Abstract
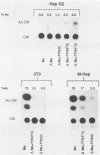
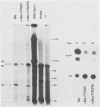
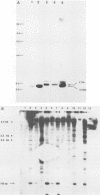
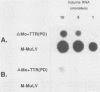
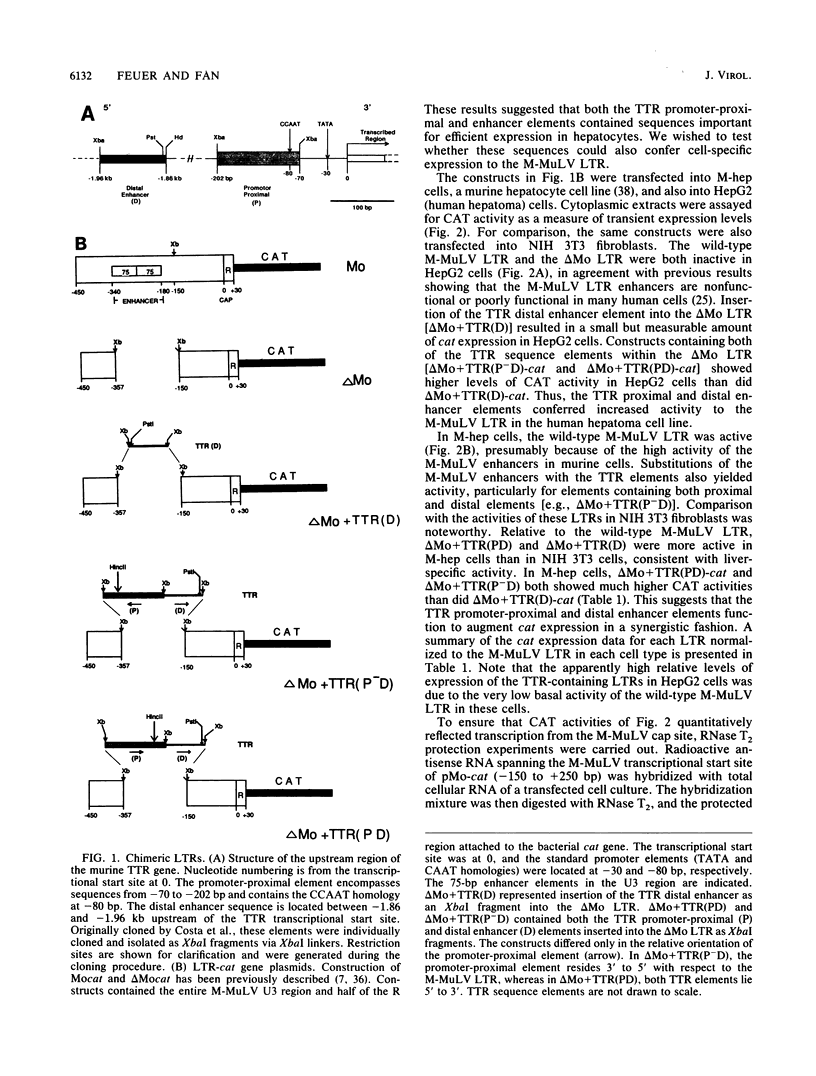
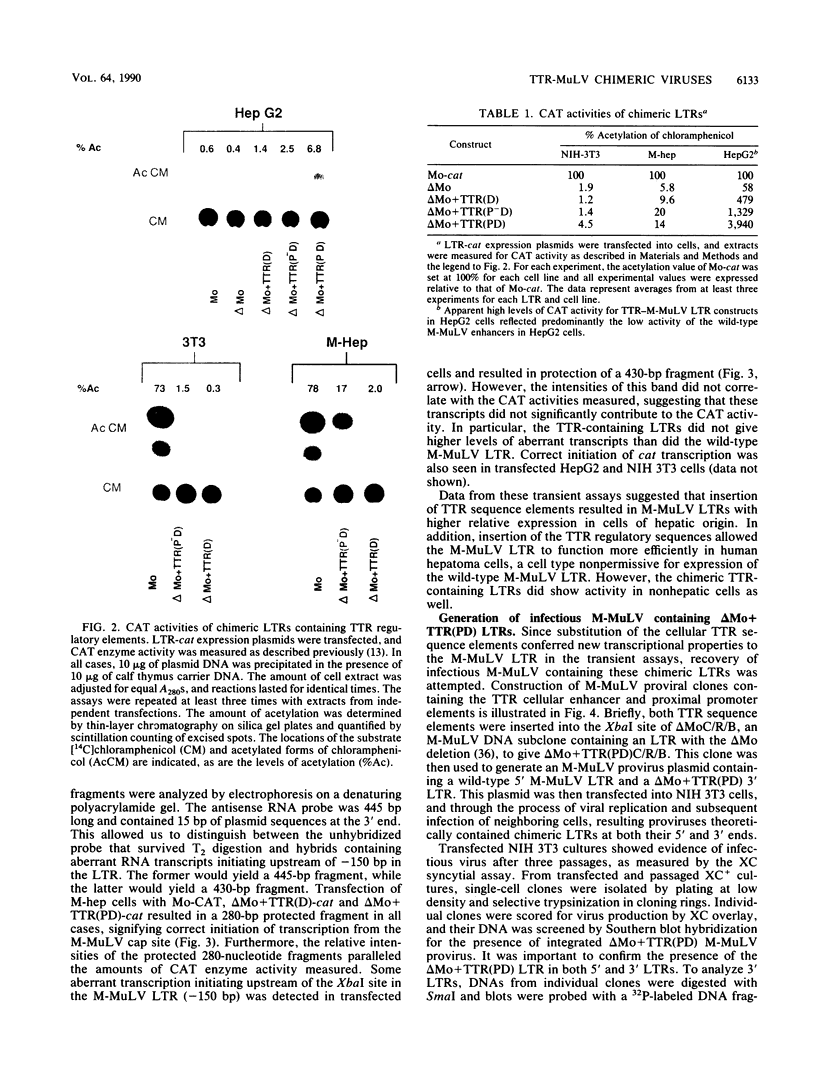
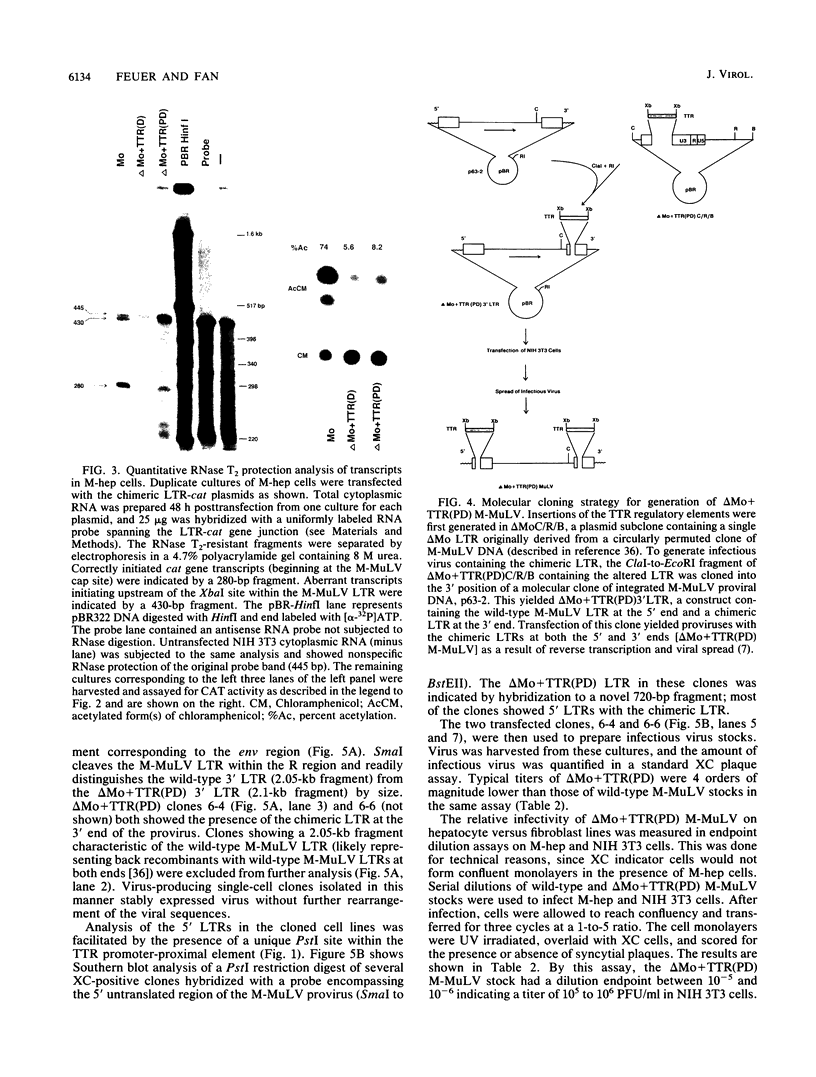
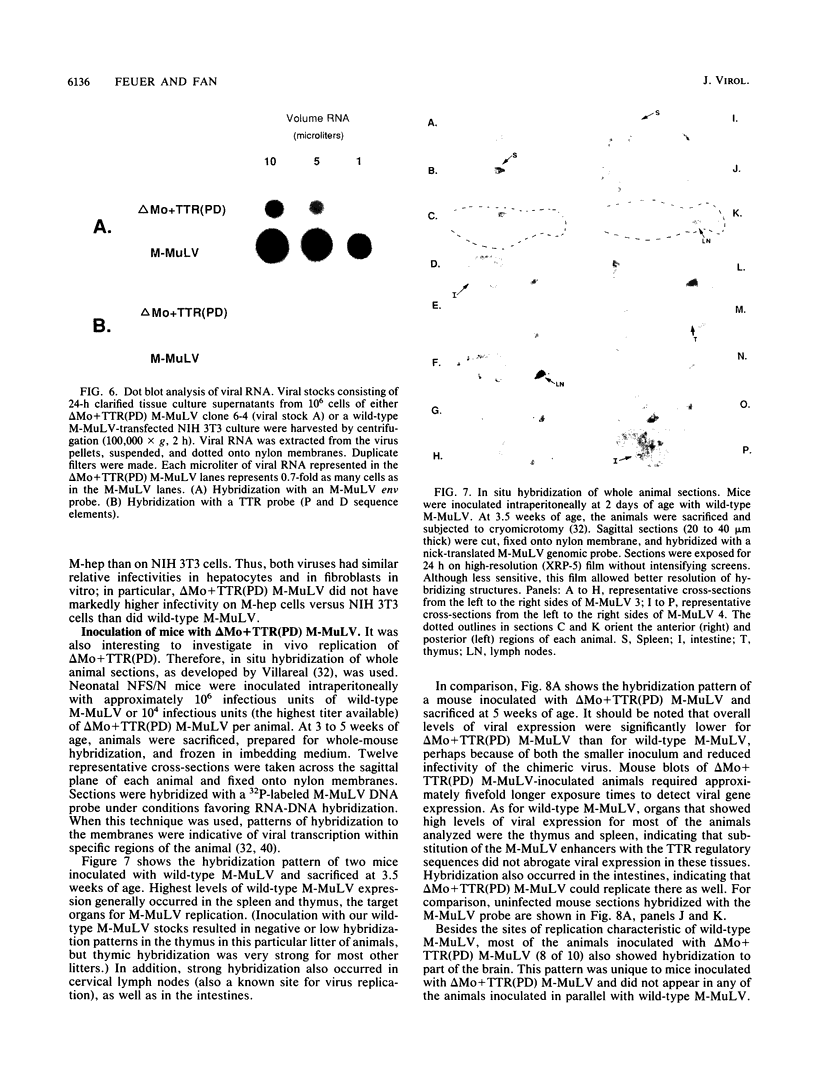
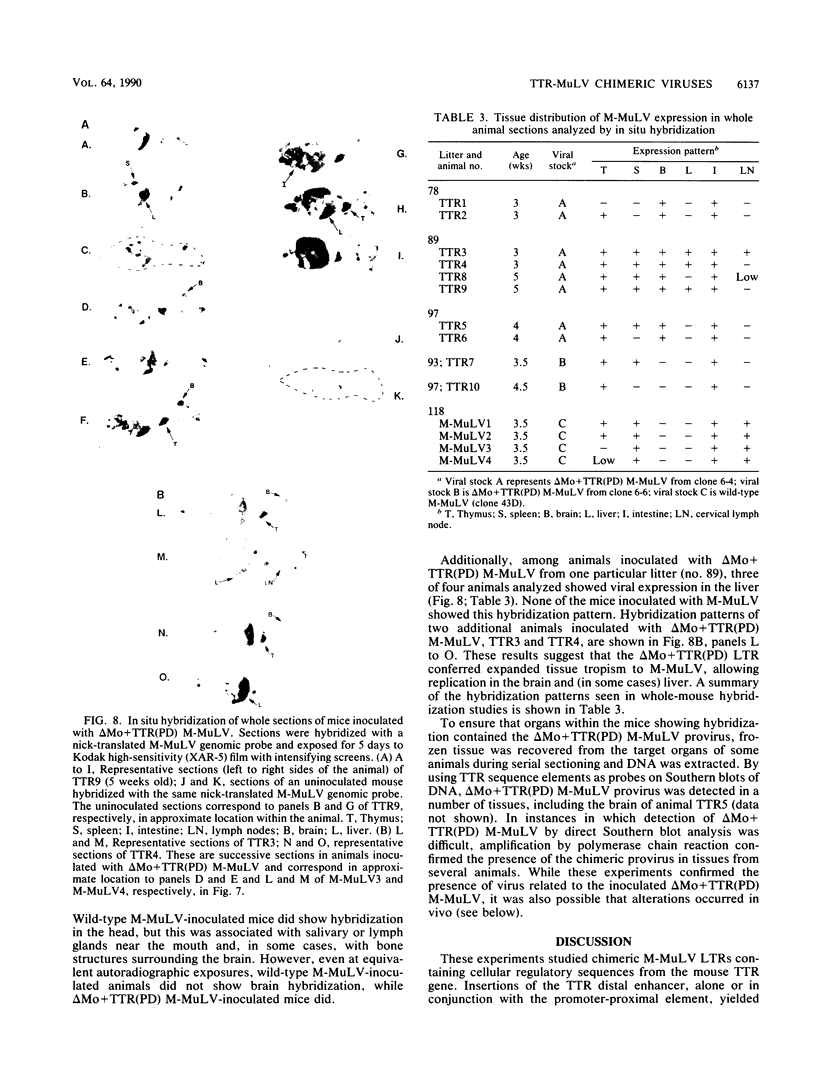
The effects of inserting cellular regulatory sequences from the murine transthyretin (TTR) gene into the Moloney murine leukemia virus (M-MuLV) long terminal repeat (LTR) were investigated. Transthyretin is expressed predominantly in the liver and choroid plexus in adult mice, and TTR upstream regulatory elements were previously shown to potentiate transcription in liver-derived cells. The effects of inserting the TTR distal enhancer and/or promoter-proximal sequences into an M-MuLV LTR lacking its enhancers were measured in three ways. (i) Chimeric LTRs were fused to the bacterial chloramphenicol acetyltransferase gene (cat) and tested for transient gene expression by transfection into liver-derived cells or NIH 3T3 fibroblasts. (ii) Infectious M-MuLV containing an altered LTR [delta Mo + TTR(PD) MuLV) was generated, and infectivity in culture on hepatocyte lines and NIH 3T3 cells was tested. (iii) Infection of delta Mo + TTR(PD) MuLV in vivo was tested by inoculating NFS/N mice and performing in situ hybridization of whole animal sections. Chimeric LTR-cat constructs showed higher levels of cat gene expression in liver-derived cell lines than in NIH 3T3 cells, indicating increased LTR activity in these cells. However, in vitro infection did not show significantly higher infectivity in