Abstract
Understanding how oncogenic transformation sensitizes cells to apoptosis may provide a strategy to kill tumor cells selectively. We previously developed a cell-free system that recapitulates oncogene dependent apoptosis as reflected by activation of caspases, the core of the apoptotic machinery. Here, we show that this activation requires a previously identified apoptosis-promoting complex consisting of caspase-9, APAF-1, and cytochrome c. As predicted by the in vitro system, preventing caspase-9 activation blocked drug-induced apoptosis in cells sensitized by E1A, an adenoviral oncogene. Oncogenes, such as E1A, appear to facilitate caspase-9 activation by several mechanisms, including the control of cytochrome c release from the mitochondria.
Expression of oncogenes that deregulate the cell cycle can either induce apoptosis or sensitize cells to proapoptotic stimuli (1, 2). An implication of this observation is that oncogene expression generates a proapoptotic signal that is present in transformed cells but absent in normal cells. When this signal is uncoupled from the apoptotic machinery, transformed cells can survive and become resistant to chemotherapeutic drugs. In principle, restoring the link between the signal and the apoptotic machinery should selectively kill transformed cells because, although untransformed cells have the machinery, they lack the signal. Understanding how the apoptotic machinery is regulated by oncogenic transformation is the first step in testing this hypothesis.
A central component of the apoptotic machinery is a family of cysteine proteases called caspases (3). Caspases are expressed as inactive precursors that are activated by proteolytic processing. According to the current model (4), two classes of caspases, initiators and effectors, are involved in apoptosis. Proapoptotic signals activate initiator caspases, such as caspase-2, -8, and -9. This activation is autocatalytic and requires the binding of specific cofactors. Activated initiator caspases process effector caspases (caspase-3, -6, and -7), which in turn cause cell collapse by cleaving a specific set of substrates. It appears that each initiator caspase is activated in response to a subset of signals, indicating that a prerequisite for understanding how a specific signal activates apoptosis is linking this signal to a particular initiator caspase. The initiator caspase that mediates oncogene-dependent apoptosis has thus far been elusive (5, 6).
To investigate how the apoptotic machinery is regulated by oncogene expression, our laboratory previously developed a cell-free system that mimics oncogene-dependent apoptosis (7). This system is based on the observation that extracts from cells transformed with the adenoviral oncogene E1A (“transformed extracts”) spontaneously activated caspases whereas extracts from untransformed cells (“untransformed extracts”) did not, an observation consistent with the proapoptotic effect of this oncogene in cells (8). It was suggested that the activity that triggered caspase activation is induced by expression of E1A, and this activity was called oncogene-generated activity (OGA) (7). OGA partially purified from 293 cells [a transformed human epithelial kidney cell line that expresses the E1A and E1B oncogenes (9)] activated caspases when added to untransformed extracts, thus mimicking the effects of E1A expression in cells. Therefore, we suggested that OGA is a link between oncogene expression and the apoptotic machinery.
Here we have purified OGA to apparent homogeneity and have identified it as APAF-1, a cofactor required for caspase-9 activation (10). We found that caspase-9 indeed was required for caspase activation in our cell free system, as was cytochrome c (Cyt c), a second caspase-9 cofactor (10). These observations predicted that caspase-9 is the initiator caspase that mediates oncogene-dependent apoptosis in cells and that expression of oncogenes sensitizes cells to apoptosis by facilitating activation of this caspase. To test these predictions we used human primary fibroblasts (IMR90) that were sensitized to drug-induced apoptosis by expression of adenoviral oncogene E1A. Consistent with the results from the cell free system, a caspase-9 dominant negative mutant blocked drug-induced apoptosis in cells expressing E1A. This finding led us to investigate how E1A expression controlled caspase-9 activation. We conclude that this control is achieved through several mechanisms, one of which is regulating Cyt c release from mitochondria.
MATERIALS AND METHODS
Extract Preparation.
All extracts were prepared as described (7).
Purification of OGA.
