Abstract
We report the identification and cloning of a 28-kDa polypeptide (p28) in Tetrahymena macronuclei that shares several features with the well studied heterochromatin-associated protein HP1 from Drosophila. Notably, like HP1, p28 contains both a chromodomain and a chromoshadow domain. p28 also shares features with linker histone H1, and like H1, p28 is multiply phosphorylated, at least in part, by a proline-directed, Cdc2-type kinase. As such, p28 is referred to as Hhp1p (for H1/HP1-like protein). Hhp1p is missing from transcriptionally silent micronuclei but is enriched in heterochromatin-like chromatin bodies that presumably comprise repressed chromatin in macronuclei. These findings shed light on the evolutionary conserved nature of heterochromatin in organisms ranging from ciliates to humans and provide further evidence that HP1-like proteins are not exclusively associated with permanently silent chromosomal domains. Our data support a view that members of this family also associate with repressed states of euchromatin.
Keywords: transcriptional repression/heterochromatin/chromodomain
At least two distinct modes of chromatin-mediated gene inactivation exist in eukaryotic cells: silencing, a process that establishes and maintains permanently repressed chromosomal domains and transcriptional repression, a process in which housekeeping or inducible genes are packaged in such a way that they can be activated when needed. Both cases involve unique chromatin configurations orchestrated by multi-protein complexes targeted to particular regions through interactions among specific DNA-binding proteins, non-histone chromosomal proteins, and uniquely modified chromatin components (1–4).
Gene silencing is thought to result, in part, from chromatin-mediated effects that often involve condensation of the chromatin fiber. Early cytologists recognized that eukaryotic genomes exist in two distinct structural forms: (i) unstained euchromatin (EU) that contains most of the expressed, single copy sequences and is largely decondensed during interphase and (ii) darkly staining heterochromatin that is relatively gene poor and remains compacted during most of the cell cycle (5). Position-effect variegation (PEV) in Drosophila and telomeric position effect in yeast are two well studied examples of an epigenetic silencing mechanism in which affected genes can be clonally inherited and yet switch between transcriptionally active and repressed states under different growth conditions. PEV and telomeric position effect are thought to result from the mosaic inactivation of genes next to “inactivation centers”, such as centromeric heterochromatin or telomeres. In many ways, the repressive spreading phenomena inherent in these processes are common with other well studied epigenetic silencing mechanisms such as mating-type silencing in yeast, inactivation of homeotic gene clusters, imprinting, and X-inactivation in mammals (1, 6).
One of the best studied modifiers of PEV is the essential Su (var) 205 gene that encodes the heterochromatin-associated protein HP1 (1, 7–8). Conservation of HP1 in organisms ranging from flies to human (9) suggests that HP1-like proteins play important, yet presently unclear, roles in cell viability and development (10). HP1 contains a 52-aa chromodomain (for chromatin organization modifier), a domain found primarily in proteins associated with transcriptionally repressed chromatin (1). Polycomb protein (Pc), a Drosophila protein implicated in the repression of homeotic gene expression (11), contains a chromodomain, suggesting a molecular link between PEV and homeotic gene repression. Interestingly, regulatory regions necessary and sufficient for Pc-mediated repression also induce PEV (12). Although it is still an enigma whether HP1 or Pc can bind to DNA directly (e.g., see ref. 13), current models speculate that both proteins participate in the assembly of unique multi-protein complexes that establish and maintain a transcriptionally repressed state during specific stages of the cell cycle (14) and in development (15).
The ciliated protozoan, Tetrahymena thermophila, provides a useful model for unraveling relationships between silencing, transcriptional repression, and gene activation. Each vegetative cell contains two functionally distinct nuclei: (i) a germ-line micronucleus that contains highly condensed, transcriptionally silent chromatin during most of the life cycle and (ii) a somatic macronucleus that contains both active and repressed chromatin domains (16). In this study, we report the cloning of a chromodomain-containing protein from Tetrahymena that resembles HP1 from Drosophila. This protein is not found in transcriptionally silent micronuclei but is, instead, enriched in the electron-dense chromatin bodies (CBs) present in macronuclei. These findings suggest that HP1-like proteins may be involved in localized transcriptional repression within a transcriptionally active environment (i.e., the macronucleus) and are not exclusively associated with transcriptionally silent (i.e., micronuclear) chromatin.
MATERIALS AND METHODS
Cell Culture.
Genetically marked strains of Tetrahymena thermophila, CU 427 (Mpr/Mpr[6-mp-s]VI) and CU 428 (Chx/Chx-[cy-s]VII), were used in all experiments reported here. These were generously provided by Peter Bruns (Cornell University, Ithaca, NY). DNA sequences reported were derived entirely from strain CU 428.
Nuclear Protein Isolation and Gel Electrophoresis.