hepatocytes for delta Mo + TTR(PD) M-MuLV than did wild-type M-MuLV. In vivo, delta Mo + TTR(PD) MuLV showed expression in the same tissues as with wild-type M-MuLV-inoculated mice, i.e., lymphoid organs and the intestines and, additionally, two novel sites not seen in wild-type M-MuLV-inoculated animals. Of 10 mice, 8 showed viral expression in the brain and 3 showed expression in the liver. Thus, insertion of TTR elements into the M-MuLV LTR altered LTR activity both in vitro and in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brightman B. K., Pattengale P. K., Fan H. Generation and characterization of a recombinant Moloney murine leukemia virus containing the v-myc oncogene of avian MC29 virus: in vitro transformation and in vivo pathogenesis. J Virol. 1986 Oct;60(1):68–81. doi: 10.1128/jvi.60.1.68-81.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos R., Villarreal L. P. An SV40 deletion mutant accumulates late transcripts in a paranuclear extract. Virology. 1982 May;119(1):1–11. doi: 10.1016/0042-6822(82)90059-9. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Darnell J. E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989 Apr;9(4):1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Darnell J. E., Jr Transcriptional control of the mouse prealbumin (transthyretin) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol. 1986 Dec;6(12):4697–4708. doi: 10.1128/mcb.6.12.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Grayson D. R., Darnell J. E., Jr The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol Cell Biol. 1988 Jan;8(1):81–90. doi: 10.1128/mcb.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B., Linney E., Fan H. Suppression of leukaemia virus pathogenicity by polyoma virus enhancers. Nature. 