Abstract
Mutant, but not wild-type p53 binds with high affinity to a variety of MAR-DNA elements (MARs), suggesting that MAR-binding of mutant p53 relates to the dominant-oncogenic activities proposed for mutant p53. MARs recognized by mutant p53 share AT richness and contain variations of an AATATATTT “DNA-unwinding motif,” which enhances the structural dynamics of chromatin and promotes regional DNA base-unpairing. Mutant p53 specifically interacted with MAR-derived oligonucleotides carrying such unwinding motifs, catalyzing DNA strand separation when this motif was located within a structurally labile sequence environment. Addition of GC-clamps to the respective MAR-oligonucleotides or introducing mutations into the unwinding motif strongly reduced DNA strand separation, but supported the formation of tight complexes between mutant p53 and such oligonucleotides. We conclude that the specific interaction of mutant p53 with regions of MAR-DNA with a high potential for base-unpairing provides the basis for the high-affinity binding of mutant p53 to MAR-DNA.
Mutations in the p53 gene constitute the most frequent alterations in a single gene in human cancer (1). Wild-type p53 (wt p53) is a tumor suppressor whose main function is to preserve the integrity of the genome. p53 not only mediates its DNA damage responses by modulating cellular transcription, it also exhibits various other biochemical activities that are directly related to its function as a major control element in preserving the integrity of the cells’ genetic information (2). Ninety percent of all mutations in the p53 gene are single missense point mutations, the majority being located in the p53 core domain, which mediates most of the biochemical activities of wt p53. Consequently, such mutations inactivate the tumor suppressor functions of p53. However, the expression of mutant (mut) p53 is not equivalent to a p53 “null” situation (3), as genetic and biological evidence indicates that mut p53 exerts oncogenic functions of its own (4–9).
So far, the biological activities related to this postulated “gain of function” of mut p53 are far from being understood at the molecular level, but a functional transactivator domain is required, suggesting that mut p53 is able to modulate gene expression (10). Mut p53-specific transcriptional activation has been reported for various genes, such as mdr-1 (10–12), PCNA (13), VEGF (14), and the HIV long terminal repeat (15), but the underlying molecular mechanism is not clear: although some mut p53 proteins still react with subsets of wt p53-responsive elements (16–18), there is no defined mut p53-specific responsive element (18, 19). Furthermore, transcriptional up-regulation of the so-far-identified mut p53-specific target genes seems to be extremely dependent on cell type and assay conditions (20). This situation is quite different from wt p53-dependent transactivation and suggests a completely different mechanism.
Murine and human mut p53, but not wt p53, specifically bind with high affinity to a variety of nuclear matrix attachment region DNA elements (MARs) (21–23). Due to the importance of MARs in nuclear processes such as gene expression and DNA replication, we hypothesized that this activity of mut p53 possibly could form the molecular basis for the documented oncogenic potential of mut p53 (4–7, 24, 25). In this study we aimed at identifying unique sequence arrangements or distinct structural determinants on MARs that are specifically recognized by mut p53. MARs bound by mut p53 are quite different in size and sequence composition (23), but as a common feature, these MARs are AT-rich and contain variations of an AATATATTT “unwinding” motif, implicated in MAR function (26, 27). Such motifs promote structural alterations within the chromatin, including regional base-unpairing (26, 27). We show that mut p53 specifically interacts with MAR-derived oligonucleotides containing variations of this motif. Depending on sequence context, mut p53 catalyzed DNA strand separation or exerted tight binding. We propose that the specific interaction of mut p53 with MARs in regions displaying a high potential for base-unpairing forms the molecular basis for its MAR-binding activity and we suggest a model that shows how mut p53 via this activity could modulate gene expression and cellular DNA replication.
MATERIALS AND METHODS
Overexpression and Purification of Recombinant Baculoviral p53 Protein.
Method A. High Five insect cells at 80% confluency were infected with recombinant baculovirus expressing wt (murine, human) or mut p53 (murine MethA; human: 273 Arg → Pro). Forty-eight hours after infection the cells were harvested and washed four times with PBS at 4°C. p53 proteins subsequently were extracted after a three-step protocol by using extraction buffer A (10 mM Hepes, pH 7.4/1.5 mM MgCl2/5 mM KCl), buffer B (10 mM Hepes, pH 9.0/1.5 mM MgCl2/5 mM KCl), and buffer C and D (buffer B containing 0.2 or 0.5 M KCl, respectively), as described previously (28). The proteins were analyzed by SDS/PAGE and Western blotting with sheep anti-p53 antibody (Boehringer Mannheim)/peroxidase-conjugated secondary serum and anti-sheep IgG from goat (Sigma) using SuperSignal Ultra Chemiluminescent system (Pierce). p53 protein in fraction C was approximately 80% pure.
Method B.
High Five insect cells at 80% confluency were infected with recombinant baculovirus expressing wt (human) 6xHis-tagged p53 or mut 6xHis-tagged p53 proteins (human: Pro 273). Forty-eight hours after infection the cells were harvested and washed four times with PBS at 4°C, and 6xHis-tagged p53 proteins were purified under nondenaturing conditions using the Talon-Metal Affinity Resin System (CLONTECH) according to the manufacturer’s instructions. The purified proteins were >95% pure as judged by SDS/PAGE.
