exhibit differential core promoter activities in vitro that may be revealed by using an endogenous TATA-less promoter in vitro even in the absence of activators. To address this important issue, we programmed our transcription assays comprising either 4/4- or 4b/4-IID with native DNA templates that contained either the c-jun promoter, a known target of TAF4b in vivo, or a control native promoter, hdm2, that fails to respond to TAF4b in vivo (Geles et al., 2006). First we found that at the highest levels of TFIIDs, 4b/4-IID was much more effective at directing transcription from the c-jun promoter compared to 4/4-IID (Figure 3A). By contrast, transcription from the control hdm2 promoter showed that both the prototypic 4/4-IID as well as the cell type-specific 4b/4-IID direct transcription with roughly equal efficiency (Figure 3B).
Figure 3. Distinct transcriptional activities of TAF4b-target genes by 4b/4-IID.
(A) Reactions were performed and quantitated exactly as described in Figure 2B, except that the native c-jun promoter template, a control hdm2 promoter (B), or native inhibin-βA subunit promoter (C) was utilized. The stars indicate non-specific transcripts. Error bars represent the standard deviations from at least three independent experiments.
Since our previous in vivo studies strongly indicated that TAF4b is in part responsible for inducing transcription from a number of ovary-specific promoters including the inhibin-βA (Inhba) promoter (Freiman et al., 2001; Geles et al., 2006), we next determined whether 4b/4-IID could also selectively target this endogenous promoter in vitro in the absence of activators. Remarkably, even in the absence of any added activators, these in vitro transcription reactions revealed that 4b/4-IID was significantly and reproducibly more efficient than 4/4-IID at directing basal core promoter activity from the native Inhba promoter (Figure 3C). These initial in vitro transcriptional studies suggest that 4b/4-IID is inherently more effective in core promoter selectivity and preferentially targets the c-jun and Inhba templates, which are also known to be TAF4b-dependent genes in vivo.
Putative co-activator functions of the 4b/4-IID complex
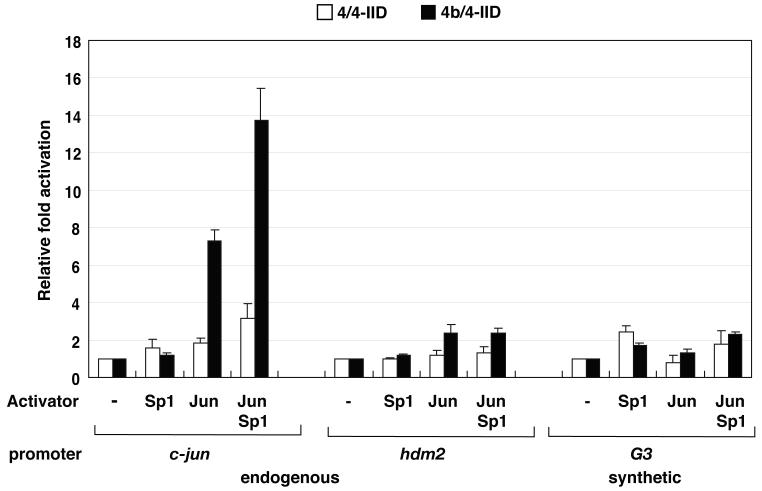
One proposed feature of cell type-specific TAFs such as TAF4b is their potential to function as gene specific transcriptional co-activators (Dikstein et al., 1996; Yamit-Hezi and Dikstein, 1998). Although our studies thus far suggest that 4b/4-IID may exhibit some intrinsic core promoter selectivity even in the absence of activators, this observed selectivity for the well-documented c-jun promoter could potentially be enhanced by the presence of gene specific activators including c-Jun itself. Previous ChIP analysis had established that both TAF4b and c-Jun are recruited to the c-jun core promoter in vivo to regulate and enhance expression of c-Jun in ovarian granulosa cells (Geles et al., 2006). Here, we find that c-Jun strongly enhanced 4b/4-IID-driven activator-dependent transcription from the c-jun template (7.3-fold). In contrast, c-Jun mediated activation was much less efficient when using 4/4-IID (1.8-fold). Strikingly, when c-Jun and Sp1 acted together, an even more robust activation of the c-jun promoter was observed when using 4b/4-IID (13.7-fold) compared to the canonical 4/4-IID (3.2-fold) (Figure 4). Unlike the c-jun promoter, transcription directed by the control hdm2 template not known to be responsive to TAF4b showed comparably modest levels of c-Jun activator dependent transcription when using either of these two distinct TFIID complexes. Similarly, the synthetic G3 template directed weak c-Jun responsive transcriptional activity using either 4/4- or 4b/4-IID as expected.
Figure 4. Putative co-activator functions of 4b/4-IID.
4b/4- or 4/4-IID (0 or 3 ng) were added to the transcription reactions containing c-jun, hdm2, or G3 promoter DNA templates in the absence or the presence of the activators c-Jun, Sp1, or both, respectively. The relative fold activation of individual TFIIDs were calculated based upon their relative activities that are measured in comparison to the activity with no addition of activators using the same promoter DNA. Error bars represent the standard deviations from at least three independent experiments.