1985 Apr 11;314(6011):550–553. doi: 10.1038/314550a0. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson P. W., Howlett G. J., Schreiber G. Rat transthyretin (prealbumin). Molecular cloning, nucleotide sequence, and gene expression in liver and brain. J Biol Chem. 1985 Jul 5;260(13):8214–8219. [PubMed] [Google Scholar]
- Fan H., Chute H., Chao E., Pattengale P. K. Leukemogenicity of Moloney murine leukemia viruses carrying polyoma enhancer sequences in the long terminal repeat is dependent on the nature of the inserted polyoma sequences. Virology. 1988 Sep;166(1):58–65. doi: 10.1016/0042-6822(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Fan H., Mittal S., Chute H., Chao E., Pattengale P. K. Rearrangements and insertions in the Moloney murine leukemia virus long terminal repeat alter biological properties in vivo and in vitro. J Virol. 1986 Oct;60(1):204–214. doi: 10.1128/jvi.60.1.204-214.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Feuer G., Taketo M., Hanecak R. C., Fan H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J Virol. 1989 May;63(5):2317–2324. doi: 10.1128/jvi.63.5.2317-2324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D. R., Costa R. H., Xanthopoulos K. G., Darnell J. E., Jr A cell-specific enhancer of the mouse alpha 1-antitrypsin gene has multiple functional regions and corresponding protein-binding sites. Mol Cell Biol. 1988 Mar;8(3):1055–1066. doi: 10.1128/mcb.8.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanecak R., Mittal S., Davis B. R., Fan H. Generation of infectious Moloney murine leukemia viruses with deletions in the U3 portion of the long terminal repeat. Mol Cell Biol. 1986 Dec;6(12):4634–4640. doi: 10.1128/mcb.6.