Characterization of p53 Protein by Immunoprecipitation Using Conformation-Specific Antibodies.
Aliquots of the purified p53 proteins were diluted with lysis buffer (50 mM Tris⋅HCl, pH 8.0/120 mM NaCl/1% Nonidet P-40/10% (vol/vol) glycerol/5 mM DTT) and mixed with 30 μl of settled protein A-Sepharose (PAS, Pharmacia). Immunoprecipitation and analysis of the immunoprecipitated p53 by SDS/PAGE subsequently were carried out as described previously (29). mAbs PAb421, PAb1620, and PAb240 were used as described in ref. 22.
Electrophoretic Mobility Shift Assay (EMSA).
Preparation of oligonucleotide probes. Synthetic oligonucleotides harboring the p53-responsive RGC-element (30) or MAR-derived MAR I (5′-AGTGTCTTTAATTTCTAATATATTTAGAAAACTGCG-3′); MAR II (5′-TTTTAACAATAATAAATTAAGTTTAAAATATTTGCG-3′); MAR III (5′-TCTAAAGAAGTTTGGATACTTCAAAAGTAAGCG-3′); MAR5 (5′GGGCCCGGG-MARI-GCGCCG-3′); MAR6 (5′-AGTGTCTTTAATTTCTACTATGCTTAGAAAACTGCG-3′); and MAR7 (5′-AGTGTCTTTAATTTCTGCTCTCTTTAGAAAACTGCG-3′) were radioactively labeled at the 5′ end using T4 polynucleotide kinase and [γ-32P]ATP and annealed with unlabeled complementary oligonucleotides in STE. Both double-stranded (ds) and single-stranded (ss) oligonucleotides were electrophoresed through neutral 12% polyacrylamide gels using 1× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). The labeled oligonucleotides were isolated as described previously (28) and counted for radioactivity, and their concentrations were determined by using the DNA DipStick kit (Invitrogen). The quality of the isolated ds and ss probes was controlled by subjecting aliquots of all isolates again to gel electrophoreses using neutral 12% polyacrylamide gels equilibrated with 1× TBE buffer. Upon reelectrophoresis the isolated ds probes migrated as ds oligonucleotides.
Preparation of the MAR-DNA probe.
The 997-bp XbaI-IgE-MAR-DNA fragment cloned into the multiple cloning site of pUC19 vector was used for gel shift experiments. After XbaI-restriction enzyme digest, the DNA fragments were dephosphorylated using alkaline phosphatase (Boehringer Mannheim) and gel-electrophoretically separated using TAE-agarose gels. The desired DNA bands were recovered from the gel using the Geneclean II kit. After photometric determination of the DNA concentration, aliquots of the fragments were 32P end-labeled using T4 polynucleotide kinase and [γ-32P]ATP by standard procedures.
Bandshift reactions.
In the binding experiments using oligonucleotide probes, the binding reaction mixtures (20 μl in total) were preincubated with or without p53 for 20 min with 2 μg of poly dI:dC (Pharmacia) in reaction buffer (10 mM Hepes, pH 7.8/50 mM KCl/1 mM EDTA/5 mM MgCl2/10% glycerol). The EMSA mixtures for the IgE-MAR-DNA-binding experiments included unlabeled BglI-pUC19 fragment as competitor DNA. PAb248 (31) was added to the preincubation mixtures as indicated. After preincubation for 20 min, radioactively labeled probe (≈1–4 × 104 cpm) was added to the mixtures and binding was allowed for 30 min at room temperature. The reaction products were analyzed by electrophoresis in 4% native polyacrylamide gels (28) and subjected to autoradiography, and the intensities of the DNA bands were quantified by using a Molecular Dynamics PhosphorImager.
RESULTS
Mutant, but Not Wild-Type p53 Interacts with MAR-Derived Oligonucleotides Containing Variations of the AATATATTT “Unwinding Motif.”