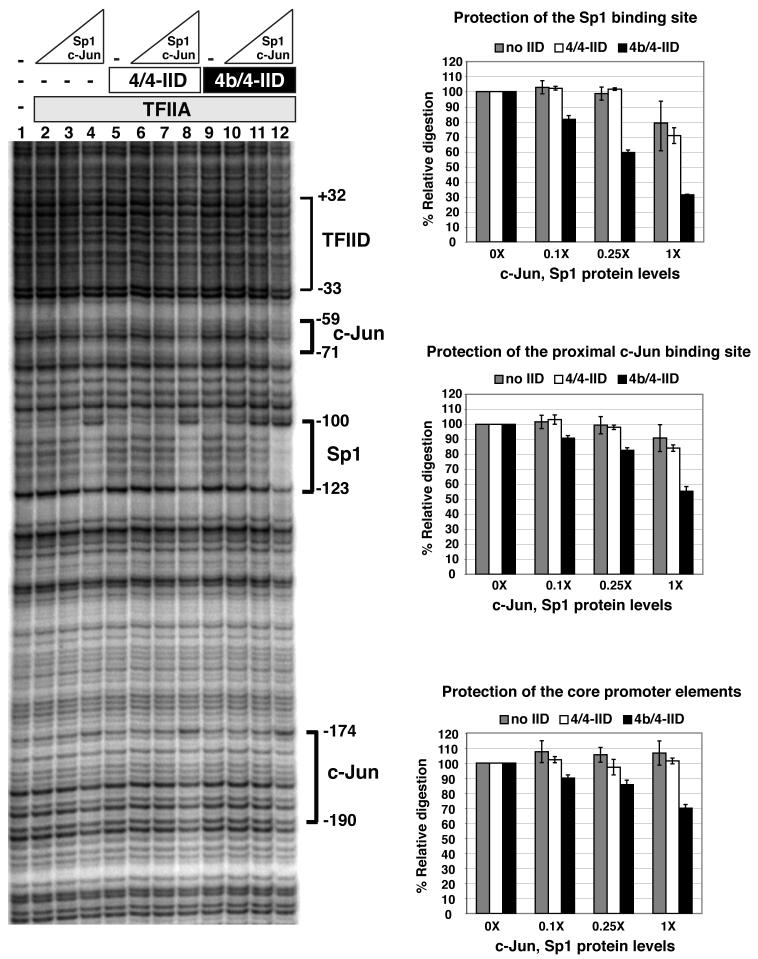
One function of co-activators is to serve as an adaptor or bridge between activators and the basal transcription machinery that could help stabilize the preinitiation complex and enhance transcriptional activation. In order to address this possibility and to complement our in vitro transcription studies, we used DNase I footprinting to determine promoter occupancy of Sp1 and c-Jun in the presence or absence of 4b/4- and 4/4-IID (Figure 5). In the absence of either 4/4- or 4b/4-IID, we saw weak binding of activators at the upstream c-Jun and Sp1 recognition sites (Figure 5, lane 4, or top two right panels). In the presence of 4b/4-IID, we observed an enhanced occupancy of the upstream c-Jun and Sp1 binding sites possibly due to direct binding by 4b/4-IID (Figure 5, compare lane 12 and 4, or right panels). By contrast, the enhanced binding of Sp1 and c-Jun at the upstream sites was not as significant when the prototypic 4/4-IID was employed (Figure 5, compare lane 12 and 8, or right panels). Since we only observed a very modest protection of the regions corresponding to the TATA box, initiator or downstream promoter element in the presence of either 4/4-IID or 4b/4-IID, we postulate that the presence of 4b/4-IID at least helps to stabilize and enhance the upstream activator contacts at this promoter rather than to promote strong downstream TFIID contacts within the promoter. This observation was surprising, given that activators bound to upstream sites of a synthetic promoter had previously been shown to induce conformational changes within TFIID that enhanced footprints downstream of the promoter (Chi and Carey, 1996). However, importantly, when using the native TATA-less c-jun promoter we observed that 4b/4-IID enhanced activator contacts upstream rather than downstream core promoter interactions. The absence of a strong footprint in vitro encompassing the TATA box and initiator was not entirely unexpected since c-jun is a TATA-less promoter and would not be expected to display the same binding pattern as that observed with the idealized super-core promoter (Juven-Gershon et al., 2006).
Figure 5. Elevated occupancy of activator binding sites in a 4b/4-IID dependent manner.
Activator occupancy on the endogenous c-jun promoter was analyzed by DNase I footprinting assay. The binding elements for various transcription factors within the promoter region are indicated. Along with 20 nM of TFIIA, c-Jun (0, 12, 30 and 120 ng) and Sp1 (0, 8, 20, and 80 ng) were incubated respectively in the absence (lanes 1-4) or the presence of either 4/4-IID (lanes 5-8) or 4b/4-IID (lanes 9-12). Data shown are representative of at least three independent experiments. Quantification of the data is shown in the right panels. The percentage of relative DNAse I digestion was calculated based upon the digestions of individual binding elements in comparison to the digestions without addition of any activators. 0X, 0.1X, 0.25X, and 1X represent the increasing amounts of c-Jun and Sp1 protein levels as listed above. Error bars represent the standard deviations from at least three independent experiments.
