Abstract
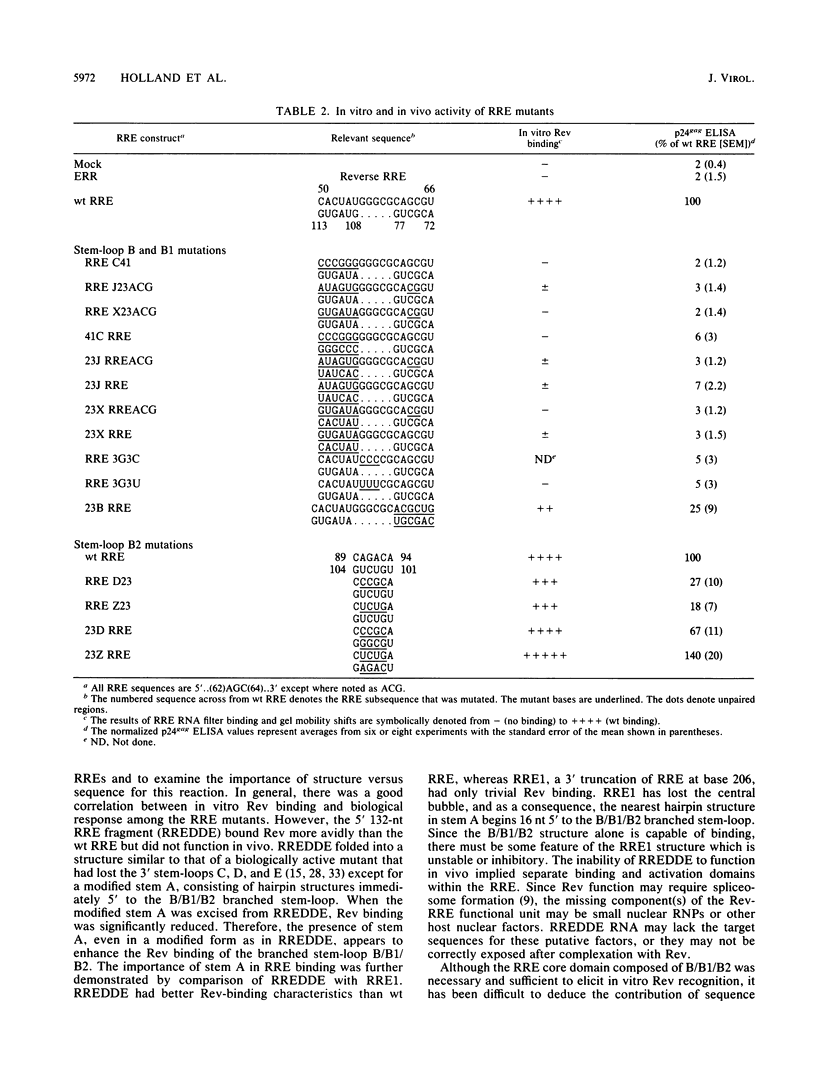
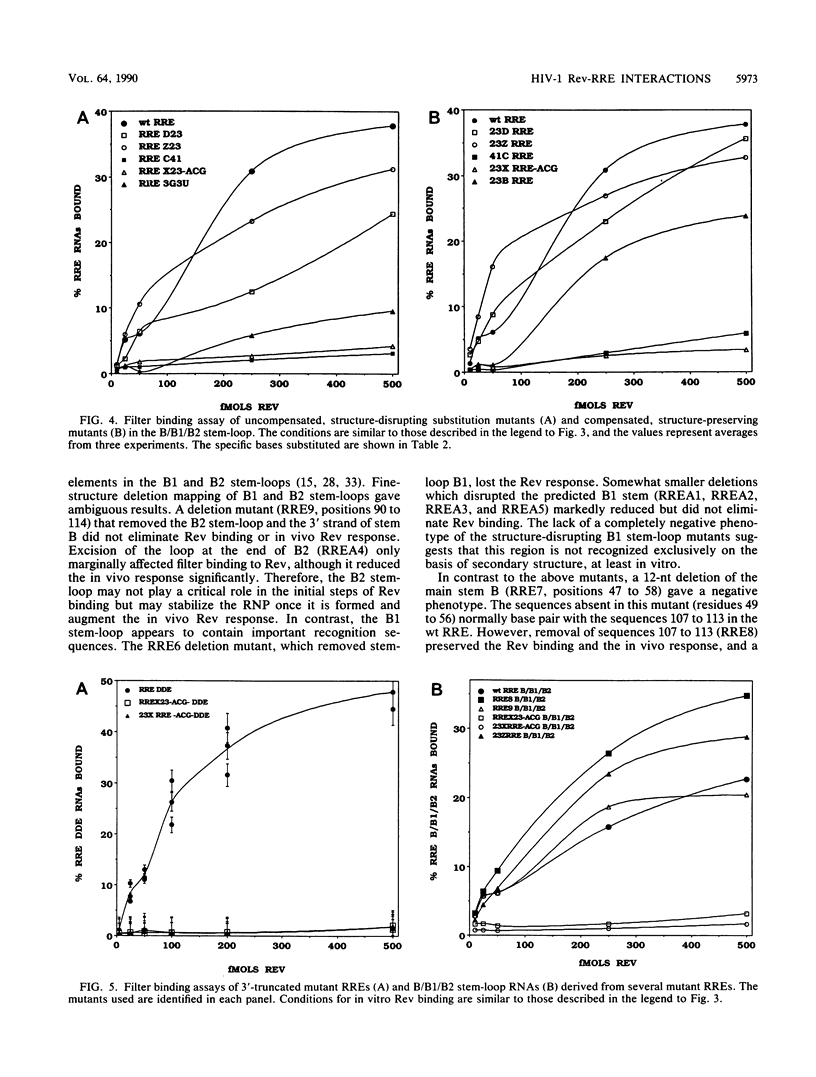
Human immunodeficiency virus type 1 Rev protein modulates the distribution of viral mRNAs from the nucleus to the cytoplasm by interaction with a highly structured viral RNA sequence, the Rev-responsive element (RRE). To identify the minimal functional elements of RRE, we evaluated mutant RREs for Rev binding in vitro and Rev response in vivo in the context of a Gag expression plasmid. The critical functional elements fold into a structure composed of a stem-loop A, formed by the ends of the RRE, joined to a branched stem-loop B/B1/B2, between bases 49 and 113. The 5' 132 nucleotides of RRE, RREDDE, which possessed a similar structure, bound Rev efficiently but were nonfunctional in vivo, implying separate binding and functional domains within the RRE. Excision of stem-loop A reduced Rev binding significantly and abolished the in vivo Rev response. The B2 branch could be removed without severe impairment of binding, but deletions in the B1 branch significantly reduced binding and function. However, deletion of 12 nucleotides, including the 5' strand of stem B, abolished both binding and function, while excision of the 3' strand of stem B only reduced them. Maintenance of the native RRE secondary structure alone was not sufficient for Rev recognition. Many mutations that altered the primary structure of the critical region while preserving the original RNA conformation were Rev responsive. However, mutations that changed a 5'..CACUAUGGG..3' sequence in the B stem, without affecting the overall structure abolished both in vitro Rev binding and the in vivo Rev response.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986 Aug;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N., Maitra R. K., Venkatesan S. Rev-induced modulation of Nef protein underlies temporal regulation of human immunodeficiency virus replication. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6111–6115. doi: 10.1073/pnas.86.16.6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N., Venkatesan S. Nef protein of HIV-1 is a transcriptional repressor of HIV-1 LTR. Science. 1988 Sep 16;241(4872):1481–1485. doi: 10.1126/science.3262235. [DOI] [PubMed] [Google Scholar]
- Allet B., Payton M., Mattaliano R. J., Gronenborn A. M., Clore G. M., Wingfield P. T. Purification and characterization of the DNA-binding protein Ner of bacteriophage Mu. Gene. 1988 May 30;65(2):259–268. doi: 10.1016/0378-1119(88)90462-3. [DOI] [PubMed] [Google Scholar]
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Christiansen J., Douthwaite S. R., Christensen A., Garrett R. A. Does unpaired adenosine-66 from helix II of Escherichia coli 5S RNA bind to protein L18? EMBO J. 1985 Apr;4(4):1019–1024. doi: 10.1002/j.1460-2075.1985.tb03733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane A. W., Chen C. H., Rosen C. A. Specific interaction of the human immunodeficiency virus Rev protein with a structured region in the env mRNA. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Hauber J., Campbell K., Sodroski J. G., Haseltine W. A., Rosen C. A. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J Virol. 1988 Jul;62(7):2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986 Sep 26;46(7):973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- Daly T. J., Cook K. S., Gray G. S., Maione T. E., Rusche J. R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989 Dec 14;342(6251):816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- Dayton E. T., Powell D. M., Dayton A. I. Functional analysis of CAR, the target sequence for the Rev protein of HIV-1. Science. 1989 Dec 22;246(4937):1625–1629. doi: 10.1126/science.2688093. [DOI] [PubMed] [Google Scholar]