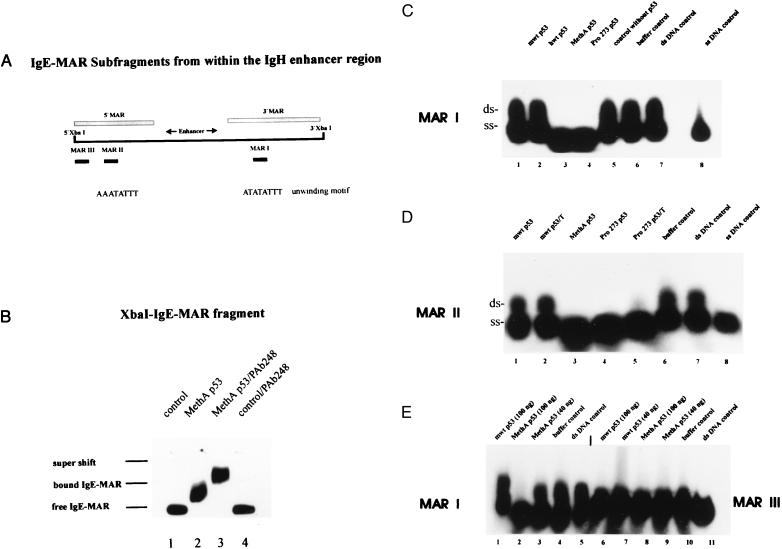
Screening the MAR-DNA fragments bound by mut p53 (23) for unique structural determinants and characteristic features revealed that they were AT-rich and contained at least one single motif of the AATATATTT-type (“unwinding motif”) that was implicated in MAR function (26, 27, 32). We therefore asked whether this motif might mediate the interaction of mut p53 with MARs and selected appropriate oligonucleotides from the 997-bp XbaI-IgE-MAR-DNA fragment located within the Ig heavy chain (IgH) gene enhancer region (33), a good binding substrate for mut p53 in a variety of different MAR-binding assays. We chose two AT-rich sequences, 36 bp in length, each containing a different variant of the “unwinding motif” (Fig. 1A, see Materials and Methods): MAR I in the 3′ flanking region and MAR II in the 5′ flanking region of the IgH enhancer and, as a control sequence, a fragment of 33-bp length (MAR III), with a slightly reduced AT-content and without an unwinding motif also in the 5′ flanking region (Fig. 1A; see Materials and Methods).
Figure 1.
Interactions of wild-type and mutant p53 with IgE-MAR-DNA and selected IgE-MAR-derived oligonucleotides. (A) Schematic representation of the XbaI-IgE-MAR-DNA fragment from the IgH enhancer region indicating the locations of the selected MAR-oligonucleotides (MAR I, II, and III). MAR I contains an unwinding motif, and MAR II contains a variant thereof. (B) Binding of murine MethA p53 to 32P-end-labeled ds IgE-MAR-DNA fragment was analyzed by EMSA in native 4% polyacrylamide gels. p53 specificity of the binding reaction was confirmed by supershifting the DNA–protein complex with the p53-specific antibody PAb248 (compare lanes 2 and 3). Lanes 1 and 4, control reactions without p53. (C) Equal amounts (100 ng) of purified murine (lane 1) or human wt p53 (lane 2), MethA mut p53 (lane 3), or human Pro273 mut p53 (lane 4) were reacted with 32P-end-labeled MAR I probe in EMSA (Materials and Methods). Control reactions were carried out with the respective fraction from lysates of insect cells not expressing p53 (lane 5) or extraction buffer C of purification method A (lane 6). Lane 7, aliquot of the isolated ds MAR I probe, consisting of ss- and dsDNA; lane 8, aliquot of ss MAR I probe. (D) Reactions of murine wt p53 (100 ng) purified according to two different purification protocols (lane 1, method A; lane 2, Talon chromatography) and of MethA p53 (100 ng) (lane 3, method A) and human Pro-273 p53 (100 ng) (lane 4, method A; lane 5, Talon chromatography) with 32P-end-labeled MAR II probes in EMSA (Materials and Methods). Lanes 6 and 7, controls without p53; lane 8, aliquot of the ss MAR II probe. (E) Reactions of murine wt p53 (lanes 1 and 6 with 100 ng) and MethA p53 (lanes 2 and 8, with 100 ng; lanes 3 and 9, with 40 ng) with 32P-end-labeled MAR I and MAR III probes in EMSA. Control reactions without p53 (MAR I, lanes 4 and 5; MAR III, lanes 10 and 11).
MAR binding by mut p53 so far had been demonstrated in a variety of different assays specifically adapted to measure the interaction of mut p53 with large DNA fragments, as represented by MARs (21–23). However, interactions of mut p53 with MAR-derived oligonucleotides could be analyzed best by EMSAs. To unequivocally relate the possible interaction of mut p53 with short MAR-derived oligonucleotides to bona fide MAR binding by mut p53, we first established conditions that allowed us to analyze MAR binding of mut p53 also by EMSA. Recombinant murine wt and MethA mut p53 proteins were purified from insect cells infected with the appropriate baculovirus and incubated with the 997-bp IgE-MAR-DNA fragment. EMSA, performed under the same conditions employed for the analysis of sequence-specific DNA binding of wt p53 (28), revealed that MethA p53 specifically bound the IgE-MAR, reflected by a mobility shift of the IgE-MAR band upon addition of approximately 20 ng of MethA p53 (Fig. 1B, lane 2). Addition of the p53-specific mAb PAb248 produced a supershift (Fig. 1B, lane 3), verifying the specific binding of mut p53 to the IgE-MAR. Under identical conditions, wt p53 did not bind the IgE-MAR (not shown), reflecting the much lower affinity of wt p53 to MAR-DNA, as demonstrated previously (22, 23).
