Abstract
Genetic and phenotypic instability are hallmarks of cancer cells, but their cause is not clear. The leading hypothesis suggests that a poorly defined gene mutation generates genetic instability and that some of many subsequent mutations then cause cancer. Here we investigate the hypothesis that genetic instability of cancer cells is caused by aneuploidy, an abnormal balance of chromosomes. Because symmetrical segregation of chromosomes depends on exactly two copies of mitosis genes, aneuploidy involving chromosomes with mitosis genes will destabilize the karyotype. The hypothesis predicts that the degree of genetic instability should be proportional to the degree of aneuploidy. Thus it should be difficult, if not impossible, to maintain the particular karyotype of a highly aneuploid cancer cell on clonal propagation. This prediction was confirmed with clonal cultures of chemically transformed, aneuploid Chinese hamster embryo cells. It was found that the higher the ploidy factor of a clone, the more unstable was its karyotype. The ploidy factor is the quotient of the modal chromosome number divided by the normal number of the species. Transformed Chinese hamster embryo cells with a ploidy factor of 1.7 were estimated to change their karyotype at a rate of about 3% per generation, compared with 1.8% for cells with a ploidy factor of 0.95. Because the background noise of karyotyping is relatively high, the cells with low ploidy factor may be more stable than our method suggests. The karyotype instability of human colon cancer cell lines, recently analyzed by Lengnauer et al. [Lengnauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature (London) 386, 623–627], also corresponds exactly to their degree of aneuploidy. We conclude that aneuploidy is sufficient to explain genetic instability and the resulting karyotypic and phenotypic heterogeneity of cancer cells, independent of gene mutation. Because aneuploidy has also been proposed to cause cancer, our hypothesis offers a common, unique mechanism of altering and simultaneously destabilizing normal cellular phenotypes.
Most cancers are clonal (1–3) yet highly heterogeneous, i.e., nonclonal with regard to the karyotypes (1, 4–9) and phenotypes of individual cancer cells. For example, individual cells of a given cancer differ widely in such phenotypic properties as metastatic capacity, transplantability, antigenic make up, drug sensitivity, growth rates, metabolism, and morphology (8–13).
In 1976, Peter Nowell postulated that this abundant heterogeneity is caused by an as-yet-poorly defined precancerous mutation that generates exceptional “genetic instability” (1) or “mutability” (13–16). The corresponding phenotype has been termed “mutator” (14). The highly mutable, “premalignant” cell would then suffer many further gene mutations, including those that cause cancer (16). For example, recently Lengnauer et al. (17) have suggested that either a mutated mismatch-repair gene or a chromosome-segregation gene would function as a mutator gene that drives the tumorigenic process toward colon cancer . The mutated chromosome-segregation gene was postulated to do this either by increasing the number of chromosomes with activated oncogenes or by decreasing those with tumor-suppression genes (16, 17). However, despite numerous investigations since 1976, there is still no consistent evidence for mutator genes in cancer cells (13, 15, 16, 18–22).
As an alternative hypothesis, we propose that genetic instability of cancer cells is caused by aneuploidy, an abnormal number of chromosomes. Because symmetrical segregation of chromosomes depends on exactly two copies of mitosis genes, aneuploidy involving chromosomes with mitosis genes will destabilize the karyotype (12, 23–25). Once aneuploid, cells will continue to be subject to asymmetric chromosome segregation every time they divide, a process that has been termed “chromosome error propagation” (12). The phenotypic heterogeneity of individual, aneuploid cancer cells described above would be an inevitable consequence of this genetic instability.
The aneuploidy–genetic instability hypothesis predicts that the degree of genetic instability is proportional to the degree of aneuploidy. In other words, it should be difficult, if not impossible, to maintain the particular karyotype of a highly aneuploid cancer cell on clonal propagation. To test this prediction, we have analyzed the chromosomes of clonal cultures of chemically transformed Chinese hamster embryo (CHE) cells with various degrees of aneuploidy. The results of these experiments demonstrate that chromosome number instability in cancer cells is indeed proportional to their degree of aneuploidy. We have also applied our hypothesis to the relationship between genetic instability and aneuploidy of human colon cancer cells, based on a recent study by Lengnauer et al. (17). This analysis confirmed and extended to human cancer cells our result that genetic instability is proportional to the degree of aneuploidy.
RESULTS
Clones of CHE Cells Derived from Transformed Cultures with Heterogenous Karyotypes.
To test the hypothesis that karyotypic instability is proportional to the degree of aneuploidy, we set out to analyze the karyotypes of clonal cultures derived from chemically transformed CHE cells with different degrees of aneuploidy.