OGA was purified by using a reconstitution and a complementation assay. In the reconstitution assay, an aliquot from each fraction (4 μl) was mixed with the flow through from Fast Flow Q chromatography (5 μl) and ATP (1 μl, 1 mM final concentration). In the complementation assay, an aliquot from each fraction (4 μl) was mixed with extract from IMR90 cells (5 μl) and ATP. In both assays, the mixture was incubated at 37°C for 15 min, and then caspase activity was measured by using a fluorogenic substrate, DEVD.afc (7), and was expressed as picomoles of free afc generated per minute per milligram of extract.
Cell extracts (293 extracts; 2 g from 80 liters of cell suspension) in buffer A (50 mM Pipes, pH 7/10 mM KCl/5 mM EGTA/1 mM MgCl2/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride) were loaded onto a Fast Flow Q column (250-ml bed vol). The flow through (FT) was concentrated by ultrafiltration and was used for the reconstitution assay. The 0.1 M KCl eluate (designated F1) was collected, potassium phosphate was added to 10 mM, and the solution was loaded onto a hydroxyapatite column (60-ml bed vol). The activity was eluted with 0.4 M potassium phosphate, was dialyzed against 100 mM Hepes (pH 7.5), 25 mM KCl, 5 mM EGTA, 1 mM MgCl2, and 1 mM DTT and was loaded onto POROS HS (8-ml bed vol). The activity was eluted with 25–500 mM KCl gradient and was supplemented with ammonium sulfate (30% saturation), and the soluble protein was loaded onto POROS PH (1.6-ml bed vol.). The activity was eluted with a 0–30% ammonium sulfate linear gradient, was loaded onto a Superdex 200 column, and eluted in buffer A supplemented with 100 mM KCl. Eluted fractions were dialyzed against buffer A and were assayed for activity.
Peptide Sequencing and Mass Spectrometry.
Protein sequencing was performed as described (11). The sequence of the peptides obtained corresponded exactly to residues 64–81, 82–98, 267–277, 421–437, 640–644, 774–791, 951–963, 1015–1023, and 1169–1176 of APAF-1. In addition, the masses of an additional eight peptides were determined by mass spectrometry and are predicted to derive from APAF-1.
mAbs.
Antibodies were prepared by using an the N-terminal fragment (amino acids 1–134) of caspase-9 and the full length caspases 7 and 8 as antigens. None of the antibodies crossreacted with all caspases tested, which were 3, 6, 7, and 8 for the caspase-9 antibody; 3, 6, 7, and 9 for caspase-8 antibody; and 3, 6, 8, and 9 for caspase 7 antibody. Antibody to APAF-1 was prepared by using a 50-kDa N-terminal fragment of the protein.
Immunodepletion of Caspase-9.
The antibodies were bound to protein A Sepharose (Pharmacia) according to the manufacturer’s instructions. Ten milligrams of 293 cell extract (300 μl) was incubated with either the bound antibody or just protein A Sepharose for 3 hours at 4°C. Aliquots were assayed for caspase activation by the fluorogenic assay and were analyzed for caspase-9 depletion by immunoblotting.
Recombinant Caspase-9.
Caspase-9 precursor with a carboxy-terminal histidine tag was expressed by using the pET-21 mch6 expression vector provided by E. Alnemri (Kimmel Cancer Institute, Philadelphia). The caspase-9 Cys287 to Ser mutant was prepared by using the Quick Change kit (Stratagene). Caspase-9 and the mutant were purified on Ni-NTA-agarose (Qiagen, Chatsworth, CA) followed by chromatography on Resource Q (Pharmacia).
Caspase-9 Gene Transfer.
cDNA encoding caspase-9 with a carboxy-terminal Flag epitope tag and the caspase Cys287 to Ser mutant were cloned into a MarxIV-puro retroviral gene transfer vector. Retrovirus was produced by transfection into LinX-A packaging cells (L. Y. Xie, D. Beach, and G.J.H., unpublished material). Medium from transfected LinX cells supplemented with 8 μg/ml Polybrene was diluted 1:2 with fresh medium and was added to plates of IMR90-E1A cells. Plates were centrifuged at 1,000 × g for 1 hour and then were incubated for 12–18 hours at 32°C. Medium then was replaced after 2 days, and infected cells were selected by using puromycin (1 μg/ml) for 4 days.