Using the same EMSA conditions, we next analyzed the interactions of wt and mut p53 with the MAR-derived oligonucleotides described above (Fig. 1A). In addition to the murine p53 proteins, recombinant human wt and mut p53 proteins Arg-273 → Pro (Pro-273) were purified from insect cells. Purity and quality of all proteins were checked by SDS/PAGE and by immunoprecipitation using mAb PAb421, wt p53 conformation-specific antibodies PAb1620 (human p53) or PAb246 (murine p53), and mut p53 conformation-specific antibody PAb240. The biologically active state of wt p53 was confirmed by EMSA by using oligonucleotides harboring the p53-responsive RGC-element (28, 30). The MAR-derived oligonucleotides were synthesized in vitro, the bottom oligonucleotides were 32P-end-labeled, and appropriate pairs of complementary top and bottom oligonucleotides were annealed. ds and ss oligonucleotides were isolated and then subjected to EMSA by using wt and mut p53 (murine MethA, human Pro-273).
The results of EMSA experiments using the MARI and MAR II oligonucleotides, containing a variant of the unwinding motif, and the MAR III oligonucleotide, not containing such a motif, are shown in Fig. 1 C (MAR I), D (MAR II), and E (MAR III). Comparison of the ds DNA control lanes (Fig. 1 C, lane 7 for MAR I; D, lane 7 for MAR II; and E, lane 11 for MAR III) already demonstrated significant differences in the physical properties between the MAR I and MAR II, and the MAR III oligonucleotides. Although all oligonucleotides were isolated as ds DNA by the same procedure, isolated MAR I and MAR II under EMSA conditions ran as mixtures of dsDNA and ssDNA (MAR I, Fig. 1C, lane 7; MAR II, Fig. 1D, lane 7), whereas MAR III under the same conditions remained double-stranded (Fig. 1E, lane 11). As judged by densitrometric evaluation, ≈70% of MAR I and MAR II were recovered as ssDNA, reflecting the intrinsic high potential of these oligonucleotides for base-unpairing.
Fig. 1C shows that incubation of the MAR I probe with either the murine (lane 1) or the human (lane 2) wt p53 protein did not change the relation of ds to ss MAR I oligonucleotide (compare with lane 7), whereas incubation with the murine MethA (lane 3) or the human Pro-273 p53 protein (lane 4) resulted in complete conversion of its ds portion to ssDNA. Neither addition of an equal volume of the corresponding protein fraction from uninfected insect cells (Fig. 1C, lane 5) nor addition of an equal volume of the corresponding extraction buffer (Fig. 1C, lane 6) had any effect on the ratio of ds to ss MAR I oligonucleotide (compare with Fig. 1C, lane 7). Furthermore, incubation of the MAR I probe in binding buffer alone (1 h) did not increase the fraction of ss MAR I generated spontaneously under EMSA conditions (not shown). Similarly, incubation of the MAR II probe with mut p53 proteins also induced conversion of its ds portion to ssDNA (Fig. 1D, lanes 3–5). Again, this activity was not seen upon incubation with wt p53 proteins (lanes 1 and 2). It is important to note that this particular interaction of mut p53 with DNA was not dependent on the purification protocol for mut p53 (Fig. 1D, lanes 3–5) and also was found with highly purified human Pro-273 p53 (Fig. 1D, lane 5), strongly suggesting that this activity is intrinsic to mut p53. This conclusion was supported further by our finding that this activity of mut p53, like MAR binding by mut p53 (22), could be blocked by the addition of mAbs PAb240, reacting with the mut p53 core domain, or PAb421, reacting with the p53 C terminus (not shown). Conversely, independent of the purification schemes used, equal amounts of purified wt p53 failed to increase the fraction of ssDNA when incubated with the respective probes (Fig. 1D, lanes 1 and 2). In contrast, addition of mut p53 to the MAR III probe did not result in any specific interaction with this oligonucleotide (Fig. 1E, lanes 8 and 9) in EMSA. As an intrinsic control, the MAR I probe, incubated with mut p53 and run on the same gel in parallel, was completely converted to ssDNA (Fig. 1E, lane 2).
Transient Interaction of Mutant p53 with MAR Oligonucleotides During DNA Strand Separation.
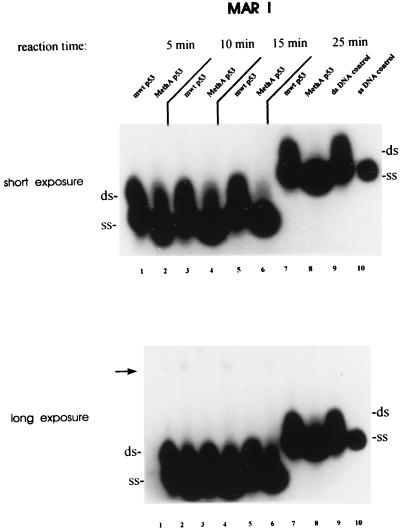
To characterize DNA strand separation of MAR-derived oligonucleotides by mut p53 in more detail, we analyzed this process in a time course experiment. Fig. 2 shows that upon incubation of the MAR I probe with MethA p53 the amount of ss MAR I increased with time, concomitant with a decrease of its dsDNA fraction. Longer exposure of Fig. 2, in addition, revealed a faint higher-molecular-weight band after short incubation times (lanes 2, 4, and 6), which decreased over time and was hardly detectable after longer incubation times (lane 8), when most of ds MAR I fraction had been converted to ssDNA. We interpret this band as a transient interaction of mut p53 with the MAR I probe (see below).