Such chemically transformed CHE cells had been prepared previously for a statistical analysis of the correlation between aneuploidy and chemical transformation. This correlation proved to be 100%, but the karyotypes of cultures derived from discrete foci of transformed CHE cells were unexpectedly heterogeneous (26). This karyotypic heterogeneity already suggested an underlying instability but did not exclude polyclonality (26).
To prepare clonal cultures, about 100 cells of a given focus-derived culture of karyotypically heterogenous, transformed CHE cells were seeded on a 10-cm culture dish and incubated for about 2 weeks as described (26). By that time, about 5–20 colonies of cells had appeared per dish. Individual colonies were then removed with a micropipet from several such dishes and grown into large cultures for analysis of their phenotypes and chromosome numbers. They were labeled like their precursors (26) and individually distinguished by hyphenated numbers. For example, clonal cultures derived from the uncloned, methylcholanthrene-transformed culture M1 were labeled M1-1, M1-2, etc., and clonal cultures derived from uncloned, dimethylbenzanthracene- and benzpyrene-transformed cultures D3 and B6 were labeled D3-1, D3-2, etc. and B6-1, B6-2, etc., respectively (Tables 1 and 2).
Table 1.
Characteristics of Chinese hamster embryo cancer cell lines
| Clone | T/F | Growth | mn | Number of cells with specific number of chromosomes
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | Over 48 | ||||
| M11-1 | T,p | f | 39/41 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 4 | 7 | 5 | 1 | 1 | ||||||||||||||||||||||||
| M11-8 | T,p | f | 40 | 1 | 1 | 1 | 3 | 2 | 1 | 2 | 7 | 2 | 2 | 1 | |||||||||||||||||||||||||
| M11-7 | T,p | f | 39 | 1 | 2 | 1 | 1 | 1 | 3 | 2 | 7 | 1 | 6 | ||||||||||||||||||||||||||
| B6-10 | T,s | n | 39 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 3 | 4 | 1 | 2 | 1 | ||||||||||||||||||||||||
| M8-5 | T,p | vf | 36–38 | 1 | 1 | 1 | 2 | 1 | 3 | 4 | 4 | 4 | 2 | 1 | 1 | 1 | 1 × 53/73 | ||||||||||||||||||||||
| M11-2 | T,s | f | 37 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | 4 | 2 | 2 | |||||||||||||||||||||||||
| M11-4 | T,p | vf | 37 | 1 | 2 | 1 | 3 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||||||||||||||||||||
| B6-4 | T,p | vf | 36/37 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 2 | 3 | 1 | 1 | 4 | 4 | 3 | 1 | ||||||||||||||||||||
| D3-1 | T,p | f | 30 | 1 | 1 | 1 | 11 | 2 | 5 | 7 | 1 | 1 × 60 | |||||||||||||||||||||||||||
| M8-11 | T,p | s | 24 | 4 | 2 | 8 | 2 | ||||||||||||||||||||||||||||||||
| B6-9 | T,s | vf | 21 | 1 | 1 | 24 | 3 | 1 | 1 | ||||||||||||||||||||||||||||||
| D3-10 | T,s | f | 21 | 1 | 1 | 1 | 21 | ||||||||||||||||||||||||||||||||
| D3-13 | T,p | n | 20 | 20 | 1 | 16 | |||||||||||||||||||||||||||||||||
| M8-1 | F,p | n | 23 | 1 | 5 | 10 | 6 | 1 | 2 | ||||||||||||||||||||||||||||||
| M8-3 | F,p | vs | 23 | 1 | 9 | 6 | 1 | ||||||||||||||||||||||||||||||||
| D3-3 | F,p | n | 22 | 1 | 16 | 1 | 1 | ||||||||||||||||||||||||||||||||
| D3-8 | F,p | n | 22 | 2 | 1 | 1 | 1 | 4 | 2 | 8 | 2 | 2 | 1 | ||||||||||||||||||||||||||
| B6-3 | F,p | f | 22 | 1 | 22 | ||||||||||||||||||||||||||||||||||
| B6-5 | F,s | vf | 22 | 1 | 8 | 15 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||
| M11-6 | F,p | n | 22 | 2 | 18 | 1 | 1 | 1 | |||||||||||||||||||||||||||||||
T, transformed; F, flat; mn, modal chromosome number; p, polygonal; s, spindle-shaped; f, fast; vf, very fast; n, normal; s, slow; vs, very slow.
Table 2.
