Abstract
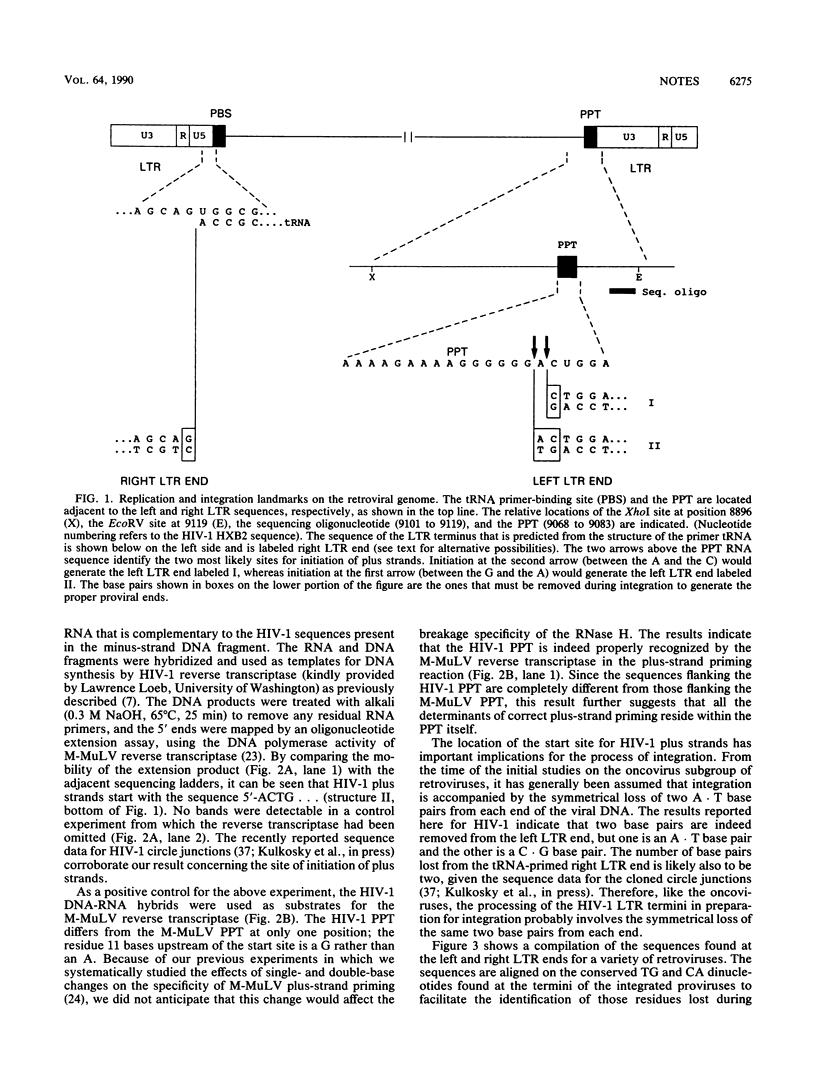
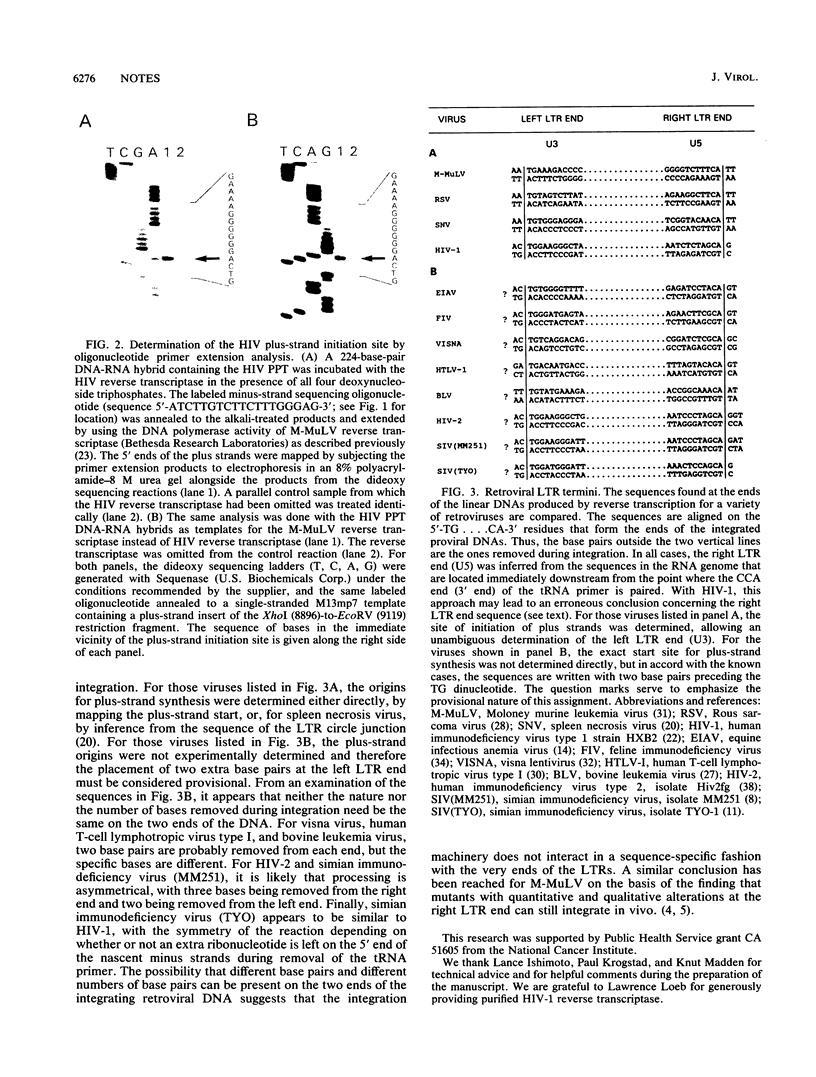
The start site for human immunodeficiency virus type 1 plus strands within the polypurine tract was mapped by an in vitro analysis to the sequence 5'-ACTG....From this result, it can be inferred that integration of human immunodeficiency virus type 1 must be accompanied by the loss of two base pairs from the polypurine tract-primed long terminal repeat end.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Been M. D., Champoux J. J. Cutting of M13mp7 phage DNA and excision of cloned single-stranded sequences by restriction endonucleases. Methods Enzymol. 1983;101:90–98. doi: 10.1016/0076-6879(83)01007-1. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finston W. I., Champoux J. J. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J Virol. 1984 Jul;51(1):26–33. doi: 10.1128/jvi.51.1.26-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini G., Gurgo C., Guo H. G., Gallo R. C., Collalti E., Fargnoli K. A., Hall L. F., Wong-Staal F., Reitz M. S., Jr Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987 Aug 6;328(6130):539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci U S A. 1989 May;86(9):3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Fukasawa M., Miura T., Hasegawa A., Morikawa S., Tsujimoto H., Miki K., Kitamura T., Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988 Jun 2;333(6172):457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Sherman L., Dahlberg J., Gazit A., Yaniv A., Tronick S. R., Aaronson S. A. Nucleotide sequence analysis of equine infectious anemia virus proviral DNA. Virology. 1987 Jun;158(2):300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Lobel L. I., Murphy J. E., Goff S. P. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J Virol. 1989 Jun;63(6):2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Omer C. A., Resnick R., Faras A. J. Evidence for involvement of an RNA primer in initiation of strong-stop plus DNA synthesis during reverse transcription in vitro. J Virol. 