Scoring Apoptosis.
The cells were harvested by combining floating cells in the media with adherent cells that were detached with EDTA (3 mM) and EGTA (3 mM) in PBS. Cells were fixed in 4% paraformaldehyde and were stained with 4′,6-diamidino-2-phenylindole (0.5 μg/ml/0.1% Triton in PBS). The percentage of cells with apoptotic nuclear morphology then was determined.
Immunofluorescence Staining of Cyt c.
Cells were grown on coverslips coated in Pronectin F Plus (Protein Polymer Technologies, San Diego), were fixed in 4% paraformaldehyde, were permeabilized in 0.2% Triton, and were blocked with 2% BSA and 2% goat serum. Cells were incubated with anti-Cyt c antibody (a gift from R. Jemmerson, University of Minnesota) at 1 μg/ml followed by a secondary antibody conjugated to Alexa 594 (Molecular Probes). Cells were stained with 4′,6-diamidino-2-phenylindole, and the coverslips were mounted by using Prolong Antifade (Molecular Probes).
RESULTS AND DISCUSSION
Previously, we partially purified OGA from 293 cell extracts by using ion-exchange chromatography. This procedure separated the extract into three fractions (F1, F2, and FT). None of these fractions had caspase activity, but combining F1 and FT reconstituted caspase activation (7). One of the fractions, F1, activated caspases when added to untransformed extracts, thereby mimicking the effect of E1A expression. We therefore concluded that F1 contained OGA, and, in this study, we used this fraction to purify OGA to homogeneity.
OGA is APAF-1.
Purification of OGA relied on two assays. In the “reconstitution” assay, we analyzed fractions obtained during purification for the ability to reconstitute caspase activation when recombined with FT. In the “complementation” assay, these fractions were tested for the ability to activate caspases in extracts from primary human fibroblasts (IMR90 cells), which, in contrast to extracts from IMR90 cells transformed with E1A, do not activate caspases spontaneously in the presence of ATP (Fig. 1E). Throughout the purification, fractions that reconstituted activation coincided with those that activated untransformed extracts.
Figure 1.
Purification of OGA. (A) OGA was purified as described in Materials and Methods. Shown are the fractions from each step with maximal activity. Size exclusion chromatography, the last step in purification, provided a single polypeptide of ≈130 kDa (B) that coeluted with activity (C and E). This polypeptide reacted with an antibody to APAF-1 (D). Purification was done twice with similar results.
OGA was purified to homogeneity by using five chromatographic steps (Fig. 1A and Materials and Methods). The last step in purification gave a single polypeptide of ≈130 kDa, as estimated by SDS/PAGE (Fig. 1B), that activated caspases in the reconstitution (Fig. 1C) and the complementation assays (Fig. 1E). Both reactions depended on ATP, consistent with the previous characterization of OGA (7).
The size of the polypeptide and its ability to activate caspases suggested OGA was APAF-1, a known cofactor for caspase-9 activation (10). Indeed, an antibody to APAF-1 (a gift from X. Wang, University of Texas Southwestern Medical Center) reacted with the purified protein (Fig. 1D). To unambiguously identify the purified protein, we obtained peptide sequence. Ten OGA peptide fragments exactly matched the APAF-1 sequence (see Materials and Methods). The finding that OGA is APAF-1 suggested that E1A-dependent apoptosis is mediated by caspase-9. We set out to test this possibility in both the cell free system and in intact cells.
Caspase-9 Is Required for Oncogene-Dependent Caspase Activation in Vitro.
APAF-1 and Cyt c were identified as cofactors for dATP-dependent activation of caspase-9 in cell extracts (10). Adding dATP activates caspases in both transformed and untransformed extracts (7) whereas adding ATP activates caspases only in transformed extracts. Hence, E1A-dependent caspase activation is revealed in the presence of ATP. Therefore, we first tested whether ATP-dependent caspase activation also requires caspase-9 and Cyt c.