3D reconstruction of the 4b/4-IID complex
Several lines of evidence provided by our in vitro transcription assays point to a differential specificity of both intrinsic core promoter binding and activator dependent transcriptional activity directed by cell type-specific versus canonical TFIID complexes. We were therefore particularly curious to see if the incorporation of TAF4b within TFIID would induce some local or possibly global conformational change to the multi-subunit assembly that could subsequently be correlated with its transcriptional specificity. Having isolated a nearly homogenous population of a newly defined TFIID complex containing both TAF4b and TAF4 (i.e. 4b/4-IID), we next set out to determine its 3D structure by EM and single particle reconstruction.
First, we optimized the negative staining conditions for protein complex preservation and image quality. We obtained the 4b/4-IID structure derived from a total of 12,851 particles employing the projection matching method using the TFIID cryo-EM structure filtered at 60Å as an initial reference (Grob et al., 2006). The final 3D structure has a resolution of 35Å as determined by Fourier-shell correlation (FSC) with a 0.5 cut-off and shows a fairly isotropic angular distribution particle dataset (Supplementary data S3). The resulting 4b/4-IID structure, filtered at 35Å, is presented with a threshold (3.2σ) for a total protein mass of ∼1.1-1.2 MDa in Figure 6. To confirm our 3D model of 4b/4-IID and to ensure that no model bias was imposed by using the TFIID cryo-EM structure as an initial reference, we independently performed reference-free 2D alignment and classification. These de novo 2D class averages of 4b/4-IID matched well with reprojections of the 3D structure, further strengthening our 3D reconstruction results (Supplementary data S4A).
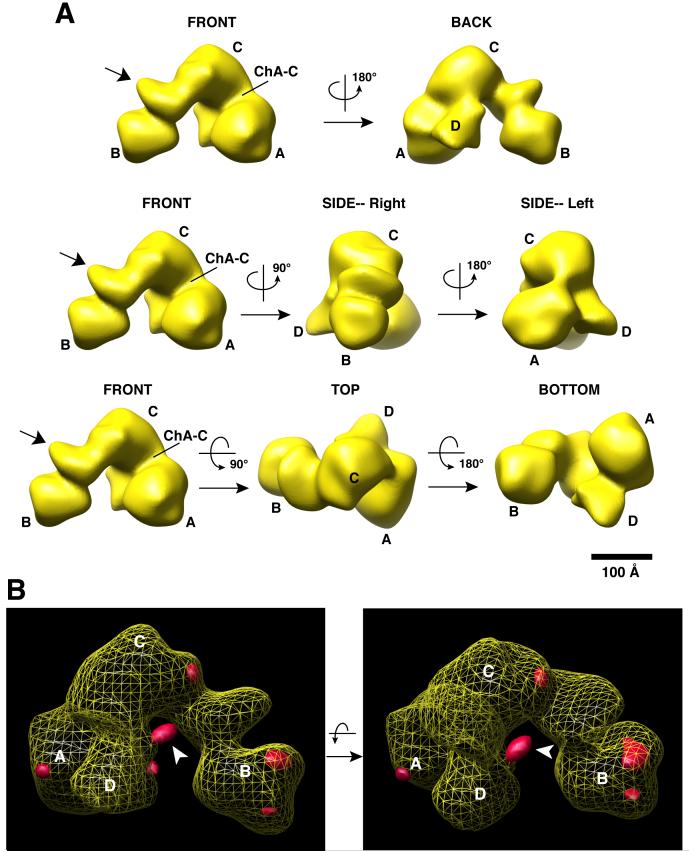
Figure 6. The 3D structure of 4b/4-IID and immunomapping of the TAF4b position within 4b/4-IID.
(A) Different views of the 4b/4-IID EM structure with rotation angles are indicated. The major lobes are labeled as A, B, and C, and a smaller domain labeled as D lobe (see “BACK” view). The channel formed between the A and C lobes is labeled ChA-C. The arrow shown in the “FRONT” view indicates the undulating region located between the B and C lobes. The scale bar represents 100Å.
(B) 3D difference density map between 4b/4-IID-anti-TAF4b and 4b/4-IID is shown in two different views. The yellow mesh corresponds to the 3D structure of 4b/4-IID with the A, B, C, and D lobes indicated. The extra densities (i.e. positive density differences) are shown in solid red. The most prominent difference region (indicated by the arrow) is located in the D lobe, close to C lobe and the central cavity, and was interpreted as the TAF4b antibody bound to 4b/4-IID.