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanecak R., Pattengale P. K., Fan H. Addition of substitution of simian virus 40 enhancer sequences into the Moloney murine leukemia virus (M-MuLV) long terminal repeat yields infectious M-MuLV with altered biological properties. J Virol. 1988 Jul;62(7):2427–2436. doi: 10.1128/jvi.62.7.2427-2436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Piasecki A., Paul D. Proliferation of fetal rat hepatocytes in response to growth factors and hormones in primary culture. J Cell Physiol. 1989 Jun;139(3):654–662. doi: 10.1002/jcp.1041390328. [DOI] [PubMed] [Google Scholar]
- Hollon T., Yoshimura F. K. Mapping of functional regions of murine retrovirus long terminal repeat enhancers: enhancer domains interact and are not independent in their contributions to enhancer activity. J Virol. 1989 Aug;63(8):3353–3361. doi: 10.1128/jvi.63.8.3353-3361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Moloney leukemia virus gene expression and gene amplification in preleukemic and leukemic BALB/Mo mice. Virology. 1979 Feb;93(1):80–90. doi: 10.1016/0042-6822(79)90277-0. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Retroviruses and embryogenesis: microinjection of Moloney leukemia virus into midgestation mouse embryos. Cell. 1980 Jan;19(1):181–188. doi: 10.1016/0092-8674(80)90399-2. [DOI] [PubMed] [Google Scholar]