Caspase-9 is processed in transformed extracts earlier than is caspase-3, the effector caspase responsible for the bulk of the detected caspase activity (A. Doseff and Y.A.L., unpublished material). To test whether caspase-9 was required for caspase activation, it was depleted from extracts (Fig. 2A). Depletion greatly reduced caspase activation whereas mock depletion had a much lesser effect (Fig. 2B), consistent with caspase-9 being required for caspase activation in this system.
Figure 2.
Oncogene-dependent caspase activation in the cell-free system requires caspase-9 and Cyt c. (A) Depletion of caspase-9. The 293-extract (Input) was depleted of caspase-9 by using protein A Sepharose bound to a caspase-9 antibody (α-casp9) or was mock-depleted by omitting the antibody. Caspase-9 remaining in extracts (S) and bound to the Sepharose (P) was assessed by immunoblotting by using the same antibody. The two additional bands are IgG. (B) Depletion of caspase-9 abolished caspase activation, which was restored by adding recombinant caspase-9. Data are typical of three experiments. (C) Depletion of Cyt c. The FT fraction (input), used in the reconstitution assay, was depleted of Cyt c by passing it over a phosphocellulose column. The resultant flow through (FT-P) had no Cyt c detectable by immunoblotting. (D) Cyt c is required for caspase activation. FT-P was mixed with ATP (1 mM) and APAF-1 (≈1 fmol) with or without Cyt c (≈20 pmol) as indicated. Results shown are typical of two experiments.
To test that the effect of depletion was caused by the removal of caspase-9, we complemented depleted extracts with recombinant pro-caspase-9. The recombinant protein reconstituted caspase activation (Fig. 2B), although the amount required exceeded endogenous levels by at least 20-fold, perhaps because of the differences between endogenous and recombinant proteins. The complementation effect of caspase-9 was ATP-dependent and required APAF-1, indicating that this effect is caused by processing of caspase-9 rather than to a contamination with the active enzyme (data not shown).
To test whether Cyt c was required for ATP-dependent caspase activation, we depleted this protein from FT, the fraction that complements APAF-1 in the reconstitution assay, by using phosphocellulose (Fig. 2C). The resulting fraction, FT-P, failed to reconstitute caspase activation when combined with APAF-1 (Fig. 2D). Activation was restored by adding either exogenous Cyt c or by adding the eluate from phosphocellulose (Fig. 2D and data not shown). Hence, ATP-dependent caspase activation in our cell-free system did require Cyt c and was mediated by caspase-9.
Caspase-9 Mediates Oncogene-Dependent Apoptosis in Cells.
The finding that OGA is APAF-1 and the observation that oncogene-dependent caspase activation in vitro is mediated by caspase-9 predicted that the proapoptotic effect of E1A in cells is achieved through caspase-9. Therefore, we tested this prediction by using a dominant negative mutant of caspase-9 (caspase-9 DN) (12). Caspase-9 DN can bind cofactors but cannot undergo autocatalytic processing because of a mutation that substitutes serine for the catalytic Cys287. As a result, caspase-9 DN can compete for binding to cofactors with the endogenous enzyme, thereby preventing activation. Consistent with the requirement for caspase-9 in our cell-free system, caspase-9 DN inhibited caspase activation when added to the 293 extract (data not shown).