As expected, the global architecture of the 4b/4-IID structure is rather similar to the TFIID cryo-EM structure (Grob et al., 2006) with its signature “horseshoe” shape consisting of a central cavity connected by three lobes (i.e. A, B, and C lobes) that are best seen from the “FRONT” view (Refer to movie in Supplementary data S4B). There is a smaller but clearly discernable D lobe evident when viewed from the “BACK” (Figure 6), while the smaller C1 and C2 lobes reported in the cryo-TFIID studies are not as clearly resolved. The undulating region linking lobes B and C likely corresponds to a shifted C2 lobe (Figure 6, FRONT view, arrow). Interestingly, our previous cryo-EM study had revealed significant movements of the three lobes relative to each other in the canonical holo-TFIID structure (Grob et al., 2006). One of the most mobile regions was seen to occur at the tip of lobe B and the region between lobes B and C, which coincides well with the undulated domain we observed here (Figure 6A).
Next, we attempted to independently determine the location of TAF4b within 4b/4-IID by immunomapping and single particle reconstruction. To obtain near stoichiometric amounts of a TAF4b monoclonal antibody (mAb) bound to a 4b/4-IID complex, we modified our tandem immunopurification protocol to include a TAF4b mAb loading step prior to elution of the complex. Briefly, 4b/4-IID bound to protein G resin containing covalently conjugated TAF4 mAb was washed extensively to remove loosely associated proteins and any remaining TAF4b peptide from the initial TAF4b immunopurication step. A 250 fold molar excess of TAF4b mAb was then incubated with the 4b/4-IID complex while still bound to the TAF4 mAb/protein G resin. Unbound TAF4b mAb was removed by extensive washing of the TAF4 affinity resin. A complex containing near stoichiometric amounts of TAF4b mAb bound to 4b/4-IID (4b/4-IID-anti-TAF4b) was eluted from the TAF4 antibody affinity column using the specific peptide recognized by the TAF4 mAb (data not shown).
This highly purified 4b/4-IID-anti TAF4b complex was subsequently subjected to EM analysis and 3D reconstruction using a total of 10,024 particles. For the 3D reconstruction, we employed essentially the same procedures as described above for the 4b/4-IID structure, except that the initial reference for the first cycle of the reconstruction was the 4b/4-IID 3D structure filtered to 50Å. A 3D density difference map was then calculated between the newly refined 4b/4-IID and 4b/4-IID-anti-TAF4b structures and both were both filtered to 35Å (Figure 6B). The position of the anti-TAF4b antibody can be assigned to a prominent extra density located in lobe D, close to the C lobe and the central cavity (see a movie in Supplementary data S5A). To further support this finding, reference-free 2D class averages were also obtained. Four de novo 2D class averages showed more clearly extra density for the TAF4b mAb (corresponding to views where the antibody does not superimpose on the 4b/4-IID density), which localized to the same region as shown in Figure 6B (Supplemental data S5B). Since the TAF4b antibody specifically recognizes the N-terminus of TAF4b (178-204 amino acids), these data suggest that TAF4b is likely located at the A-C-D junction region (Supplemental data S5C). A small extra density at lobe A was also observed that is likely due to some minor conformational changes (Figure 6B).
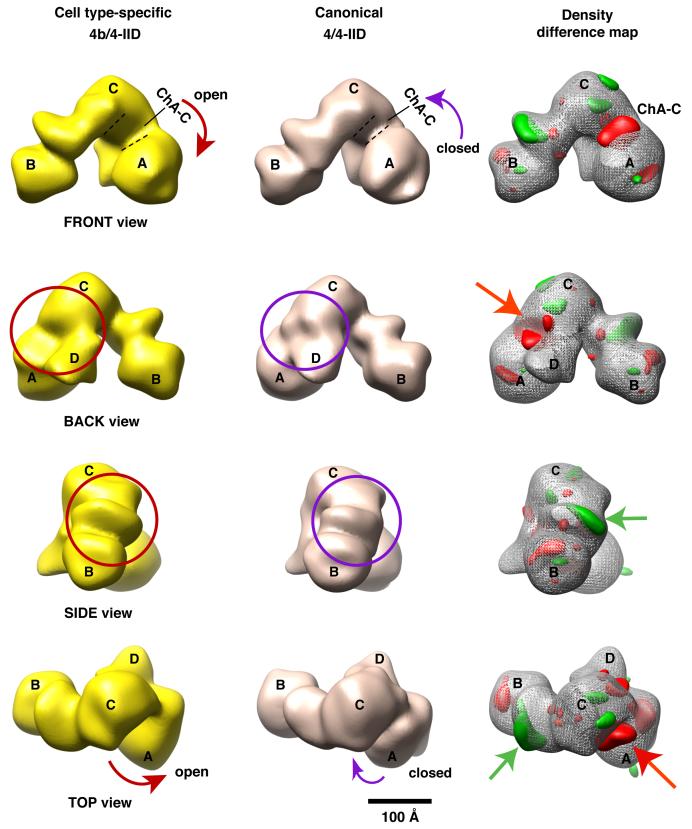
In order to map potential structural differences between the canonical 4/4-IID and the cell type-specific 4b/4-IID, we adopted the same protein purification procedures, staining conditions, and 3D reconstruction strategy, to obtain the 3D structure of the canonical holo-TFIID using 14,000 particles (Figure 7). The newly refined 4/4-IID structure was filtered to 35Å with a threshold (3.2σ) for a protein mass of ∼1.1-1.2 MDa. Like the 4b/4-IID, the angular distribution of the particles used for this TFIID structure was mostly isotropic (data not shown). The overall structure of 4/4-IID is very similar to the 4b/4-IID structure (4/4-IID movie in the supplementary data S6), indicating that incorporating one subunit of TAF4b into the TFIID complex does not lead to dramatic global structural changes. However, a careful comparison of these two large structures revealed three prominent features that differ. First, the channel that bridges lobe A and C (ChA-C) in the 4/4-IID structure is considerably smaller than the one seen in the 4b/4-IID structure (Figure 7, FRONT view). As a result, 4/4-IID largely exhibits a more “closed” conformational state on average, whereas 4b/4-IID clearly displays a considerably more “open” conformational state (FRONT view). Notably and intriguingly, there is a substantial amount of density missing in the junction between the lobes A, C, and D in 4b/4-IID (Figure 7, BACK view). We also observed that the undulating region located between lobes B and C of 4b/4-IID is significantly distinct from the equivalent domain in 4/4-IID (SIDE view, bottom panel).