Characteristics of Chinese hamster embryo cancer cell lines
| Clone | T/F | Growth | mn | Number of cells with specific number of chromosomes
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | Over 48 | ||||
| B6-10-1 | T,s | n | 39 | 1 | 6 | 8 | 2 | 9 | 1 | 3 | 1 | 1 | 1 × 53/58 | ||||||||||||||||||||||||||
| B6-10-2 | T,s | n | 38 | 1 | 1 | 2 | 5 | 2 | 6 | 5 | 3 | 5 | |||||||||||||||||||||||||||
| B6-10-3 | T,s | n | 38 | 1 | 1 | 3 | 4 | 6 | 5 | 4 | 1 | 1 | 2 | 1 | 1 × 73 | ||||||||||||||||||||||||
| B6-10-4 | T,s | n | 38 | 1 | 2 | 8 | 8 | 11 | 4 | 5 | 3 | 1 × 49 | |||||||||||||||||||||||||||
| B6-10-5 | T,s | n | 39 | 1 | 6 | 4 | 7 | 4 | 2 | ||||||||||||||||||||||||||||||
| B6-10-6 | T,s | n | 37 | 2 | 1 | 2 | 1 | 4 | 3 | 9 | 3 | 2 | 1 | 1 | 1 | ||||||||||||||||||||||||
| B6-10-7 | T,s | n | 36/39 | 3 | 8 | 4 | 4 | 8 | 2 | 1 | |||||||||||||||||||||||||||||
| B6-10-8 | T,s | n | 36/37 | 1 | 2 | 5 | 5 | 3 | 2 | 2 | 1 | ||||||||||||||||||||||||||||
| B6-10-9 | T,s | n | 34 | 2 | 1 | 1 | 2 | 6 | 3 | 2 | 5 | 2 | 3 | 1 | 1 | 1 | |||||||||||||||||||||||
| B6-10-10 | T,s | n | 38 | 1 | 1 | 1 | 5 | 10 | 6 | 3 | 1 | 2 | |||||||||||||||||||||||||||
| B6-10-11 | T,s | n | 39 | 1 | 1 | 1 | 1 | 1 | 3 | 5 | 6 | 3 | 2 | 1 | 1 | ||||||||||||||||||||||||
| B6-10-12 | T,s | n | 38 | 2 | 1 | 3 | 10 | 4 | 9 | 1 | |||||||||||||||||||||||||||||
| B6-10-13 | T,s | n | 34/36 | 1 | 1 | 2 | 1 | 2 | 1 | 6 | 6 | 1 | 5 | 3 | 1 | ||||||||||||||||||||||||
| B6-10-14 | T,s | n | 38/39 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 4 | 5 | 5 | 3 | 3 | ||||||||||||||||||||||||
| B6-4-1 | T,p | vf | 38/42 | 1 | 1 | 1 | 3 | 1 | 7 | 6 | 1 | 7 | 1 | 1 | |||||||||||||||||||||||||
| B6-4-2 | T,p | vf | 39 | 1 | 1 | 3 | 1 | 5 | 2 | 2 | 14 | 1 | |||||||||||||||||||||||||||
| B6-4-3 | T,p | vf | 41 | 1 | 1 | 1 | 3 | 1 | 7 | 3 | 8 | 3 | 1 | 1 | |||||||||||||||||||||||||
| B6-4-4 | T,p | vf | 37 | 1 | 1 | 1 | 1 | 3 | 7 | 11 | 2 | 2 | 1 | ||||||||||||||||||||||||||
| B6-4-5 | T,p | vf | 36 | 1 | 1 | 1 | 3 | 3 | 1 | 3 | 6 | 5 | 1 | 1 × 54 | |||||||||||||||||||||||||
| B6-4-6 | T,p | vf | 34/42 | 1 | 2 | 5 | 2 | 2 | 5 | 3 | 6 | 1 | |||||||||||||||||||||||||||
| B6-4-7 | T,p | vf | 35 | 1 | 1 | 2 | 5 | 7 | 5 | 6 | 1 | 5 | 1 | 1 × 60/76 | |||||||||||||||||||||||||
| B6-4-8 | T,p | vf | 34 | 3 | 5 | 1 | 2 | 1 | 1 | ||||||||||||||||||||||||||||||
| B6-4-9 | T,p | vf | 32 | 1 | 3 | 1 | 7 | 1 | 3 | 1 | 1 | 5 | 1 | ||||||||||||||||||||||||||
| B6-4-10 | T,p | vf | 39 | 1 | 5 | 2 | 7 | 11 | 3 | 2 | |||||||||||||||||||||||||||||
| B6-4-11 | T,p | vf | 32 | 1 | 1 | 1 | 7 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | ||||||||||||||||||||||||
| B6-4-12 | T,p | vf | 42 | 1 | 3 | 1 | 3 | 4 | 2 | 9 | 3 | 2 | 3 | ||||||||||||||||||||||||||
| B6-4-13 | T,p | vf | 40 | 1 | 1 | 1 | 4 | 5 | 3 | 1 | |||||||||||||||||||||||||||||
| B6-4-14 | T,p | vf | 40 | 2 | 3 | 3 | 2 | 11 | 7 | 2 | |||||||||||||||||||||||||||||