- Kitado H., Chen I. S., Shah N. P., Cann A. J., Shimotohno K., Fan H. U3 sequences from HTLV-I and -II LTRs confer pX protein response to a murine leukemia virus LTR. Science. 1987 Feb 20;235(4791):901–904. doi: 10.1126/science.3027896. [DOI] [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980 Jul 25;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Koyanagi Y., Miles S., Mitsuyasu R. T., Merrill J. E., Vinters H. V., Chen I. S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987 May 15;236(4803):819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E., Davis B., Overhauser J., Chao E., Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. 1984 Mar 29-Apr 4Nature. 308(5958):470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- Lipkin W. I., Villarreal L. P., Oldstone M. B. Whole animal section in situ hybridization and protein blotting: new tools in molecular analysis of animal models for human disease. Curr Top Microbiol Immunol. 1989;143:33–54. doi: 10.1007/978-3-642-74425-9_4. [DOI] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M. A., Sharp P. A., Hopkins N. Nuclear factors that bind to the enhancer region of nondefective Friend murine leukemia virus. J Virol. 1989 Oct;63(10):4210–4223. doi: 10.1128/jvi.63.10.4210-4223.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhauser J., Fan H. Generation of glucocorticoid-responsive Moloney murine leukemia virus by insertion of regulatory sequences from murine mammary tumor virus into the long terminal repeat. J Virol. 1985 Apr;54(1):133–144. doi: 10.1128/jvi.54.1.133-144.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskind M. P., Weinberg R. A., Baltimore D. Dependence of Moloney murine leukemia virus production on cell growth. Virology. 1975 Sep;67(1):242–248. doi: 10.1016/0042-6822(75)90421-3. [DOI] [PubMed] [Google Scholar]