Figure 7. Comparison of the 4b/4- and 4/4-IID 3D structures.
Comparison from four different views (FRONT, BACK, SIDE, and TOP) of the 3D reconstructions of cell type-specific 4b/4-IID (left) and the canonical 4/4-IID (center). The distinct feature seen in the FRONT view is the ChA-C channel, which in 4/4-IID is significantly smaller than in 4b/4-IID (the curved arrows highlight this structural change). There is some missing density in 4b/4-IID located at the junction of A-C-D lobes (circles in the BACK view). Similarly, missing density was observed in 4/4-IID in the undulating region between B and C lobes (circles in the SIDE view). The right column shows the difference map between 4/4- and 4b/4-IID, with “red” representing positive differences and “green” representing negative differences. The “red” and “green” arrows point to the most significant positive and negative differences, respectively. The scale bar represents 100Å.
In order to be more quantitative in mapping the structural alterations detected between the 4/4- and 4b/4-IID structures, we calculated 3D density difference maps using both volumes (Figure 7, right column). The structural features shown in red represent the positive density difference when the 3D volume of 4b/4-IID is subtracted from the reference volume of 4/4-IID (Figure 7, FRONT, SIDE, and TOP views). The negative part of the density difference is represented in green at the same density threshold. Consistent with our description above, one major gain in density (red) of the 4/4-IID structure was mapped to the flexible region between lobes A and C, i.e. ChA-C (Figure 7, FRONT view). In addition, the other major gain in density within 4/4-IID is located in the junction of the A, C, and D lobes. One distinct green area includes a loss in density within the undulating region of 4b/4-IID located between lobes B and C. Interestingly, a small negative difference is also located in the C domain.
In order to assess the significance of these differences between both reconstructions, we performed two additional analyses using FSC calculations with a combination of 4/4- and 4b/4-IID data sets (Supplemental Experimental Procedures). The results confirmed that the two structures obtained were significantly different at the resolution of 35Å and that the differences that were mapped are significantly above the level of background noise (Supplemental data S7). Taken together, these results support the notion that the conformational differences observed between these two related TFIID complexes represent distinct and likely functionally relevant structural states, which correlate with the promoter selective transcriptional properties of 4b/4-IID that we have uncovered here.
Discussion
The architectural consequences of TAF4b incorporation into TFIID
The 35Å structure of 4b/4-IID shows that 4b/4-IID exhibits a distinctively more open conformation than the 4/4-IID complex, with a significant widening of the A-C channel (CHA-C) of the A lobe domain (Figures 6 and 7). The observed “open” and “closed” conformations appear to be achieved by the relative positions of lobes C and A. We previously found that TFIIA interacts with lobe A and spans the A-C channel of TFIID (Andel et al., 1999). TFIIA is known to stabilize the binding of TFIID to promoter DNA and to serve as a bridge between activators and TFIID (Guermah et al., 2001; Kobayashi et al., 1995). Recent label transfer protein crosslinking experiments indicate that TFIIA contacts Sp1, TAF4 and TAF4b within the intact 4b/4-IID complex (R. Coleman unpublished data). Additional label transfer protein crosslinking experiments also show that Sp1 can target TAF4 within intact TFIID complexes, consistent with a co-localization of TFIIA, TAF4, and Sp1 within lobe A of TFIID (R. Coleman unpublished data). In addition, Hann and co-workers have found that TFIIA crosslinks to TAF4 in pre-assembled PICs in yeast (Warfield et al., 2004). Thus, multiple independent lines of evidence suggest that lobe A of TFIID is involved in bridging TFIIA to various activators. Interestingly, the canonical TFIID is predominantly in the “closed” conformation on average, while 4b/4-IID, by contrast, is largely in the “open” conformation resulting from the movement of lobe A. Based on our in vitro transcription data, we speculate that the incorporation of TAF4b to the TFIID assembly induces an open conformation of lobe A that facilitates contacts with TFIIA and activators, thereby potentiating TAF4b-dependent transcriptional synergy.