Figure 2.
Mutant p53-mediated DNA strand separation is dependent on the reaction time. MAR I probe was incubated with equal amounts of wt and MethA p53 (100 ng) for 5, 10, 15, or 25 min. While the first three reaction mixtures were loaded at the same time on a native 4% polyacrylamide gel, the 25-min reaction was added 10 min later. After shorter incubation times (5–15 min) in addition to strand separation, faint shifted MAR I bands were observed (lanes 2, 4, and 6; see long exposure, indicated by an arrow), which were not detectable after longer incubation times (>25 min; see long exposure, lane 8). Lanes 1, 3, 5, and 7, incubation of the MAR I probe with wt p53 for the same periods of time; 9 and 10, aliquots of ss and ds MAR I probe after incubation for 25 min without p53.
It is important to note that mut p53 did not interact with separated ssDNA. This can be deduced from the observation that ss MAR I and MAR II oligonucleotides were not retarded in EMSA (see also Fig. 1 C and D). To exclude that the strand separation activity of mut p53 is mediated by its ability to bind ssDNA (34, 35), we checked the binding of MethA p53 to isolated radioactive labeled top or bottom strand of the MAR I under identical EMSA conditions, but were unable to detect any binding (not shown).
To further demarcate the mut p53-specific strand separation activity from the nonspecific ssDNA-binding activity of the p53 C terminus (36–38), we performed competition experiments using increasing amounts of unlabeled ss MAR I substrate (bottom strand) (not shown). Strand separation catalyzed by mut p53 was not affected by a low molar excess of MAR I bottom oligonucleotide (10- to 50-fold), indicating that ssDNA binding as such is not sufficient to explain this strand separation. In contrast, a very high excess of unlabeled ss MAR I oligonucleotide (1,000-fold) strongly reduced strand separation. This probably reflects the blocking of the nonspecific DNA-binding domain of p53 by the MAR I bottom oligonucleotide and is in accordance with our finding that the addition of PAb 421, recognizing a C-terminal epitope on p53, also blocked this reaction.
Controls.
Dissolution of the ds MAR I and MAR II probes after incubation with mut p53 could result from a variety of different enzymatic activities, such as helicases (39) or strand-specific nucleases (40). Since our assay did not include ATP or any other energy-providing source required for helicase activity (39), we can exclude an intrinsic or associated helicase activity of mut p53. Furthermore, our mut p53 preparation did not exert a strand-specific nuclease activity. Mut and wt p53 were incubated with MAR I probes, alternatively containing the 32P label either in the bottom or the top strand. Incubation of either MAR I probe with mut p53 resulted in increased levels of labeled ssDNA (not shown).
Influence of Sequence Context on Mutant p53-Mediated Strand Separation of MAR Oligonucleotides.
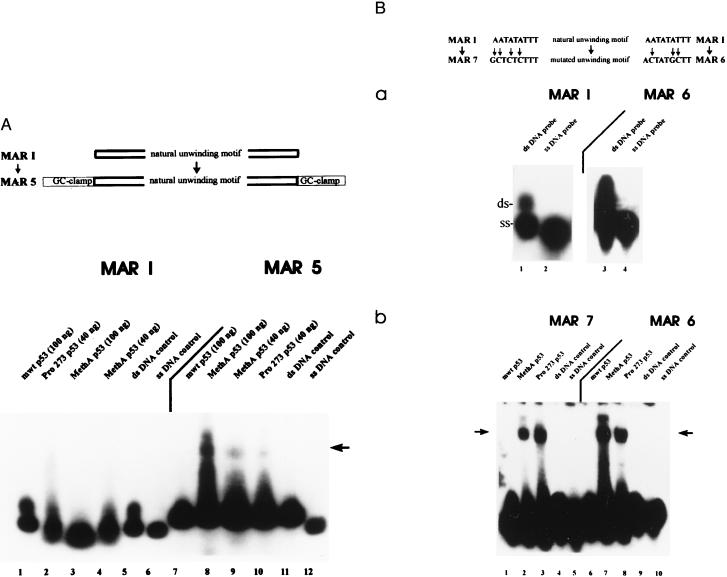
The shifted MAR I band observed during kinetic analysis of mut p53 catalyzed DNA strand separation (Fig. 2) suggested a transient interaction of mut p53 with the MAR I oligonucleotide, with mut p53 transiently binding to opening intermediates. If this were true, it should be possible to stabilize such intermediates by adding GC-clamps to the ds MAR I. Mut p53 should still be able to partially open the double strand, but then should remain bound to the opening intermediates. The results of EMSA using ds MAR 5 probe, created by adding GC-clamps to the MAR I oligonucleotide, are shown in Fig. 3A. The addition of GC-clamps to the MAR I oligonucleotide strongly reduced spontaneous strand separation, as the majority of the MAR 5 probe was recovered as dsDNA after EMSA. The presence of GC-clamps also abolished strand separation. However, mut p53 now bound the MAR 5 probe, resulting in a bandshift (Fig. 3A, lanes 8–10). Comparing the interactions of mut p53 with the MAR I and the MAR 5 probes revealed a close relationship between mut p53-induced strand separation of the MAR I probe and its ability to bind the MAR 5 probe: addition of lower amounts of MethA p53 or of human Pro-273 p53 resulted in only partial strand separation of the MAR I probe and also in a reduced amount of shifted MAR 5 probe (compare lanes 2–4 with lanes 8–10).