1984 May;50(2):465–470. doi: 10.1128/jvi.50.2.465-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Quinn T. P., Grandgenett D. P. Genetic evidence that the avian retrovirus DNA endonuclease domain of pol is necessary for viral integration. J Virol. 1988 Jul;62(7):2307–2312. doi: 10.1128/jvi.62.7.2307-2312.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Champoux J. J. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J Mol Biol. 1989 Aug 5;208(3):445–456. doi: 10.1016/0022-2836(89)90508-1. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Champoux J. J. The role of Moloney murine leukemia virus RNase H activity in the formation of plus-strand primers. J Virol. 1987 Sep;61(9):2843–2851. doi: 10.1128/jvi.61.9.2843-2851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick R., Omer C. A., Faras A. J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984 Sep;51(3):813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P. L., Goff S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989 Jul 14;58(1):47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Sorge J., Hughes S. H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982 Aug;43(2):482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbott R. L., Sparger E. E., Lovelace K. M., Fitch W. M., Pedersen N. C., Luciw P. A., Elder J. H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J., Sharmeen L. Retrovirus genome replication: priming specificities of plus-strand DNA synthesis. J Cell Sci Suppl. 1987;7:189–195. doi: 10.1242/jcs.1987.supplement_7.14. [DOI] [PubMed] [Google Scholar]
- Whitcomb J. M., Kumar R., Hughes S. H. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990 Oct;64(10):4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagury J. F., Franchini G., Reitz M., Collalti E., Starcich B., Hall L., Fargnoli K., Jagodzinski L., Guo H. G., Laure F. Genetic variability between isolates of human immunodeficiency virus (HIV) type 2 is comparable to the variability among HIV type 1. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5941–5945. doi: 10.1073/pnas.85.16.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]