| M8-5-1 | T,p | n | 36 | 1 | 1 | 7 | 8 | 6 | 4 | 2 | 1 | ||||||||||||||||||||||||||||
| M8-5-2 | T,p | f | 37 | 4 | 1 | 6 | 10 | 2 | 2 | 1 | 2 | 1 | 1 | ||||||||||||||||||||||||||
| M8-5-3 | T,p | n | 38 | 1 | 1 | 1 | 1 | 3 | 1 | 5 | 9 | 1 | 1 | ||||||||||||||||||||||||||
| M8-5-4 | T,p | f | 36 | 1 | 1 | 4 | 7 | 1 | 2 | 4 | 3 | 2 | 1 | ||||||||||||||||||||||||||
| M8-5-5 | T,p | f | 37 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 10 | 4 | 4 | 2 | ||||||||||||||||||||||||
| M8-5-6 | T,p | n | 39 | 1 | 4 | 2 | 1 | 3 | 2 | 1 | 1 | 2 | 1 | 3 | 5 | 2 | 2 | 4 | 1 | 1 × 54/64/66 | |||||||||||||||||||
| M8-5-7 | T,p | n | 34 | 1 | 2 | 3 | 1 | 11 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||
| M8-5-8 | T,p | f | 34 | 1 | 1 | 3 | 2 | 1 | 7 | 1 | 1 | 2 | 3 | 2 | 1 | ||||||||||||||||||||||||
| M8-5-9 | T,p | n | 35 | 4 | 1 | 10 | 4 | 6 | 1 | 2 | 1 × 56 | ||||||||||||||||||||||||||||
| D3-1-1 | T,p | n | 31 | 1 | 2 | 2 | 1 | 4 | 12 | 2 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||
| D3-1-2 | T,p | n | 34/36 | 1 | 1 | 1 | 2 | 4 | 9 | 9 | 3 | ||||||||||||||||||||||||||||
| D3-1-3 | T,p | f | 31 | 5 | 8 | 1 | 10 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||
| D3-1-4 | T,p | f | 31 | 3 | 2 | 10 | 1 | 4 | 6 | 3 | 2 | 2 | 3 | 1 × 54 | |||||||||||||||||||||||||
| D3-1-5 | T,p | n | 31/34 | 1 | 1 | 1 | 2 | 7 | 3 | 5 | 7 | 2 | 1 | ||||||||||||||||||||||||||
| D3-1-6 | T,p | n | 32 | 1 | 2 | 2 | 8 | 14 | 2 | 1 | |||||||||||||||||||||||||||||
| D3-1-7 | T,p | n | 31 | 2 | 3 | 3 | 4 | 21 | 14 | 5 | 4 | 1 | 1 | 1 × 57 | |||||||||||||||||||||||||
| D3-1-8 | T,p | f | 32 | 3 | 2 | 2 | 1 | 15 | 8 | 10 | 1 | ||||||||||||||||||||||||||||
| D3-1-9 | T,p | f | 34 | 1 | 2 | 4 | 5 | 7 | 6 | 4 | 1 | ||||||||||||||||||||||||||||
| D3-1-10 | T,p | f | 34 | 2 | 1 | 7 | 10 | 2 | 4 | 3 | 1 | ||||||||||||||||||||||||||||
| D3-1-11 | T,p | f | 32 | 1 | 9 | 5 | 5 | 5 | 3 | 1 | 1 | ||||||||||||||||||||||||||||
| D3-1-12 | T,p | f | 32 | 2 | 1 | 9 | 5 | 4 | 3 | ||||||||||||||||||||||||||||||
| D3-1-13 | T,p | n | 34 | 2 | 4 | 7 | 10 | 3 | 5 | 2 | |||||||||||||||||||||||||||||
| D3-1-14 | T,p | f | 29/30 | 2 | 2 | 2 | 7 | 7 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | ||||||||||||||||||||||||
| B6-9-1 | T,s | vf | 21 | 1 | 2 | 24 | 1 | 1 | 2 | ||||||||||||||||||||||||||||||
| B6-9-2 | T,s | vf | 21 | 1 | 2 | 12 | 15 | 1 | |||||||||||||||||||||||||||||||
| B6-9-3 | T,s | vf | 38 | 1 | 1 | 1 | 2 | 3 | 8 | 4 | 4 | 3 | 1 | 2 | |||||||||||||||||||||||||
| B6-9-4 | T,s | vf | 22 | 1 | 1 | 1 | 2 | 8 | 16 | 4 | |||||||||||||||||||||||||||||
| B6-9-5 | T,s | vf | 21 | 1 | 2 | 4 | 2 | 10 | 3 | 2 | |||||||||||||||||||||||||||||
| B6-9-6 | T,s | vf | 21 | 1 | 5 | 4 | 2 | 3 | 12 | 1 | 1 | ||||||||||||||||||||||||||||
| B6-9-7 | T,s | vf | 21 | 1 | 4 | 15 | 23 | 5 | |||||||||||||||||||||||||||||||
| B6-9-8 | T,s | vf | 21 | 2 | 2 | 21 | 7 | 3 | |||||||||||||||||||||||||||||||