Isolation of macro- and micronuclei as well as extraction and precipitation of acid-soluble proteins were performed as described (17). Proteins were electrophoresed on a SDS polyacrylamide gel and stained by Coomassie blue (Sigma).
HPLC Purification and Peptide Sequencing of Hhp1p.
Perchloric acid (PCA)-insoluble macronuclear proteins were separated by reverse-phase (RP) HPLC by using a C8 analytical column (Aquapore Octyl-RP300; Brownlee Lab) with a linear gradient of 0–60% acetonitrile and 0.1% trifluoroacetic acid over 120 min. Fractions containing Hhp1p (p28) were detected by Western analysis with α-phosphorylated H1 antibodies (17), pooled, subjected to SDS gel electrophoresis, and immobilized on Immobilon PSQ (Millipore) membrane. Pure proteins, generated by cutting an individual band from the membrane, were processed for microsequencing by using standard methods. Cyanogen bromide peptides of Hhp1p also were generated and sequenced as described by Lu et al. (17).
Generation and Characterization of Polyclonal Antibodies Against Hhp1p.
HPLC-partially purified Hhp1p were separated on an SDS gel, transferred to nitrocellulose, and excised for immunizing rabbits by standard methods. Approximately 200 μg of Hhp1p protein was used for two separate injections. Preimmune serum was obtained from the same animal before immunization. In immunoblotting experiments, crude serum was used at 1:2,000 dilution, and immunoreactivity was detected by alkaline phosphatase (AP)-conjugated secondary antibodies or by enhanced chemiluminesence and autoradiography as indicated.
Gene Cloning and Sequencing of HHP1.
Two nested degenerate oligonucleotide primers were designed against portions of the Hhp1p amino-terminal peptide, with consideration given to Tetrahymena codon usage frequency (18). Their sequences are: HHP1–5(1) [5′-ACYAARGTYTAYGARGTYG-3′] and HHP1–5(2) [5′-GTYGAARAAYATYATYGGTCAYAG-3′] (degenerate bases in oligonucleotide primers are: r = A + G, Y = C + T). Total cDNA was obtained from growing cells as described (19), and reverse transcriptase–PCR was performed by using Moloney Murine Leukemia Virus (MMLV) reverse transcriptase with oligo (dT) primers and two nested degenerate primers HHP1–5(1) and HHP1–5(2) as described above. Two gene-specific primers: HHP1-S [5′-GGGAAGTTAGATTGAATACC-3′] and HHP1-A [5′-AATCTAATCAGCGGATTAGC-3′] were designed from the above cDNA sequence and used to amplify the genomic sequence of HHP1. Locations of two introns were determined by comparing the cDNA sequence and genomic sequence in the coding region. Inverse-PCR (20) was then used to obtain the 5′ end of the gene including the upstream region. Whole cell genomic DNA was restriction digested with SpeI religated at low DNA concentration and finally amplified by using inverse primers, HHP1-A1 [5′-AGAGTAAGCCTTTCCATTAGATATGG-3′] and HHP1-S1 [5′-GCTAAGTATGCTTCTCCCG-3′]. All PCR products were cloned and sequenced as described (19).
AP Treatment.
RP-HPLC purified Hhp1p was dried and resuspended in 50 μl of 20 mM Tris, pH 8.0, and 1 unit of Escherichia coli AP (Sigma) was added. The reaction mixture was incubated at 37°C for 1–2 hr before boosting with another 1 unit of enzyme, after which the incubation was continued for another 1–2 hr. After the reaction, AP was separated from Hhp1p by RP-HPLC.
In Vitro Phosphorylation.
Hhp1p partially purified by RP-HPLC was mixed, with or without 2 units of p34cdc2/cyclin B (New England Biolabs), in 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, 100 μM ATP, and [γ-32P]ATP to a final specific activity of 120 μCi/μmol. Reaction mixes were incubated at 30°C for 30 min, subjected to acid-urea gel electrophoresis on a 15%, 30-cm polyacrylamide gel, and analyzed by immunoblotting and autoradiography.
Glu-C Partial Digestion and Peptide Mapping.
RP-HPLC purified Hhp1p (phosphorylated or unmodified) was resolved by SDS/PAGE, excised from the gel, and subjected to limited Glu-C digestion as described (21). The resulting peptides were resolved on a 22% SDS polyacrylamide gel, transferred to poly(vinylidene difluoride) membrane, and analyzed by autoradiography and immunoblotting by using α-phosphorylated H1 antiserum.
Indirect Immunofluorescence, Confocal Microscopy, and Immunocytochemistry.
For immunofluorescence analyses, vegetative cells were fixed in periodate-lysine-paraformaldehyde (PLP) according to previously described methods (22) for 20 min at room temperature. Fixative was removed by centrifugation and pellets were washed with 37 mM potassium phosphate buffer, dehydrated with methanol at room temperature, and processed for indirect immunofluorescence as described (23). α-Hhp1p antibodies were used at 1:100. Confocal images were captured by using the Leica TCS NT confocal system.