Location of TAF4b within TFIID
Previous biochemical and immunomapping studies of yeast TFIID revealed the presence of two copies of TAF4 within TFIID, one in lobe C and a second in yeast lobe B (yeast nomenclature), which is called Lobe A in human TFIID, consistent with the finding that both yTAF4 and hTAF4 are direct targets of TFIIA (Guermah et al., 2001; Leurent et al., 2004; Warfield et al., 2004). Previous biochemical studies together with our recent label transfer experiments also established that TAF4b specifically interacts with TFIIA (Dikstein et al., 1996). Here, we have carried out immunomapping experiments to independently confirm that the N-terminus of TAF4b is most likely located at the lobe C-D junction (Figure 6B). Furthermore, the size and shape of the A lobe within 4b/4- and 4/4-IID appear identical (Figure 7). These complementary experiments collectively indicate that TAF4b is very likely located at the junction between A, C, and D lobes of the 4b/4-IID complex (Supplemental data S3C). However, we cannot exclude the possibility that a very minor population of the 4b/4-IID complex contains TAF4b and TAF4 in a reciprocal location.
Mechanistic underpinnings of TAF4b-induced promoter recruiting properties
Our in vitro reconstitution assays suggest that TAF4b regulates transcription, at least in part, through two distinct mechanisms. First, in the absence of activators, 4b/4-IID appears to be intrinsically more active in driving TAF4b-dependent promoters than the canonical 4/4-IID, relative to the control promoter (Figure 3). These results suggest that TAF4b incorporation into TFIID may bias the intrinsic promoter selectivity of 4b/4-IID towards select target promoters independent of activators. A previous report found that the TAF4b subunit had some DNA binding capacity when incorporated into TFIID (Shao et al., 2005), although it remains unclear whether TAF4b contacts DNA in a sequence-specific manner. Thus, the modest but significant intrinsic promoter selectivity of 4b/4-IID could be attributed either to direct DNA binding or to TAF4b-induced conformational changes that alter critical protein:protein interactions during PIC formation. Upon examination of promoter sequences, we found that the c-jun promoter contains a strong Inr element but no DPE element, while the inhibin-βA promoter lacks an Inr element and DPE element. Thus, it seems that TAF4b dependence does not correlate with any known core promoter elements. Both the c-jun and inhibin-βA promoters share similar features in that each promoter contains binding sites for AP-1 family proteins.
More importantly, we see that TAF4b-directed target promoter selectivity may be substantially enhanced by activators such as c-Jun and Sp1 at a bona fide target gene, c-jun. Previous studies reported that TAF4 can directly contact Sp1 (Rojo-Niersbach et al., 1999), suggesting that the presence of at least one copy of TAF4 in 4b/4-IID may contribute to its transcriptional specificity, perhaps via direct interactions between TAF4 and Sp1.
Certain combinations of different activators work in concert to regulate transcription in response to signaling pathways (Hess et al., 2004). For example, c-Jun and Sp1 together modulate the expression of many genes in response to stress and other physiological cues. Often, this collaboration between c-Jun and Sp1 involves cooperative binding to the core transcription machinery at the promoter. However, there has been little precedence for c-Jun and Sp1 to utilize alternative, cell type-specific core components to control networks of transcription. Here we found that Sp1 and the c-Jun homodimer selectively enhances transcription by 4b/4-IID relative to the canonical 4/4-IID at the native c-jun promoter (Figure 4). Strikingly, we observed a concomitant increased occupancy of c-Jun and Sp1 at their cognate binding sites in the presence of 4b/4-IID but not 4/4-IID (Figure 5). These findings strongly suggest that 4b/4-IID facilitates or stabilizes the binding of transcriptional activators like c-Jun and Sp1 to specific target promoters. Given this unconventional relationship between 4b/4-IID, c-Jun and Sp1, it would be intriguing to investigate whether 4b/4-IID associates with activators prior to or after binding to promoter DNA. c-Jun was found to interact with TAF7 and the N-terminus of TAF1 (Munz et al., 2003; Lively et al., 2001). Thus, it may not be unreasonable to propose that 4b/4-IID could bind c-Jun directly in the absence of DNA. Efforts are currently underway to obtain structures of 4b/4-IID bound to various activators to visualize potential structural consequences of an activator bound 4b/4-IID.