| B6-9-9 | T,s | vf | 21 | 1 | 2 | 17 | 3 | 3 | 3 | ||||||||||||||||||||||||||||||
| B6-9-10 | T,s | vf | 21 | 1 | 1 | 3 | 5 | 5 | 15 | 9 | 1 | ||||||||||||||||||||||||||||
| B6-9-11 | T,s | vf | 22 | 1 | 1 | 2 | 5 | 8 | 26 | 7 | 2 | 1 | |||||||||||||||||||||||||||
| B6-9-12 | T,s | vf | 21 | 1 | 2 | 1 | 1 | 7 | 5 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| B6-9-13 | T,s | vf | 19 | 3 | 14 | 9 | 7 | 2 | |||||||||||||||||||||||||||||||
| B6-9-14 | T,s | vf | 21 | 2 | 6 | 12 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||
| B6-9-15 | T,s | vf | 21 | 3 | 7 | 27 | 6 | 1 | |||||||||||||||||||||||||||||||
| B6-9-16 | T,s | vf | 21 | 3 | 2 | 5 | 16 | 4 | |||||||||||||||||||||||||||||||
| B6-9-17 | T,s | vf | 21 | 2 | 2 | 4 | 3 | 8 | 2 | 2 | |||||||||||||||||||||||||||||
| D3-10-1 | T,s | vf | 21 | 1 | 1 | 8 | 23 | 2 | 1 | ||||||||||||||||||||||||||||||
| D3-10-2 | T,s | vf | 20/21 | 1 | 3 | 3 | 1 | 7 | 7 | 1 | 3 | 1 | |||||||||||||||||||||||||||
| D3-10-3 | T,s | vf | 21 | 1 | 5 | 2 | 16 | 2 | 1 | 1 | |||||||||||||||||||||||||||||
| D3-10-4 | T,s | vf | 20/21 | 1 | 4 | 5 | 16 | 17 | 2 | ||||||||||||||||||||||||||||||
| D3-10-5 | T,s | vf | 21 | 1 | 1 | 2 | 15 | 3 | 2 | 1 | |||||||||||||||||||||||||||||
| D3-10-6 | T,s | vf | 20 | 1 | 3 | 9 | 14 | 6 | 3 | 1 | |||||||||||||||||||||||||||||
| D3-10-7 | T,s | vf | 21 | 1 | 3 | 5 | 13 | 2 | 1 | 1 | |||||||||||||||||||||||||||||
T, transformed; F, flat; mn, modal chromosome number; p, polygonal; s, spindle-shaped; f, fast; vf, very fast; n, normal; s, slow; vs, very slow.
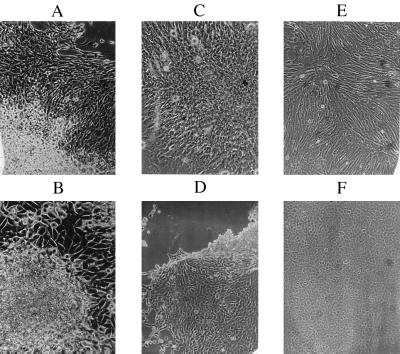
Based on their social properties and morphology, these clonal colonies fell into a major class of 13 transformed clones and a minor class of 7 flat clones (Table 1). In contrast to normal CHE cells, the transformed clones grew three-dimensionally into several layers (Fig. 1 A–D; Table 1). They also displayed various transformed shapes, including polygonal and spindle-shaped variants (Fig. 1; Table 1). The growth rates of 10 of the 13 transformed clones were fast or very fast, those of two others, B6-10 and D3-13, were normal, and that of one, M8-11, was slow compared with the growth rate of normal CHE cells (Table 1). Thus, the transformed cells were heterogeneous with regard to growth rates and morphology.
Figure 1.
The appearance of clonal cultures of chemically transformed, highly aneuploid (A and B) and little aneuploid or near-diploid (C and D) CHE cells and of clones of flat CHE cells with the normal modal chromosome number 22 (E and F). The highly aneuploid clone B6-4 (A) and the near-diploid clone D3-10 (C) consist of spindle-shaped cells. The highly aneuploid clone M11-1 (B) and the little aneuploid clone M8-11 (D) consist of polygonal cells. The flat clone B6-5 (E) consists of spindle-shaped cells, and the flat clone D3-8 (F) consists of polygonal cells.