For electron microscopic analyses, cells (2 × 106) were washed with 40 mM Hepes buffer (pH 7.5) and fixed in periodate-lysine-paraformaldehyde (PLP) fixative and 0.05% glutaraldehyde overnight at 4°C. The samples were then dehydrated, embedded in London Resin white resin and polymerized at 55°C as described in Lending (24). Ultrathin sections were collected on copper grids and subject to immunocytochemical analyses as described (25). Primary antibody concentrations were 1:25 and 1:100, for α-Hhp1p and α-H2A antibodies, respectively. Goat anti-rabbit antibodies conjugated to 10-nm gold beads were used for detection of the primary antibodies (25). Sections were then either poststained with uranyl acetate for 10 min followed by lead citrate for 5 min, or were only poststained with lead citrate for 5 min and examined with a Hitachi H-7000 electron microscope (Hitachi Scientific Instruments, Gaithersburg, MD) operated at 75 KV. Quantitative analyses on the relative distribution of gold particles (number of particles/unit area) were performed from photographic enlargements of electron micrographs as described (23). In each analysis, ≈500 gold particles were scored. Multiple attempts were made to increase the α-Hhp1p signals in our immunocytochemical analyses. These were not successful, perhaps due to sensitivity of Hhp1p epitopes to aldehyde fixation, a feature which also has been reported for other HP1-like proteins (26).
RESULTS
p28 Is a PCA-Insoluble Polypeptide Unique to Macronuclei.
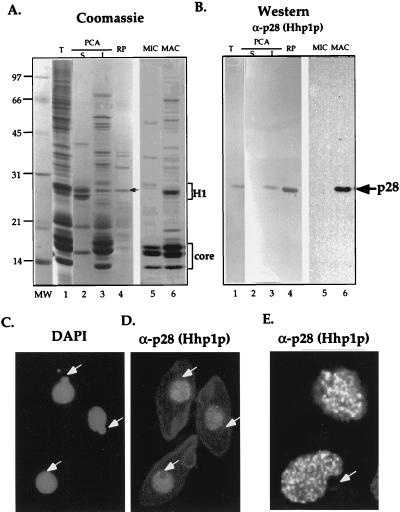
A series of immunoblotting experiments, aimed at characterizing linker histone (H1) phosphorylation in Tetrahymena macronuclei, detected a polypeptide with an apparent molecular mass of 28 kDa (p28) that reacted strongly with several phosphorylation-specific as well as general linker H1 antibodies (data not shown). However, unlike all other known H1 molecules, p28 was insoluble in PCA, a hallmark feature of most H1 molecules. Due to its H1-like mobility on SDS/PAGE and cross-reactivity with several H1 antisera (17), we reasoned initially that p28 may represent a H1 variant. To investigate this possibility further, a polyclonal antiserum was raised against gel-purified p28 after initial enrichment by RP-HPLC. Immunoblotting analyses demonstrated that this antiserum reacted specifically with p28 in total macronuclear extracts (Fig. 1A–B, lane 1) and, as expected, p28 partitioned preferentially into PCA-insoluble fractions (Fig. 1A–B, lane 3). However, the p28 antiserum failed to react with macronuclear H1, demonstrating that p28 is immunologically distinct from H1 in this assay (Fig. 1A–B, lane 2).
Figure 1.
p28 is specific to somatic macronuclei and missing from germ-line micronuclei. Total macronuclear proteins (T, lane 1), PCA soluble (S, lane 2), and insoluble (I, lane 3) proteins from equivalent amount of macronuclei and RP-HPLC-purified p28 (RP, see arrow in lane 4) were electrophoresed in a 12% SDS gel and either stained with Coomassie (A) or were blotted onto nitrocellulose and reacted with α-p28 (Hhp1p) antibodies followed by detection with AP (B, lanes 1–4). As well, comparable loads of acid-soluble micro- (lane 5) and macronuclear (lane 6) proteins, from unit gravity-purified nuclei, were subjected to a similar analyses except that enhanced chemiluminesence was used for detection (B, lanes 5–6,). The position of linker histone H1 and core histones is indicated by brackets (A); arrows denote p28 (A and B). Vegetatively growing cells also were fixed and incubated with p28 (Hhp1p) antibodies. In situ reactions were detected indirectly with rhodamine-conjugated secondary antibodies (C); nuclei in the same cells were detected with 4′,6-diamidino-2-phenylindole (D). A close inspection of p28 (Hhp1p) staining was taken by using confocal microscopy (E). White arrows point to micronuclei, that are consistently not stained with HHP1p antibodies.