Figure 3.
Influence of sequence context on mutant p53-mediated strand separation of MAR-oligonucleotides. (A) The MAR 5 oligonucleotide was created by the addition of GC-clamps to both ends of the MAR I oligonucleotide, and the interactions of wt and different amounts of mut p53 with the 32P-end-labeled MAR I or MAR 5 probes were compared in EMSA. Reactions with MAR I were carried out in the presence of different amounts of MethA p53 (lane 3, 100 ng; lane 4, 40 ng), human Pro-273 p53 (lane 2, 40 ng), or wt p53 (lane 1, 100 ng). The MAR 5 probe was incubated with wt p53 (lane 7, 100 ng), MethA p53 (lane 8, 100 ng; lane 9, 40 ng), or human Pro-273 p53 (lane 10, 40 ng). Lanes 5 and 11, control reactions with MAR I (lane 5) or MAR 5 (lane 11) in the absence of p53; 6 and 12, aliquot of the ss MAR I and MAR 5 probe. (B) MAR 7 and MAR 6 oligonucleotides were created by inserting mutations into the unwinding motif of MAR I (indicated by arrows) and subjected to EMSA in the absence or presence of wt p53 or mut p53. (a) Aliquots of the isolated ss and ds MAR I and MAR 6 were subjected to EMSA in the absence of p53 to analyze their ability for spontaneous strand separation under EMSA conditions. (b) Binding reactions were performed with 32P-end-labeled MAR 7 probe in the presence of equal amounts (100 ng) of murine wt p53 (lane 1), murine MethA p53 (lane 2), human Pro-273 p53 (lane 3), or in the absence of protein (lane 4). Lane 5, aliquot of the ss MAR 7 probe. The MAR 6 probe was subjected to EMSA with murine wt p53 (lane 6), murine MethA p53 (lane 7), and human Pro-273 p53 (lane 8), or in the absence of p53 (lane 9). Lane 10, aliquot of the ss MAR 6 probe.
Influence of Mutations in the Unwinding Motif on the Interaction of Mutant p53 with the MAR I Oligonucleotide.
MAR I and MAR II, containing an unwinding motif, had only a slightly higher AT content than MAR III without such a motif. However, AT richness as such is not sufficient for the interaction of mut p53 with MARs (22, 23). Furthermore, MAR III remained ds under EMSA conditions, whereas MAR I and MAR II displayed a high intrinsic potential for base-unpairing. Therefore, we wanted to determine the roles of the unwinding motif and of intrinsic structural instability in the specific interaction of mut p53 with the MAR I and the MAR II probes. As introduction of only three point mutations into this motif resulted in a reduction of its property to promote base-unpairing (26, 32), we introduced such point mutations into the unwinding motif of MAR I (MAR 6 and MAR 7). Although this increased the structural stability of these ds oligonucleotides, as indicated by a significant reduction of spontaneous strand separation under EMSA conditions (Fig. 3B, a, compare lanes 1 and 3), a significant fraction of MAR 6 and MAR 7 still was recovered as ssDNA under EMSA conditions. Incubation of the MAR 6 and MAR 7 probes (Fig. 3B, b) with mut p53 did not noticeably increase the fraction of ssDNA. However, the mut p53 proteins now strongly bound the MAR 7 (Fig. 3B, b, lanes 2 and 3) and MAR 6 probes (Fig. 3B, b, lanes 7 and 8), as reflected by a significant band shift in EMSA, indicating a tight interaction of mut p53 with partially opened MAR 6 and MAR 7 probes. Binding to MAR 6 and to MAR 7 probes was specific for mut p53, as incubation of the same probes with wt p53 (Fig. 3B, b, lanes 1 and 6) or with buffer alone (Fig. 3B, b, lanes 4 and 9) did not result in bandshifts. Mut p53 was unable to bind the isolated ss MAR 6 and MAR 7 top- or bottom-strand oligonucleotides (not shown), verifying that a specific structural feature of these oligonucleotides was responsible for the tight binding by mut p53, and not single-strandedness of the DNA, as such.
DISCUSSION
To further characterize the interaction of mut p53 with MARs and to get a clue as to its possible function, we searched for an MAR entity specifically recognized by mut p53 and found that mut p53 specifically interacts with MARs in regions with a high potential for base-unpairing. Mut p53, but not wt p53, specifically interacted with AT-rich MAR-oligonucleotides containing variations of an AATATATTT unwinding motif, a sequence that previously was shown to promote regional base-unpairing in the context of MAR-DNA (26, 27). With the relatively short MAR-oligonucleotides used in this study, this interaction promoted complete strand separation. This activity is intrinsic to mut p53 and is not mediated by an associated helicase (39) or strand-specific nuclease (40).