The seven flat clones were essentially contact-inhibited, growing two-dimensionally into confluent monolayers like normal CHE cells (Fig. 1 E and F; Table 1). They also differed from each other in their morphology, some being more polygonal and others more spindle-shaped (Fig. 1). Their growth rates were either faster than normal or normal, except for M3-8, which grew more slowly than normal cells (Table 1).
Karyotypes of Clonal Cultures of Transformed CHE Cells.
Despite being derived from clones of single cells, the karyotypes of individual cells from 9 of the 13 transformed CHE clones—M11-1, M11-8, M11-7, B6-10, M8-5, M11-2, M11-4, B6-4, and D3-1—were highly heterogeneous with regard to the modal chromosome number (mn) of each clone (Table 1). The mns of these 9 clones were also highly abnormal, ranging between 30 and 40 compared with the 22 chromosomes of normal Chinese hamsters (Table 1). As a quantitative measure of karyotypic abnormality, we introduce the ploidy factor (pf), which is the quotient of the mn divided by the normal number of chromosomes of the species. For example, M11-8 with a mn of 40 has a pf of 1.8.
By contrast to the clones with high pfs, the four transformed clones with pfs close to 1—M8-11, B6-9, D3-10, and D3-13—were almost homogeneous with regard to the karyotypes of individual cells (Table 1). Thus there was a correlation between the degree of karyotypic heterogeneity and the mn, or the pf, of each clone: the higher the pf, the more heterogeneous the corresponding karyotype (Table 1).
In an effort to extend the statistical and biological basis of the relationship between aneuploidy and karyotype heterogeneity, we analyzed subclones of six primary clones of transformed CHE cells with distinct pfs—B6-10, B6-4, M8-5, D3-1, B6-9, and D3-10 (Table 1). Preparation of the subclones was as described above for the primary clones. The subclones shared most of their phenotypic properties with their parents, including the transformed phenotype, but some differed from their parents and from each other in their growth rates (Tables 1 and 2).
Karyotypic heterogeneity was measured as the percentage of cells with nonmodal chromosome numbers. The relationship between the average pf and the average karyotypic heterogeneity of all subclones from a given clone is reported in Table 3. The results of this experiment indicated again that karyotypic heterogeneity is proportional to the pf: the higher the pf, the higher the heterogeneity.
Table 3.
Characteristics of chemically transformed Chinese hamster embryo cells
| Subclones | pf | mn | non-mn, % | Karyotype change per generation, % |
|---|---|---|---|---|
| B6-10-1 to -14 | 1.7 | 37.5 | 72 | 3.1 |
| B6-4-1 to -14 | 1.7 | 37.5 | 70.5 | 3.05 |
| M8-5-1 to -9 | 1.65 | 36 | 70 | 3.04 |
| D3-1-1 to -14 | 1.45 | 32 | 64 | 2.8 |
| B6-9-1 to -17 | 1 | 22 | 54 | 2.3 |
| D3-10-1 to -7 | 0.95 | 21 | 43 | 2.1 |
The karyotypic instability, or the rate of karyotype change per cell generation, of a given clone was estimated by dividing the percentage of nonmodal karyotypes by the number of cell generations separating the clonal culture analyzed from its founder cell (Table 3). Because clonal cultures of about 10 million cells were required for our analysis, our cultures were at least 23 generations removed from the founder cell. On this basis, it can be estimated that 3.1% of the cells of clone B6-10 change their chromosome number per generation, because the karyotypes of 72% of its cells were nonmodal after about 23 generations (Table 3). By contrast, only 1.8% of D3-10 cells change their karyotype per generation (Table 3). (These calculations assume that the unknown karyotype of the founder cell is the same as the average mn of a clone.) Because the background noise of karyotyping is relatively high as a result of losses and overlaps of chromosomes (4, 9, 27), the karyotypes of cells with low pfs may be more stable than our estimate suggests.
We can draw two additonal conclusions from these results. First, the karyotypic heterogeneity of the original, uncloned cultures of transformed CHE cells studied previously (26) was predominantly the product of karyotypic instability rather than polyclonality, because most of its clonal derivatives were unstable. Second, our new results confirm and extend the perfect statistical correlation between chemical transformation and aneuploidy described previously (26). However, 2 of the 72 subclones of transformed CHE cells, namely B6-9-4 and B6-9-11, had the normal modal chromosome number of 22 (Table 2). Nevertheless, these are probably pseudodiploid cells (with an abnormal chromosome composition but a normal chromosome number) based on the following. Because these two clones were derived from B6-9 with the mn 21 (Table 1), the odds are only 1:21 that the unpaired chromosome of their B6-9 parent, rather than a paired chromosome, was doubled to generate the subclones with 22 chromosomes. It is also possible that the 10 chromosome pairs of the B6-9 parent were already unbalanced, since it came from a CHE culture that had been exposed repeatedly to aneuploidy-inducing carcinogens (28, 29) to generate transformants (26). A preliminary analysis of B6-9-4 has indeed suggested that it is pseudodiploid. Thus, we have yet to find the first truly diploid or euploid transformant among the chemically transformed CHE cells described here and previously (26).