Interestingly, p28 was observed only in extracts from macronuclei and was not detected in micronuclei (Fig. 1A–B, cf. lanes 5 and 6). Consistent with these results, indirect immunofluorescence analyses showed that only macronuclei, but not micronuclei (see white arrows in Fig. 1C–E), are stained by p28 antiserum. Confocal microscopy revealed that the p28 antiserum stained macronuclei in a nonuniform fashion; numerous small punctate dots were detected at all focal levels through macronuclei (Fig. 1E). Taken together, these results demonstrate that p28 is localized to specific domains within macronuclei (see below) and is absent from transcriptionally silent micronuclei.
Isolation of the Gene Encoding p28 Predicts an HP1-Like Protein.
Amino acid sequence was obtained from amino terminal and internal (≈20 kDa peptides, generated by cyanogen bromide cleavage) fragments of membrane-purified p28 by standard microsequencing techniques. Degenerate oligonucleotide primers were then designed from these peptide sequences and used in multiple PCR techniques to clone the gene encoding p28, as well as upstream and downstream DNA. Southern blot analysis of total genomic DNA revealed that the gene encoding p28 is present in single copy (data not shown).
The predicted translation of the longest ORF of the p28 cDNA, beginning at the first in-frame AUG codon, is shown in Fig. 2. This ORF is believed to be correct for several reasons: First, both peptide sequences obtained from p28 (including its amino-terminal sequence) are found in the predicted coding region (underlined in Fig. 2). Second, the calculated molecular mass of the predicted protein (21 kDa) is in reasonable agreement with the apparent mass of the polypeptide (28 kDa) in SDS/PAGE analysis. Third, the predicted noncoding regions are highly AT-rich, which is a characteristic of most known Tetrahymena genes (27).
Figure 2.
HHP1-derived protein sequence. Translation of the longest ORF of the HHP1 gene is shown from the presumed initiator methionine. Protein sequences unambiguously identified by direct sequencing of the intact polypeptide and the fragment generated by cyanogen bromide are underlined. The amino-terminal chromodomain and carboxy-terminal chromoshadow domain are shown in bold. The positions of two potential phosphorylation sites for Cdc-2 kinase were indicated by asterisks.
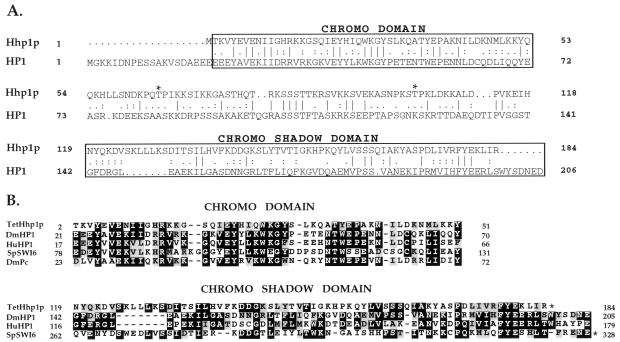
Comparison of p28 amino acid sequence with available protein databases suggests that p28 is a unique polypeptide. However, alignments reveal that p28 is most highly related to the heterochromatin-associated protein 1 (HP1) from Drosophila (probability of matching chance, P < 10−10, see Fig. 3A and ref. 28). Like other HP1-like proteins, p28 contains a highly conserved amino-terminal chromodomain and a carboxy-terminal chromoshadow domain (see Fig. 3B and ref. 29). Recent studies have demonstrated that both of these domains are important for the association of HP1 with heterochromatin and for gene silencing, presumably through protein–protein interactions (1). The homology between HP1 and p28 raises the intriguing possibility that p28 plays a role in transcriptional repression mediated through heterochromatin formation in what is a transcriptionally active nucleus (16).
Figure 3.
Sequence alignment among Tetrahymena HHP1p, Drosophila HP1, and selected chromodomain family members (A and B). DmHP1, D. melanogaster (28); HuHP1, human (40); SpSWI6, Schizosaccharomyces pombe (35); DmPc, D. melanogaster (11). The alignment was made by the program clustal w (http://alfredo.wustl.edu/msa/clustal.cgi) and fine adjusted manually. The shading was then made by the program boxshade (http://ulrec3.unil.ch:80/software/BOX_form.html). Gaps in sequence alignments are indicated by dashes. Domain positions in the protein sequences are indicated by numbers; domains that end at the C terminus of the protein are marked with asterisks.
In agreement with the cross-reactivity observed initially with several H1 antisera, alignment of p28 with Tetrahymena H1 shows that the central region between the amino-terminal chromodomain and carboxy-terminal chromoshadow domain (amino acids 64–116) exhibits H1 characteristics in being lysine/serine/threonine-rich and containing several putative phosphorylation sites including two putative Cdc2 kinase recognition sites (TPIK/TPK, see asterisks in Figs. 2 and 3A). Phosphorylation-specific H1 antibodies that recognize phosphorylated TPVK motifs in macronuclear H1 (17) cross-react strongly with p28 (Figs. 4B), suggesting that p28 is phosphorylated, at least on one or both of these proline-directed sites (see below). Because the p28 gene encodes a polypeptide that contains features of both HP1 and H1, we have renamed it Hhp1p (for H1/HP1-like protein).