During DNA strand separation mut p53 only transiently interacted with the MAR I probe, as revealed by our time-course experiment, and also did not bind to the resulting ssDNA. This provides hints to a possible mechanism. We propose that mut p53 binds with high affinity to junctions of ss- to dsDNA, most likely generated in or around the unwinding motif. In regions of high structural instability, this binding will further destabilize the DNA helix and promote base-unpairing. As mut p53 does not bind to the resulting ssDNA with the same affinity as to the DNA junction, it will fall off from the now ssDNA region, but rebind to the migrated junction. If the entire DNA fragment is highly structurally unstable, these interactions are faster than any reannealing process and will result in complete strand separation. This interpretation is strongly supported by our finding that addition of GC-clamps to MAR I not simply abolished strand separation, but resulted in binding of mut p53 to the modified oligonucleotide. This model could explain the tight binding of mut p53 to MARs. MARs that are tightly bound by mut p53 (23) typically contain several regions with a high potential for base-unpairing, characterized by AT richness and the presence of variations of an AATATATTT unwinding motif that are flanked by structurally more stable DNA. We propose that mut p53 interacts with such regions in MARs and promotes local base-unpairing. Because of the presence of regions of structurally stable DNA in these MARs, mut p53 will remain bound to ss-/dsDNA junctions, leading to the previously established high-affinity binding (Kd ≈ 10−10 M) of MARs by mut p53 (21). However, whereas DNA strand separation seemed to depend on an intact unwinding motif, binding of mut p53 to MAR-derived oligonucleotides was as dependent on structural instability of the MAR DNA as it was on the presence of an intact unwinding motif. This can be deduced from our finding that mut p53 remained bound to partially opened DNA but was unable to induce strand separation in the MAR 6 and MAR 7 oligonucleotides, where base-unpairing at the unwinding motif was impaired by introduction of mutations into this motif. As a mechanism for the postulated interaction of mut p53 with ss-/dsDNA junctions, we suggest that the mutated core domain recognizes the junction and the p53 C terminus stabilizes this interaction by binding to the adjacent ssDNA. This model would be in line with the observation that MAR binding and DNA strand separation both require the mutated core domain and the C-terminal nonspecific DNA-binding domain of mut p53.
Our finding of a mut p53-mediated DNA strand separation activity, which depends on a sequence context typically found in MARs, might provide a clue for understanding mut p53-specific transcriptional modulation. MARs organize the cellular chromatin into topologically independent loops, thereby providing a structural basis for the independent spatial and temporal regulation of gene expression and initiation of DNA synthesis. Such a regulation is thought to form a higher-order regulatory mechanism for controlling development and differentiation (41–44) and will render MARs important targets for regulatory proteins. A prominent example is SATB1, a homeodomain protein primarily expressed in thymocytes (45), which was shown to participate in negative regulation of tissue-specific gene expression (46). SATB1 preferentially binds to AT-rich regions in MARs containing variations of the unwinding motif (46). SATB1 directly recognizes this motif and supposedly blocks regional base-unpairing and strand separation starting from this motif (47). In analogy we propose that mut p53, by binding to such regions in MARs, possibly could modulate gene expression. As the p53 N-terminal transactivator domain is required for mut p53-specific transactivation (10), interaction of mut p53 with MARs might also serve to recruit transcriptional complexes on these elements. Clearly, these considerations still are speculative, but the identification of a mut p53-specific activity that possibly could be involved in the modulation of chromatin structure and function might provide clues to better understand the activities of mut p53 related to its “gain of function” properties.
Acknowledgments
We thank Dr. J. Bode, Gesellschaft für Biotechnologische Forschung, Braunschweig, for helpful discussion. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and by the Bundesministerium für Gesundheit.