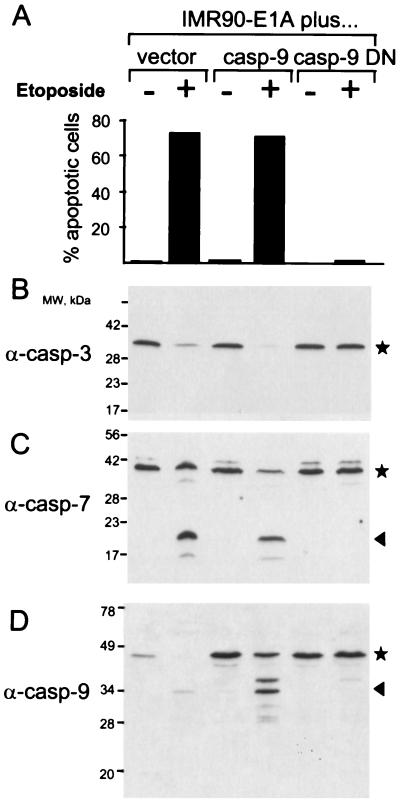
To test the effect of caspase-9 DN on oncogene-dependent apoptosis in cells, we used IMR90 fibroblasts transformed with E1A. In these cells, drug-induced apoptosis is oncogene-dependent because untransformed IMR90 cells are resistant to apoptosis induced by chemotherapeutic drugs but IMR90 cells expressing E1A are sensitive (13). When IMR90-E1A cells were treated with the chemotherapeutic drug etoposide, the cells underwent apoptosis as judged by cell morphology and chromatin condensation (Fig. 3A and Fig. 4A), by detection of caspases 3 and 7 processing (Fig. 3 B and C), by Annexin V staining, and by DFF cleavage (data not shown). Expression of caspase-9 DN abolished all of these changes, consistent with the notion that oncogene-dependent sensitization is mediated by caspase-9 (Fig. 3A and data not shown).
Figure 3.
Oncogene-dependent apoptosis and caspase-9 processing are blocked in cells by caspase-9 DN. (A) Caspase-9 DN prevents apoptosis and caspase activation in IMR-90-E1A cells that were infected with either caspase-9 DN, caspase-9, or vector alone by using a retroviral gene transfer vector. After 24 hours of treatment with etoposide (50 μM), apoptotic cells were scored as described in Materials and Methods. Processing of caspase-3 (B), -7 (C), and -9 (D) was assessed by immunoblotting. The caspase precursors are indicated by asterisks, and the processed caspases are indicated by arrows. Note that the fully processed caspase-3 is not detected because the epitope for the anti-caspase-3 antibody used (Transduction Laboratories, Lexington, KY) lies in the prodomain (11).
Figure 4.
E1A facilitates Cyt c release. (A) After drug treatment, Cyt c is released in transformed but not normal cells. IMR90 (top panels), IMR90-E1A (middle panels), and IMR90-E1A/casp9DN (bottom panels) cells were incubated with (“treated”) or without (“not treated”) 50 μM etoposide for 24 hours. The release of Cyt c was visualized by immunofluorescence. White arrows indicate cells with released Cyt c. The chromatin structure was visualized by staining with 4′,6-diamidino-2-phenylindole. The white star indicates a mitotic cell. (B) After etoposide treatment, the rate of apoptosis in IMR90-E1A cells is similar to the increase in the number of IMR90-E1A-casp9DN cells with released Cyt c. (C) Caspase-8 processing is not required for Cyt c release. Cells were treated as in Fig. 3; caspase-8 processing was assessed by immunoblotting. The two species of caspase-8 precursor revealed are probably previously reported isoforms (21).
Although etoposide treatment caused caspase-9 processing in cells infected with either vector alone or with caspase-9, this processing was not detectable in cells expressing caspase-9 DN (Fig. 3D). This observation is consistent with caspase-9 not being activated by other caspases but by autocatalysis, as is expected if caspase-9 is acting as an initiator caspase.
As cells expressing caspase-9 DN manifested no apoptotic changes after drug treatment, the question arose regarding whether the signals leading to apoptosis were sent at all. We reasoned that caspase-9 DN would not affect drug-induced changes that precede caspase-9 activation because this mutant is thought to prevent caspase activation by scavenging APAF-1 and/or endogenous caspase-9 (12). Because Cyt c should be released from mitochondria to participate in caspase-9 activation (14, 15), we set out to determine whether Cyt c was released in cells expressing caspase-9 DN.
Staining for Cyt c in untreated IMR90 E1A-caspase-9 DN cells revealed a characteristic mitochondrial pattern, consistent with the location of this protein in live cells (Fig. 4A). In contrast, a fraction of treated cells showed a diffuse staining, indicating that Cyt c was released from mitochondria (16), as it was in apoptotic IMR90 E1A cells (Fig. 4A). Therefore, the release of Cyt c was independent of caspase-9 activation and could precede it. Consistent with this conclusion, the time course of Cyt c release in cells expressing caspase-9 DN was similar to the time course of apoptosis in IMR90-E1A cells (Fig. 4B). Thus, our data indicate that, in cells expressing caspase-9 DN, etoposide initiated the signals leading to caspase activation but that the activation did not take place. Of interest, the treated cells were not viable and died in a few days without any signs of apoptotic morphology. Whether this death is caused by the failure to survive with altered mitochondria or is a caspase-independent part of the apoptotic process remains to be investigated.