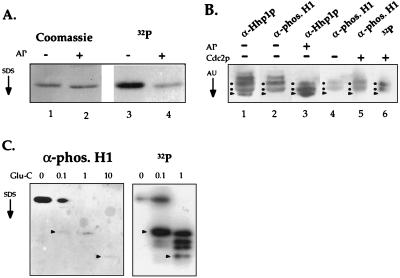
Figure 4.
HHP1p is multiply phosphorylated by using two potential Cdc2 kinase phosphorylation sites. (A) HHP1p is phosphorylated in vivo. Vegetative cells were labeled continuously during growth with 32P-orthophosphate. RP-HPLC-purified HHP1p was treated with AP (+) or buffer only (−), resolved by 12% SDS/PAGE, and analyzed by Coomassie staining and autoradiography. (B) Hhp1p was incubated in the presence (+) or absence (−) of AP. As well, AP-treated Hhp1p was incubated with (+) or without (−) Cdc2 in a standard kinase reaction. All samples were then resolved on a long (30 cm) acid-urea gel and analyzed by autoradiography (where appropriate) or by immunoblotting by using α-Hhp1p or α-phosphorylated H1 antisera. Dots indicate the putative isoforms phosphorylated by Cdc2 kinase, and the arrow heads point to the faster migrating, dephosphorylated isoform. (C) Gel-purified, unlabeled Hhp1p (Left) as well as in vitro-phosphorylated Hhp1p (Right) were partially digested by Glu-C protease, resolved on a 22% SDS/PAGE, and analyzed by immunoblotting by using the phosphorylated H1 antiserum (Left) or autoradiography (Right). Arrow heads point to peptides that are detected in both cases. Reactivity of Hhp1p peptides with the phosphorylated H1 antiserum is reduced after Glu-C digestion for reasons that remain unclear. The amount (in micrograms) of Glu-C used in each reaction is indicated at the top of each lane.
Phosphorylation Status of Hhp1p During Vegetative Growth.
Phosphorylation of HP1 correlates with the formation of heterochromatin in Drosophila (30). The presence of several putative phosphorylation sites in Hhp1p encouraged us to explore whether it also was phosphorylated in vivo. Vegetative cells were labeled continuously during growth with 32P-orthophosphate before purifying Hhp1p by RP-HPLC. The recovered Hhp1p was then analyzed by Coomassie staining and autoradiography after SDS/PAGE. As shown in Fig. 4A, Hhp1p is extensively phosphorylated under these conditions, and to a large extent, the incorporated 32P-label is removed by AP treatment.
To enhance the resolution of phosphorylated isoforms, Hhp1p was separated on long acid-urea gels and analyzed by immunoblotting with α-Hhp1p and phosphorylation-specific H1 antisera (Fig. 4B). With this increased resolution, four to five Hhp1p isoform bands are typically detected depending on individual protein preparations. The phosphorylation-specific H1 antiserum (one that recognizes only phosphorylated isoforms of macronuclear H1; ref. 17) cross-reacts strongly with all but the fastest-migrating isoform (Fig. 4B, see arrowheads in lanes 1 and 2), suggesting that this fastest-migrating band represents the dephosphorylated isoform of Hhp1p and that the collection of slower-migrating bands is comprised of phosphorylated isoforms. This interpretation was supported by the fact that several of the slower-migrating bands disappear with AP treatment (lane 3, Fig. 4B).
To test whether Hhp1p can be phosphorylated by Cdc2 kinase as suggested by the presence of two Cdc2 consensus motif, Hhp1p was first treated with AP before incubation with [32P]ATP in the presence or absence of Cdc2 kinase. The reaction products were then resolved on a long acid-urea gel and detected by immunoblotting. As shown in Fig. 4B (lanes 5 and 6), incorporation of 32P is clearly detected in two separable isoforms (see dots along the side of each lane) demonstrating that Hhp1p is a substrate for Cdc2 kinase in vitro. In addition, the phosphorylated H1 antiserum cross-reacts more strongly with Hhp1p after treatment with Cdc2 kinase (Fig. 4B, cf. lanes 4 and 5).