- Emerman M., Vazeux R., Peden K. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989 Jun 30;57(7):1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- Feinberg M. B., Jarrett R. F., Aldovini A., Gallo R. C., Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986 Sep 12;46(6):807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- Felber B. K., Derse D., Athanassopoulos A., Campbell M., Pavlakis G. N. Cross-activation of the Rex proteins of HTLV-I and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol. 1989 Dec;1(3):318–328. [PubMed] [Google Scholar]
- Felber B. K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G. N. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M., Felber B. K., Cladaras C., Athanassopoulos A., Tse A., Pavlakis G. N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989 Mar;63(3):1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanly S. M., Rimsky L. T., Malim M. H., Kim J. H., Hauber J., Duc Dodon M., Le S. Y., Maizel J. V., Cullen B. R., Greene W. C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989 Oct;3(10):1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- Heaphy S., Dingwall C., Ernberg I., Gait M. J., Green S. M., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990 Feb 23;60(4):685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Böhnlein S., Fenrick R., Le S. Y., Maizel J. V., Cullen B. R. Functional comparison of the Rev trans-activators encoded by different primate immunodeficiency virus species. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8222–8226. doi: 10.1073/pnas.86.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M. H., Böhnlein S., Hauber J., Cullen B. R. Functional dissection of the HIV-1 Rev trans-activator--derivation of a trans-dominant repressor of Rev function. Cell. 1989 Jul 14;58(1):205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Tiley L. S., McCarn D. F., Rusche J. R., Hauber J., Cullen B. R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990 Feb 23;60(4):675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Olsen H. S., Nelbock P., Cochrane A. W., Rosen C. A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990 Feb 16;247(4944):845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Newby R. F., Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- Rimsky L., Hauber J., Dukovich M., Malim M. H., Langlois A., Cullen B. R., Greene W. C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature. 1988 Oct 20;335(6192):738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Sodroski J., Goh W. C., Rosen C., Dayton A., Terwilliger E., Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986 May 22;321(6068):412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- Sodroski J., Rosen C., Wong-Staal F., Salahuddin S. Z., Popovic M., Arya S., Gallo R. C., Haseltine W. A. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985 Jan 11;227(4683):171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Uhlenbeck O. C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987 Dec 15;26(25):8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Green M. R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989 Dec 7;342(6250):714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]