E1A Facilitates Cyt c Release from Mitochondria.
Because our results indicated that caspase-9 is involved in E1A-dependent apoptosis and because Cyt c is required for caspase-9 activation, we asked whether E1A sensitizes cells to drug-induced apoptosis by facilitating Cyt c release. We found that, although etoposide induced Cyt c release in cells expressing E1A irrespective of other manifestations of apoptosis, it did not have this effect in normal IMR90 cells, except in the small fraction that underwent apoptosis after etoposide treatment (<5% of the whole population; Fig. 4A and data not shown). Thus, it appears that E1A controls drug-induced apoptosis, at least in part, by regulating Cyt c release and thereby facilitating caspase-9 activation. This facilitation may be a biochemical mechanism underlying oncogene-induced sensitization of cells to apoptosis.
It has been suggested that the release of Cyt c can be induced by caspase-8 activity (17). In addition, effects on Fas-mediated apoptosis, which proceeds through caspase-8 activation, has been proposed as the mechanism of oncogene-dependent sensitization (6). Therefore, we tested whether caspase-8 is involved in drug-induced cytochrome release in IMR90-E1A cells. As caspases have to be processed to become active, we investigated whether processing of caspase-8 is required for Cyt c release. Caspase-8 was processed in IMR90 E1A cells but not in the cells expressing caspase-9 DN (Fig. 4C). However, Cyt c was released in both cell lines, indicating that this release did not require caspase-8 activity, consistent with evidence that cells unable to undergo Fas-mediated apoptosis still are sensitized to apoptosis by oncogenes (5). How E1A regulates the release of Cyt c remains to be elucidated. One way can be through induction of proteins that release Cyt c such as bax (16, 18). Considering that E1A-dependent bax expression is controlled by p53 (19, 20), one can speculate that the function of p53 in oncogene-dependent apoptosis is to induce proteins required for Cyt c release.
Multiple Effects of E1A on the Caspase-9 Activation Complex.
Although E1A can control caspase-9 activation by facilitating Cyt c release, our findings in vitro suggest that additional mechanisms are involved. In the cell-free system, transformed and untransformed extracts have similar concentrations of Cyt c, because breaking mitochondria during extract preparation bypasses any mechanism that regulates Cyt c release in cells (7). However, caspases become active only in transformed extracts, which led us to propose that an activity, OGA, accounts for the difference. Yet, OGA, which was identified here as APAF-1, is also present in both extracts, as is caspase-9.
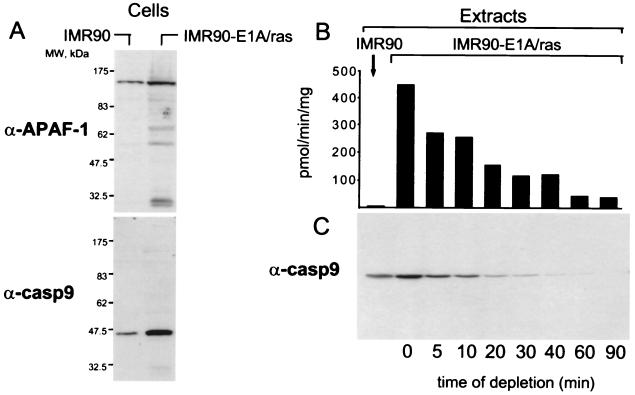
We noticed that, in cells that express E1A, the levels of APAF-1 and caspase-9 are increased (≈2- and 6-fold, respectively; Fig. 5A), indicating that oncogene expression regulates not only Cyt c release but also affects other components of the caspase-9 activating complex. As in cells, the caspase-9 and APAF-1 levels in transformed extracts were also higher than those in untransformed extracts (Fig. 5C and data not shown). To test whether this difference is responsible for oncogene-dependent caspase activation, we used immunodepletion to decrease the caspase-9 level in a transformed extract to the level found in untransformed extracts (Fig. 5 B and C; 10-min depletion). This depletion decreased the rate of caspase activation by about half, which could not account for the 75-fold difference in the rate between transformed and untransformed extracts. Thus, the increase in the caspase-9 level alone is not sufficient to explain oncogene-dependent caspase activation, at least in the cell free system. Whether the effect of oncogenes is limited to quantitative changes of APAF-1 and caspase-9 or yet-unidentified factors are involved remains to be investigated. In summary, our observations indicate that oncogene expression sensitizes cells to drug-induced apoptosis by facilitating caspase-9 activation. This regulation is likely to be achieved by several mechanisms, one of which is control of Cyt c release.