We next set out to map the phosphorylation sites in Hhp1p in an attempt to determine whether one or both of the two predicted Cdc2 consensus sites (see asterisks in the sequence alignments in Figs. 2 and 3B) are phosphorylated in vivo. Hhp1p, directly isolated from growing cells (i.e., in vivo phosphorylated), or an AP-treated Hhp1p subsequently phosphorylated in vitro by Cdc2 kinase (i.e., in vitro phosphorylated), were subjected to cleavage by Glu-C followed by resolution of the resulting peptides on SDS/PAGE. The resulting phosphopeptides were detected by immunoblotting with α-phosphorylated H1 antiserum (in vivo phosphorylated) or by autoradiography (in vitro phosphorylated) and then were directly compared. As shown in Fig. 4C, two peptides, each phosphorylated by Cdc2 kinase were detected by autoradiography that were common between the in vitro- and in vivo-phosphorylated samples. These data indicate that at least one (and potentially both) Cdc2 site(s) is phosphorylated in vivo. Consistent with these data, recent studies have demonstrated that p34Cdc2 is present in macronuclei but is missing in micronuclei (31, 32).
In Situ Distribution of Hhp1p in Macronuclei: Enrichment in Chromatin Bodies.
The fact that Hhp1p contains both a chromodomain and a chromoshadow domain found in many heterochromatin-associated proteins suggests that Hhp1p may be involved in the formation and/or maintenance of heterochromatin in macronuclei. Close inspection of the confocal data presented in Fig. 1E shows that Hhp1p is not uniformly distributed within macronuclei. Previous ultrastructural analyses have shown that macronuclear chromatin also is not uniform with respect to the distribution of condensed and decondensed chromatin (23, 33). The punctate staining exhibited by Hhp1p antisera suggested that Hhp1p might be enriched in electron-dense CBs that punctuate macronuclei (shown in the low magnification micrograph in Fig. 5A; ref. 33).
Figure 5.
HHP1p is enriched in electron-dense CBs in macronuclear chromatin. (A) Ultrastructure of a macronucleus: Numerous condensed CBs as well as surrounding EU and peripheral nucleoli (N) are evident. Antibodies against Hhp1p (B and C) and H2A (D) were used in parallel immunogold analyses. (Bar = 0.2 micrometer; A and B, and C and D are in the same magnification). (E) Statistical analysis of gold particle distribution in CBs and EU by using α-Hhp1p or α-H2A antisera.
Immunocytochemical analyses, using Hhp1p antibodies, indicate that Hhp1p is largely confined to the electron-dense regions of the CBs (Fig. 5B). Verification of Hhp1p enrichment in CBs was provided by statistical analyses of immunogold localizations similar to what is shown in Figs. 5C and D (see Fig. 5E). The vast majority of Hhp1p gold particles was distributed within the electron-dense CBs (Fig. 5B and 5E, 86% are CB-associated). Relatively few gold particles were observed in the surrounding EU (15%, Fig. 5E) or in peripherally located nucleoli (<1%). Antibodies against nucleolar marker proteins (not shown) and core histone H2A (Fig. 5D) were used to control for the specificity of our staining reactions. Moreover, as histone H2A associates with DNA in a constant ratio, it provided us with an internal measure of the amount of DNA in CBs relative to the surrounding, DNA-poor EU (see ref. 46). As expected, both euchromatic and heterochromatic regions are stained with H2A antibodies in a relative ratio of one to three, respectively. Relative to Hhp1p, in which 86% of the gold particles are CB-associated, a smaller fraction of H2A gold particles are CB associated (73%, Fig. 5E). These data suggest a modest enrichment of Hhp1p in CB over that of a core histone consistent with the potential role of Hhp1p in the formation and/or maintenance of heterochromatin in macronuclear CBs.
DISCUSSION
This report describes the identification and cloning of a phosphoprotein, Hhp1p, from Tetrahymena macronuclei. Western blot analyses and immunolocalization experiments demonstrate that this polypeptide is preferentially localized to the condensed CBs of macronuclear chromatin and is not present in transcriptionally silent micronuclei. Analysis of the protein sequence reveals that Hhp1p contains an amino-terminal chromodomain and a extended carboxy-terminal chromoshadow domain, linked together by a serine/threonine-rich “hinge” region that is likely to contain multiple sites of phosphorylation.
Hhp1p has several features in common with the Drosophila heterochromatin-associated protein HP1, suggesting that it may function like HP1 in heterochromatin formation and gene repression. Similar to HP1 and several other known silencing proteins [e.g., Polycomb (11), Swi6 (35, 36)], Hhp1p contains an N-terminal chromodomain, a region essential for the gene silencing function of both HP1 (37) and Pc (38). Moreover, two amino acids in HP1, Y24 (Y4 in Hhp1p), and V26 (V6 in Hhp1p) that are critical for the intact tertiary structure of chromodomain (39) and the function of HP1 in PEV (37), are conserved in Hhp1p. Hhp1p also contains a C-terminal chromoshadow domain, which to date has been found only in HP1-like proteins (29), and the distance between these two domains in Hhp1p (67 aa) is very similar to that in Drosophila HP1 (71 aa). Moreover, in both proteins, this central region contains multiple putative phosphorylation sites. Because of these similarities, Hhp1p is considered to be a member of the HP1-like family of proteins.