Since there is some evidence that TAF4b-containing TFIID can contact DNA directly (Shao et al., 2005), it is conceivable that the 4b/4-IID complex binds to the core promoter first, followed by subsequent activator recruitment. Indeed, the hsp70 (Lebedeva et al., 2005) and TNF-α responsive gene A20 promoters (Ainbinder et al., 2002) have been reported to bind TFIID before specific activators are recruited. Intriguingly, in each of these cases (c-jun, hsp70, and A20), external signals were required to stimulate recruitment of specific activators. In addition, binding of Sp1 to the cyclin D1 promoter in vivo is dependent on a functional TAF1, further supporting the notion that the activity of TAFs within TFIID can affect activator recruitment to the promoter (Hilton et al., 2005). Perhaps the open conformation of 4b/4-IID favors constitutive binding to the c-jun promoter thereby facilitating the rapid initiation of transcription in response to extracellular signals. This scenario is consistent with the finding that TAF4b-containing TFIID is constitutively bound to the A20 promoter and poised for immediate initiation upon TNF-α stimulation. In support of this concept, both A20 and c-jun are immediate early genes. A previous taf4 knockout study revealed that TAF4b mediates transforming growth factor-β (TGF-β) signaling in a more efficient manner compared to TAF4 (Mengus et al., 2005). Perhaps 4b/4-IID utilizes the similar mechanism to activate gene expression in TGF-β signaling pathway.
Additional TAF4b targets that may be regulated by c-Jun and Sp1 in vivo
Recent studies demonstrated that Sp1 and c-Jun are both required for the expression of vimentin (Wu et al., 2003), which also turned out to be a TAF4b target gene identified in our previous in vivo studies (Geles et al., 2006). We therefore speculate that specific induction of such genes in granulosa cells could be in part attributed to the high levels of 4b/4-IID in these cells. The involvement of 4b/4-IID could either enhance occupancy of c-Jun and Sp1 at the promoter, induce more expression of c-Jun, or a combination of both events. Thus, the mechanisms described in this paper for regulation of the c-jun promoter by c-Jun and Sp1 in concert with 4b/4-IID may be more widespread and conserved in other cell types.
Experimental Procedures
Plasmids, reagents and protein purification
Details of constructs and reagents along with protein purification of all complexes and factors used in this study are described in the Supplemental experimental procedures.
In vitro reconstituted transcription
Reactions were performed as previously described with minor changes (Naar et al., 1998). Reactions contained 70 ng of linearized DNA templates, 40 nM rIIA, 5 nM rIIB, 20 nM rIIE, 20 nM rIIF, 2 nM RNA Pol II, 0.1 nM TFIIH, and 0-100 ng of TFIIDs in a final volume of 25 μl. 30 ng of Sp1 and 20 ng of c-Jun were used as listed in the figures. Activators were preassembled with the general factors at 30°C for 5 min before addition of the DNA templates. Each reaction was repeated several times with the representative data shown.
DNaseI footprinting assay
Reactions contained 50 ng of 4b/4- or 4/4-IID, 8-80 ng of Sp1, 12-120 ng of c-Jun, 20 nM of rTFIIA, and approximately 30 fmols of 32P end-labeled DNA probe in a final volume of 50 μl of Buffer A (12.5 mM Hepes, pH 7.9, 5% glycerol, 6.25 mM MgCl2, 0.05 mM EDTA, 50 mM KCl, 4 mM Spermidine, 5 μg/ml BSA, 14 μM [by nucleotides] poly(dG-dC), and 1 mM DTT). Reactions were incubated at 30°C for 30 minutes followed by incubation with 50 μl of DNAse I solution (containing 0.9 ng of DNAse I [Worthington], 10 mM MgCl2, and 5 mM CaCl2) for an additional 1 min. Reactions were stopped with a solution containing 1% SDS, 0.2 M NaCl, 20 mM EDTA and 0.2 mg/ml glycogen. Samples were extracted with phenol/chloroform/Isoamyl alcohol, precipitated with ethanol, electrophoresed on a 8% polyacrylamide gel with 7 M Urea and visualized using a Typhoon Phosphorimager (Amersham Biosciences). Results were quantitated using ImageQuant (Amersham Biosciences). To account for loading or digestion differences between individual lanes, signals corresponding to the indicated binding sites were normalized against the signal from a region outside of the c-jun promoter whose digestion pattern showed fairly little variance under all of the conditions tested.
Antibody labeling of 4b/4-IID for immunomapping
An antibody labeling step was added prior to elution of the complex from our TAF4 mAb affinity resin. TAF4b mAb (50 μg) was incubated (2 hrs) with 2 μg of the washed 4b/4-IID complex bound to 250 μl of protein G sepharose beads containing covalently conjugated TAF4 mAb. The 4b/4-IID-anti-TAF4b complex was washed 3 times with 0.3 M KCl/HEMG buffer (containing 0.05% NP40 and 10 μM leupeptin) and eluted with a peptide recognized by the TAF4 mAb as described in Supplemental experimental procedures. The eluates were concentrated with a microcon-10 concentrator, immediately applied to grids, and negatively stained as described below for the 4b/4- and 4/4-IID complexes.