- Paul D., Höhne M., Pinkert C., Piasecki A., Ummelmann E., Brinster R. L. Immortalized differentiated hepatocyte lines derived from transgenic mice harboring SV40 T-antigen genes. Exp Cell Res. 1988 Apr;175(2):354–362. doi: 10.1016/0014-4827(88)90199-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rochford R., Campbell B. A., Villarreal L. P. A pancreas specificity results from the combination of polyomavirus and Moloney murine leukemia virus enhancer. Proc Natl Acad Sci U S A. 1987 Jan;84(2):449–453. doi: 10.1073/pnas.84.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Swift G. H., Hammer R. E., MacDonald R. J., Brinster R. L. Tissue-specific expression of the rat pancreatic elastase I gene in transgenic mice. Cell. 1984 Oct;38(3):639–646. doi: 10.1016/0092-8674(84)90258-7. [DOI] [PubMed] [Google Scholar]
- TODARO G. J., GREEN H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963 May;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. D., Edlund T., Boulet A. M., Rutter W. J. Cell-specific expression controlled by the 5'-flanking region of insulin and chymotrypsin genes. Nature. 1983 Dec 8;306(5943):557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Wolff J. A., Yee J. K., Skelly H. F., Moores J. C., Respess J. G., Friedmann T., Leffert H. Expression of retrovirally transduced genes in primary cultures of adult rat hepatocytes. Proc Natl Acad Sci U S A. 1987 May;84(10):3344–3348. doi: 10.1073/pnas.84.10.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. C., Morley B. J., Campbell R. D. Cell-specific expression of the human complement protein factor B gene: evidence for the role of two distinct 5'-flanking elements. Cell. 1987 Jan 30;48(2):331–342. doi: 10.1016/0092-8674(87)90436-3. [DOI] [PubMed] [Google Scholar]