Karyotypes of Clonal Cultures of Flat CHE Cells.
The seven flat clones obtained from uncloned cultures of transformed cells fell into two groups based on their mns (Table 1). The larger group of 5 clones had the same mn as normal CHE cells (22). Such clones were expected from the presence of diploid or pseudodiploid cells in the uncloned cultures studied previously (26).
The smaller group of two flat clones, i. e., M8-1 and M8-3, had near-diploid but aneuploid mns of 23 (Table 1). It is argued below that there must be a threshold of aneuploidy for transformation and that, therefore, the extra chromosome of both M8-1 and M8-3 may not be sufficient to alter the morphological phenotype (see Discussion).
Correlation Between the Degree of Aneuploidy and Genetic Instability of Human Colon Cancer Cells.
We next apply our hypothesis that genetic instability is caused by aneuploidy to an independent analysis of the karyotype stability of human colon cancer cell lines published by Lengnauer et al. (17). As in our analysis of CHE cells, several colon cancer lines that differed in their degrees of aneuploidy, or pfs, were investigated. But whereas the mns of our transformed CHE cells ranged only from near-diploid to hypotetraploid (Tables 1 and 2), those of the human colon cancer lines ranged from hypodiploid to hypertetraploid (17). Genetic instability of these lines was determined from the percentage of cells that had gained or lost a given chromosome per 25 generations. Chromosomes were identified by hybridization of interphase cells with chromosome-specific fluorescent DNA probes (17).
Our hypothesis predicts that the degree of genetic instability of the colon cancer cells should be proportional to their degree of aneuploidy. Thus, diploid, tetraploid, and octaploid cells, with ploidy factors 1, 2, and 4, respectively, should be stable, because they share a normal balance of mitosis genes and proteins. But cells with other ploidies should be unstable in proportion to the deviation of their ploidy from normal. Below, we show that this was indeed the case.
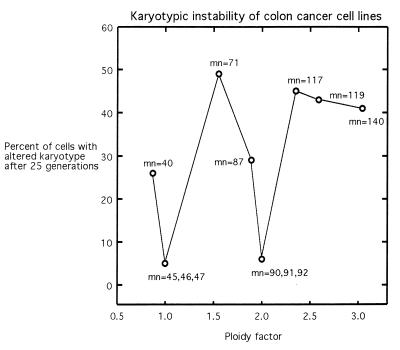
To cover the wide range of the mns of the colon cancer lines, the relationship between genetic instability and pfs was recorded graphically in Fig. 2. The pfs were obtained by dividing the mns by 46, which is the normal complement of human chromosomes. It can be seen in Fig. 2 that two highly aneuploid colon cancer cell lines with mns of 71 (HT29) and 119 (SW480) and pfs of 1.54 and 2.54, respectively, and two highly aneuploid artificial hybrid lines with mns of 117 (DLD1+HT29) and 140 (2× HT29) and pfs of 2.53 and 3.04, respectively, also were highly unstable. After 25 cell generations, about 50% of the cells of these clones had reportedly changed the ploidy of a given chromosome (ref. 17; Fig. 2).
Figure 2.
The relationship between the genetic instability and the pf of human colon cancer cell lines (17). The pf is the quotient of the mn of a cancer line divided by 46, the chromosome number of normal human cells. Genetic instability was determined from the percentage of cells that gained or lost a given chromosome per 25 generations. The karyotypes of cells with near-diploid and tetraploid karyotypes, i.e., with pfs close to 1 and 2, respectively, are stable. The karyotypes of aneuploid cells are unstable, in proportion to their degree of aneuploidy, or to the deviation of their ploidy factors from 1 and 2.
Two moderately aneuploid colon cancer cell lines with mns of 40 (SW837) and 87 (LoVo) and pfs of 0.87 and 1.89 were moderately unstable. After 25 cell generations, about 30% of the cells had changed the ploidy of a given chromosome (Fig. 2).