Figure 5.
E1A expression increases APAF-1 and caspase-9 levels. (A) Levels of APAF-1 and caspase-9 in transformed and untransformed cells were assessed by immunoblotting by using 25 μg of total cell lysate. (B and C) The difference in caspase-9 levels is not responsible for oncogene-dependent caspase activation in vitro. Caspase-9 was immunodepleted from a transformed extract to the level present in untransformed extracts. The partial depletion was achieved by varying the time of incubation with the antibody and monitoring the rate of caspase activation in the extract (B) and the level of caspase-9 (C).
Acknowledgments
We thank Jeanne Wiggins for cell culture, Jonathan Hoffman for extract preparation, Scott Lowe for IMR90-E1A/ras cells, James Chong for helpful discussion, and Lin Ying Zie for LinX cells and for help with gene transfer. We thank Kim Wanat for help with peptide sequencing. H.O.F. is supported by a Seligson Fellowship. J.R. is a Howard Hughes Medical Institute predoctoral fellow. Y.A.L. and G.H. are Pew Scholars. This work was supported by National Institutes of Health Grant CA 13106–25 to Y.A.L.
ABBREVIATION
- OGA
oncogene-generated activity
- FT
flow through
- caspase-9 DN
dominant negative mutant of caspase-9
- Cyt c
cytochrome c
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.White E. Proc Soc Exp Biol Med. 1993;204:30–39. doi: 10.3181/00379727-204-43631. [DOI] [PubMed] [Google Scholar]
- 2.Harrington E A, Fanidi A, Evan G I. Curr Opin Genet Dev. 1994;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J Y. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 5.Yeh W C, Pompa J L, McCurrach M E, Shu H B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, et al. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 6.Hueber A O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 7.Fearnhead H O, McCurrach M E, ONeill J, Zhang K, Lowe S W, Lazebnik Y A. Genes Dev. 1997;11:1266–1276. doi: 10.1101/gad.11.10.1266. [DOI] [PubMed] [Google Scholar]
- 8.Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 10.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X D. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 11.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan G H, O’Rourke K, Dixit V M. J Biol Chem. 1998;273:5841–5845. doi: 10.1074/jbc.273.10.5841. [DOI] [PubMed] [Google Scholar]
- 13.Samuelson A V, Lowe S W. Proc Natl Acad Sci USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Liu X S, Bhalla K, Kim C N, Ibrado A M, Cai J Y, Peng T I, Jones D P, Wang X D. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 15.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 16.Rosse T, Olivier R, Monney L, Rager M, Conus S, Fellay I, Jansen B, Borner C. Nature (London) 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 17.Kuwana T, Smith J J, Muzio M, Dixit V, Newmeyer D D, Kornbluth S. J Biol Chem. 1998;273:16589–16594. doi: 10.1074/jbc.273.26.16589. [DOI] [PubMed] [Google Scholar]
- 18.Jurgensmeier J M, Xie Z H, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCurrach M E, Connor T M F, Knudson C M, Korsmeyer S J, Lowe S W. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin C Y, Knudson C M, Korsmeyer S J, Van Dyke T. Nature (London) 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 21.Scaffidi C, Medema J P, Krammer P H, Peter M E. J Biol Chem. 1997;272:26953–26958. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]