However, several differences exist between HP1 and Hhp1p preventing a definitive assignment. All known HP1-like proteins are relatively acidic, with a pI of ≈5. In contrast, Hhp1p is considerably more basic with a pI of 10.0 (determined by compute pi/mw toolin the ExPASy Molecular Biology Server), and we note that a cluster of glutamic acid residues found immediately before chromodomain in several HP1-like proteins (29) is missing in Hhp1p. Although the function of this acidic stretch in these polypeptide is unclear, an interaction with the basic tail domains of core histones has been suggested (9).
In flies, HP1 is multiply phosphorylated, and this phosphorylation appears to be important in the assembly and maintenance of heterochromatin (30). Similarly, Hhp1p is a phosphoprotein and, like HP1, most of the potential phosphorylation sites are concentrated in the central domain of the protein linking the chromodomain and chromoshadow domain. Whether this region acts as a “hinge” that is regulated by phosphorylation-induced conformational changes remains an attractive possibility (30). Among the potential phosphorylation sites in Hhp1p, two potential Cdc2 sites are present in the ciliate protein that are not found in other known HP1s. Our results demonstrate that Hhp1p is a substrate for Cdc2 kinase in vitro (Fig. 4B) and suggest that at least one of the two Cdc2 sites in the central region is phosphorylated in vivo. In support, antibodies specific to phosphorylated H1, which are thought to recognize phosphorylated Cdc2 sites on linker histone H1 (17), strongly react with phosphorylated Hhp1p isolated from growing cells. However, as four to five isoforms of Hhp1p are well resolved on acid urea gels, it seems likely that additional phosphorylation events and/or other post-translational modifications occur on Hhp1p.
In analogy to Drosophila HP1, Hhp1p may play a role in heterochromatin assembly and/or gene silencing. Our finding that Hhp1p is missing from inactive micronuclei reinforces the view that HP1-like proteins do not exist exclusively in “silent” chromatin and is in agreement with studies in mammalian systems that have shown that HP1-like proteins also participate in generating “repressed” states of EU (1, 40). It seems likely that genomic silencing in Tetrahymena involves either separate mechanisms altogether (i.e., specialized linker histones, see ref. 32) or a distinct class of micronuclear-specific, chromodomain-containing proteins.
In general, members of this chromodomain-containing superfamily are being classified on the basis of single or multiple chromodomains with distinctive sequence features (29). As far as we are aware, Hhp1p is the second chromodomain-containing protein identified in Tetrahymena. Interestingly, the first such protein, Pdd1p, is a specialized heterochromatin-associated protein, containing multiple chromodomains (41, 42), that is functionally linked to programmed DNA degradation during the sexual pathway in this organism (reviewed in ref. 43). Ultrastructural studies have documented the timing of the appearance of CBs during macronuclear development (≈14 hrs; 44), and in situ analyses demonstrate that Hhp1p is present in the newly formed CBs during this stage (H.H. and D.A, unpublished observations). Pdd1p-based DNA elimination structures also form during this period of conjugation (45). Interestingly, both types of structures, although distinct in size and morphology, are electron-dense and coexist during a brief period macronuclear development (12–16 hr). Thus, these two chromodomain-containing proteins appear to play distinct roles in organizing macronuclear heterochromatin for what is presumably distinct functions.
During vegetative growth, a significant fraction of the Tetrahymena macronuclear genome (80–90%) exists in CBs (46). Because ≈50% of the macronuclear genome is expressed during vegetative growth (46), it seems likely that these specialized chromatin domains contain both active as well as repressed sequences. Based on recent results obtained with the disruption of macronuclear H1 genes (47), it is becoming clear that the appropriate chromatin environment affects both transcriptional activation and repression in macronuclei. As one of the first molecular markers for electron-dense CBs in macronuclei, it is of interest to determine the phenotypic consequence of disruption of the single copy HHP1 gene in a nucleus specialized for a high level of transcriptional output.
Acknowledgments
We are grateful to Dr. Richard Cook (Baylor College of Medicine, Houston) for help with peptide sequencing and Dr. Ron Pearlman (York University; Ontario) for assistance in DNA sequencing. We also acknowledge Drs. David Landsman and Steven Hennikoff for their valuable comments on sequencing alignments. We also are grateful to Drs. Martin Gorovsky, Richard Levy, John Belote, Eleanor Maine, and members of the Allis laboratory group for their technical advice and helpful discussion during all stages of this work. This research was supported by grants from the National Institutes of Health to C.D.A. (GM53512).
ABBREVIATIONS
- Hhp1p
H1/HP1-like protein
- EU
euchromatin
- PEV
position-effect variegation
- AP
alkaline phosphatase
- PCA
perchloric acid
- Pc
polycomb protein
- RP-HPLC
reverse-phase HPLC
- CBs
chromatin bodies
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AF079405).