ABBREVIATIONS
- wt
wild type
- MAR
matrix attachment region DNA element
- mut
mutant
- ds
double stranded
- ss
single stranded
- IgH
Ig heavy chain
- EMSA
electrophoretic mobility shift assay
References
- 1.Hainaut P, Soussi T, Shomer B, Hollstein M, Greenblatt M, Hovig E, Harris C C, Montesano R. Nucleic Acids Res. 1997;25:151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deppert W. J Cell Biochem. 1996;62:172–180. doi: 10.1002/(sici)1097-4644(199608)62:2<172::aid-jcb5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler K W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 4.Deppert W, Buschhausen-Denker G, Patschinsky T, Steinmeyer K. Oncogene. 1990;5:1701–1706. [PubMed] [Google Scholar]
- 5.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 6.Michalovitz D, Halevy O, Oren M. J Cell Biochem. 1991;45:22–29. doi: 10.1002/jcb.240450108. [DOI] [PubMed] [Google Scholar]
- 7.Levine A J, Wu M C, Chang A, Silver A, Attiyeh E F, Lin J, Epstein C B. Ann NY Acad Sci. 1995;768:111–128. doi: 10.1111/j.1749-6632.1995.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 8.Shaulsky G, Goldfinger N, Rotter V. Cancer Res. 1991;51:5232–5237. [PubMed] [Google Scholar]
- 9.Pohl J, Goldfinger N, Radler-Pohl A, Rotter V, Schirrmacher V. Mol Cell Biol. 1988;8:2078–2081. doi: 10.1128/mcb.8.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Teresky A K, Levine A J. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 11.Chin K V, Ueda K, Pastan I, Gottesman M M. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 12.Strauss B E, Haas M. Biochem Biophys Res Commun. 1995;217:333–340. doi: 10.1006/bbrc.1995.2781. [DOI] [PubMed] [Google Scholar]
- 13.Deb S, Jackson C T, Subler M A, Martin D W. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieser A, Weich H A, Brandner G, Marm D, Kolch W. Oncogene. 1994;9:963–969. [PubMed] [Google Scholar]
- 15.Subler M A, Martin D W, Deb S. Oncogene. 1994;9:1351–1359. [PubMed] [Google Scholar]
- 16.Ory K, Legros Y, Auguin C, Soussi T. EMBO J. 1994;13:3496–3504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park D J, Nakamura H, Chumakov A M, Said J W, Miller C W, Chen D L, Koeffler H P. Oncogene. 1994;9:1899–1906. [PubMed] [Google Scholar]
- 18.Park D J, Chumakov A M, Miller C W, Pham E Y, Koeffler H P. Mol Carcinogen. 1996;16:101–108. doi: 10.1002/(SICI)1098-2744(199606)16:2<101::AID-MC6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Chen J Y, Funk W D, Wright W E, Shay J W, Minna J D. Oncogene. 1993;8:2159–2166. [PubMed] [Google Scholar]
- 20.Alvarez-Salas L M, Velazquez A, Lopez-Bayghen E, Woodworth C D, Garrido E, Gariglio P, DiPaolo J A. Cancer Lett. 1995;91:85–92. doi: 10.1016/0304-3835(95)03721-8. [DOI] [PubMed] [Google Scholar]
- 21.Weissker S, Müller B, Homfeld A, Deppert W. Oncogene. 1992;7:1921–1932. [PubMed] [Google Scholar]
- 22.Müller B, Paulsen D, Deppert W. Oncogene. 1996;12:1941–1952. [PubMed] [Google Scholar]
- 23.Will K, Warnecke G, Albrechtsen N, Boulikas T, Deppert W. J Cell Biochem. 1998;69:260–270. doi: 10.1002/(sici)1097-4644(19980601)69:3<260::aid-jcb4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Nakamura K, Wendel E, Colburn N. Proc Natl Acad Sci USA. 1993;90:2827–2831. doi: 10.1073/pnas.90.7.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. Am J Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- 26.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 27.Mielke C, Kohwi Y, Kohwi-Shigematsu T, Bode J. Biochemistry. 1990;29:7475–7485. doi: 10.1021/bi00484a017. [DOI] [PubMed] [Google Scholar]
- 28.Kim E, Albrechtsen N, Deppert W. Oncogene. 1997;15:857–869. doi: 10.1038/sj.onc.1201412. [DOI] [PubMed] [Google Scholar]
- 29.Staufenbiel M, Deppert W. Eur J Cell Biol. 1983;31:341–348. [PubMed] [Google Scholar]
- 30.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 31.Yewdell J W, Gannon J V, Lane D P. J Virol. 1986;59:444–452. doi: 10.1128/jvi.59.2.444-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohwi-Shigematsu T, Kohwi Y. Biochemistry. 1990;29:9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- 33.Cockerill P N, Yuen M-H, Garrard W T. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- 34.Steinmeyer K, Deppert W. Oncogene. 1988;3:501–507. [PubMed] [Google Scholar]
- 35.Oberosler P, Hloch P, Ramsperger U, Stahl H. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foord O, Navot N, Rotter V. Mol Cell Biol. 1993;13:1378–1384. doi: 10.1128/mcb.13.3.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayaraman L, Prives C. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 39.Thommes P, Hubscher U. J Biol Chem. 1990;265:14347–14354. [PubMed] [Google Scholar]
- 40.Maryon E, Carroll D. Mol Cell Biol. 1989;9:4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berezney R. J Cell Biochem. 1991;47:109–123. doi: 10.1002/jcb.240470204. [DOI] [PubMed] [Google Scholar]
- 42.Gasser S M, Laemmli U K. Trends Genet. 1987;3:16–22. [Google Scholar]
- 43.Herbomel P. New Biol. 1990;39:937–945. [PubMed] [Google Scholar]
- 44.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Nature (London) 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 45.Dickinson L A, Dickinson C D, Kohwi-Shigematsu T. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Dudley J P. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Dickinson L A, Koivunen E, Ruoslahti E, Kohwi-Shigematsu T. J Biol Chem. 1995;270:23239–23242. doi: 10.1074/jbc.270.40.23239. [DOI] [PubMed] [Google Scholar]