Electron microscopy and 3D reconstruction
4 μl (10-20 ng in total amount) of the fresh 4b/4- or 4/4-IID was applied directly onto a thin carbon film supported by holey carbon on a 400-mesh copper grid (Electron Microscopy Sciences), which was freshly glow-discharged. After exchange with 3% Trehalose in 0.3 M KCl/TEM buffer (50 mM Tris-HCl, 0.1 mM EDTA, 2 mM MgCl2 [final pH 7.9]) the sample grid was stained in five successive 75 μl drops of 1% Uranyl formate.
The image data was collected with a Tecnai 12 transmission electron microscope (FEI) operated at 120KeV and at a magnification of 30,000x with a defocus range of -0.85 μm to -1.5 μm. All the images were collected on SO-163 films (Kodak) and developed with D-19 developer (Kodak). The micrographs were digitized with a Super Coolscan 8000 scanner (Nikon) at a pixel size of 12.7 μm, resulting in a 4.23Å/pixel size at the specimen scale.
A total of 12,851 particles (for 4b/4-IID), 14,000 particles (for 4/4-IID), or 10,024 particles (for 4b/4-IID-anti-TAF4b) were manually selected using Boxer (EMAN) (Ludtke et al., 1999) and then extracted to a particle window size of 119 × 119 pixels and converted to the “SPIDER” format for further data processing by SPIDER (System for Processing of Image Data from Electron microscopy and Related fields). The particles were then low-pass band filtered and normalized prior to the 3D reconstruction using SPIDER’s projection matching approach (Frank et al., 1996). Details of the 3D reconstructions and representations are described in the Supplemental experimental procedures.
Supplementary Material
Acknowledgement
We thank S. Zheng for TAF4b antibody generation work, the Tjian lab tissue culture facility technicians, C. Inouye for purifying the basal factors, M. Haggart for assistance, and S. Zhou (HHMI, UC-Berkeley) for mass spectrometry. We also thank S. Lipscomb, A. Leschziner, H. Wang, and B. Siridechadilok for helpful advice and technical support. We are grateful to D. Reinberg, J. A. Goodrich, Z. Zhang, and U. Schulze-Gahmen for critical comments of the manuscript. This work was supported by NIH National Cancer Institute PO1 CA112181 (R.T. and E.N.), NIH General Medical Sciences RO1 GM63072 (E.N.), and by the U.S. Department of Energy in contact with the University of California (E.N.). R.T. and E.N. are Howard Hughes Medical Institute Investigators. R.T. is Director of the Li Ka-Shing Center for Biomedical and Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainbinder E, Revach M, Wolstein O, Moshonov S, Diamant N, Dikstein R. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-kappaB. Mol Cell Biol. 2002;22:6354–6362. doi: 10.1128/MCB.22.18.6354-6362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- Chi T, Carey M. Assembly of the isomerized TFIIA--TFIID--TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Geles KG, Freiman RN, Liu WL, Zheng S, Voronina E, Tjian R. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc Natl Acad Sci U S A. 2006;103:2594–2599. doi: 10.1073/pnas.0510764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Guermah M, Tao Y, Roeder RG. Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol Cell Biol. 2001;21:6882–6894. doi: 10.1128/MCB.21.20.6882-6894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hilton TL, Li Y, Dunphy EL, Wang EH. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol. 2005;25:4321–4332. doi: 10.1128/MCB.25.10.4321-4332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Boyer TG, Berk AJ. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva LA, Nabirochkina EN, Kurshakova MM, Robert F, Krasnov AN, Evgen’ev MB, Kadonaga JT, Georgieva SG, Tora L. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurent C, Sanders SL, Demeny MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Mapping key functional sites within yeast TFIID. EMBO J. 2004;23:719–727. doi: 10.1038/sj.emboj.7600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively TN, Ferguson HA, Galasinski SK, Seto AG, Goodrich JA. c-Jun binds the N terminus of human TAF(II)250 to derepress RNA polymerase II transcription in vitro. J Biol Chem. 2001;276:25582–25588. doi: 10.1074/jbc.M100278200. [DOI] [PubMed] [Google Scholar]
- Munz C, Psichari E, Mandilis D, Lavigne AC, Spiliotaki M, Oehler T, Davidson I, Tora L, Angel P, Pintzas A. TAF7 (TAFII55) plays a role in the transcription activation by c-Jun. J Biol Chem. 2003;278:21510–21516. doi: 10.1074/jbc.M212764200. [DOI] [PubMed] [Google Scholar]
- Naar AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo-Niersbach E, Furukawa T, Tanese N. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, Unger T, Dikstein R. Core promoter binding by histone-like TAF complexes. Mol Cell Biol. 2005;25:206–219. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Naar AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lovasco LA, Gyuris A, Baumgartner RA, Parlow AF, Freiman RN. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 2006;303:715–716. doi: 10.1016/j.ydbio.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ranish JA, Hahn S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Zehner ZE. c-Jun and the dominant-negative mutant, TAM67, induce vimentin gene expression by interacting with the activator Sp1. Oncogene. 2003;22:8891–8901. doi: 10.1038/sj.onc.1206898. [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A, Dikstein R. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.