References
- 1.Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 3.Grunstein M. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 4.Wallrath L L. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 5.John B. In Heterochromatin: Molecular and Structure Aspects. Cambridge, U.K.: Cambridge Univ. Press; 1988. pp. 1–147. [Google Scholar]
- 6.Renauld H, Gasser S M. Trends Cell Biol. 1997;7:201–205. doi: 10.1016/S0962-8924(97)01034-9. [DOI] [PubMed] [Google Scholar]
- 7.Pirotta V. Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- 8.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 9.Singh P B, Millwe J R, Rearce J, Kothary R, Buton R D, Paro R, James T C M, Gaunt S J. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eissenberg J C, Hartnett T. Mol Gen Genet. 1993;240:333–338. doi: 10.1007/BF00280383. [DOI] [PubMed] [Google Scholar]
- 11.Paro R, Hogness D S. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fauvarque M O, Dura J M. Genes Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto K, Yamada T, Muro Y, Himeno M. J Biochem. 1996;120:153–159. doi: 10.1093/oxfordjournals.jbchem.a021378. [DOI] [PubMed] [Google Scholar]
- 14.Pak D T S, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 15.Paro R. Curr Opin Cell Biol. 1993;5:999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- 16.Gorovsky M A. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- 17.Lu M J, Dadd C A, Mizzen C A, Perry C A, Mclachlan D R, Annunziato A T, Allis C D. Chromosoma. 1994;103:111–121. doi: 10.1007/BF00352320. [DOI] [PubMed] [Google Scholar]
- 18.Martindale D W. J Protozool. 1989;36:29–34. doi: 10.1111/j.1550-7408.1989.tb02679.x. [DOI] [PubMed] [Google Scholar]
- 19.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 20.Triglia T, Peterson M G, Kemp D J. Nucleic Acids Res. 1988;16:81–86. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allis C D, Glover C V, Bowen J K, Gorovsky M A. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- 22.McLean I W, Nakane P K. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 23.Lu M J, Mpoke S S, Dadd C A, Allis C D. Mol Cell Biol. 1995;6:1077–1087. doi: 10.1091/mbc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lending C R, Larkins B A. Plant Cell. 1989;1:1011–1023. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lending C R. Protoplasma. 1996;195:68–77. [Google Scholar]
- 26.Kellum R, Raff J W, Alberts B M. J Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- 27.Prescott D M. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C R. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aasland R, Stewart A F. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eissenberg J C, Ge Y-W, Hartnett T. J Biol Chem. 1994;269:21315–21321. [PubMed] [Google Scholar]
- 31.Roth S Y, Collini M P, Draetta G, Beach D, Allis C D. EMBO J. 1991;10:2069–2075. doi: 10.1002/j.1460-2075.1991.tb07738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweet M T, Jones K, Allis C D. J Cell Biol. 1996;135:1219–1228. doi: 10.1083/jcb.135.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiske-Benner A, Eckert W A. Differentiation. 1985;28:225–236. [Google Scholar]
- 34.McGrath K E, Smothers J F, Dadd C A, Madireddi M T, Gorovsky M A, Allis C D. Mol Biol Cell. 1997;8:97–108. doi: 10.1091/mbc.8.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorentz A, Osterman K, Fleck O, Schmidt H. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 36.Ekwall K, Javerzat J-P, Lorentz A, Schmidt H, Cranston G, Allshire R. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 37.Platero J S, Hartnet T, Eissenberg J C. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messmer S, Franke A, Paro R. Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 39.Ball L J, Murzina N V, Broadhurst R W, Raine A R C, Archer S J, Stott R J, Murzin A G, Singh P B, Domaille P J, Laue E D. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders W S, Cooke C A, Earnshaw W C. J Cell Biol. 1991;115:919–931. doi: 10.1083/jcb.115.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madireddi M T, Coyne R S, Smothers J F, Mickey K M, Yao M-C, Allis C D. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- 42.Callebaut I, Courvalin J-C, Worman H J, Mornon J-P. Biochem Biophys Res Commun. 1997;235:103–107. doi: 10.1006/bbrc.1997.6748. [DOI] [PubMed] [Google Scholar]
- 43.Coyne R S, Chalker D L, Yao M-C. Annu Rev Genet. 1997;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 44.Weiske-Benner A, Eckert W E. Differentiation. 1985;28:225–236. [Google Scholar]
- 45.Smothers J F, Madireddi M T, Warner F D, Allis C D. J Eukaryotic Microbiol. 1997;44:79–88. doi: 10.1111/j.1550-7408.1997.tb05942.x. [DOI] [PubMed] [Google Scholar]
- 46.Gorovsky M A. Ph.D. thesis. Chicago: Univ. of Chicago; 1968. [Google Scholar]
- 47.Shen X, Gorovsky M A. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]