Three near-diploid colon-cancer cell lines with mns close to the normal human complement of 46 were karyotypically stable. One of these is the DLD1 line. It contains a chromosome of unknown origin instead of the normal chromosome no. 2 and therefore has a pseudonormal mn of 46 and a pf of 1. The chromosome distribution of this line ranges from 40 to 51 (30). Another is the SW48 line, with a mn of 47, with trisomy of chromosome no. 7 and two marker chromosomes of unknown origin. The chromosome distribution ranges from 38 to 50 (30). HCT 116 is a third line with a near-diploid mn of 45. It lacks a Y chromosome and includes three marker chromosomes of unclear derivation. The chromosome distribution ranges from 43 to 47 (30). Likewise, three near-tetraploid hybrid cell lines were stable, as may be expected from the diploid balance of mitosis proteins in truly tetraploid cells. They included the HCT 116 dimer with 90 chromosomes and a pf of 1.96, the DLD 1× HCT 116 heterodimer with 91 chromosomes and a pf of 1.98, and the DLD 1 dimer with 92 chromosomes and a pf of 2.
It follows that the karyotypic instability of the human colon cancer cells is directly proportional to the degree of aneuploidy (Fig. 2).
DISCUSSION
Aneuploidy Is Sufficient for Genetic Instability.
The hypothesis that aneuploidy is a sufficient cause of genetic instability was confirmed, because the karyotypic instability of transformed CHE and human cancer cells is directly proportional to the degree of aneuploidy. Therefore, we propose that the phenotypic heterogeneity of cancer cells is the result of this karyotypic instability.
Because probably not all chromosomes encode balance-sensitive mitosis proteins, our hypothesis also predicts stable, or relatively stable aneuploidies involving such chromosomes. The nonrandom aneuploidies of near-diploid cancer cells (9, 31) and the nonrandom monosomies and trisomies of congenital, noncancerous defects, like Down’s syndrome (32, 33), are consistent with this prediction. But it remains to be determined which chromosomes carry balance-sensitive mitosis proteins.
Aneuploidy Versus Gene Mutation as Causes of Genetic Instability.
Both Lengnauer et al. (17) and we have obtained nearly identical data on the relationship between chromosome-number instability and aneuploidy. Even our estimates for the rates of karyotype changes per cell generation in highly aneuploid cells are similar, i.e., 3% for any chromosome of highly aneuploid CHE cells with a pf of 1.7 versus 2% for a specific chromosome of highly aneuploid human colon cancer cells, some of which also have a pf of 1.7 (Fig. 2). But, in contrast to our interpretation, other workers blame karyotype instability on the dominant, negative mutation of a mitosis gene (16, 17, 34).
Nevertheless, the following comparison of the two proposed mechanisms favors aneuploidy as the cause of genetic instability. (i) The mutation hypothesis predicts that destabilization of the karyotype is independent of aneuploidy, but this was not observed. By contrast, the aneuploidy hypothesis correctly predicts the observation that destabilization of chromosome segregation is proportional to the degree of aneuploidy. (ii) The postulated destabilizing mutation has been found in only 2 of 19 karyotypically unstable colon cancers (16, 34). By contrast, all karyotypically unstable cells in those studies and ours were highly aneuploid, which is a consistent explanation for “dominant” genetic instability, according to our hypothesis. (iii) If a “dominant” mutation resulting in gains or losses in excess of 10−2 per chromosome per generation “drives the tumorigenic process” as Lengnauer et al. (17) argue, colon cancer should follow very soon after such a destabilizing mutation occurs. In other words, the colon cancer risk should essentially be a single-hit event, increasing linearly with age. If this were the case, a significant fraction of colon cancers should occur at a young age. By contrast, colon cancer shows a 1,000-fold bias for old age (2, 35). According to the aneuploidy hypothesis, this age bias may reflect the gradual buildup over time of the pf toward the threshold for cancer (see below). (iv) The existence of a human or animal gene that could be converted to a ”dominant“ aneuploidy gene is hard to reconcile with the evolution of multicellular organisms. Because the normal, spontaneous mutation rates are 1 in 109 nucleotides per mitosis even after proofreading, and because human cells contain about 109 nucleotides, 1 in 109 human cells would contain such a mutation (15, 26, 36). Thus, one such mutation would soon kill the organism via carcinogenesis, because cancers are clonal.
This argument does not call into question the existence of mutations that cause karyotype instability. But unlike those postulated by Cahill et al. (34), such mutations would have to be recessive, requiring a homozygous mutation to destabilize the karyotype. Because the probability of a specific base mutation per cell generation is 1 in 109 (see above), the probability of the same mutation in both sister alleles is 1 in 1018. Thus, only 1 in 100 average humans would be at risk to develop cancer from such a specific base mutation, because a human lifetime corresponds to approximately 1016 cells (2, 15, 37, 38). Indeed, there is a rare genetic defect, Bloom’s syndrome, that is caused by such a mutation (39). In accordance with our hypothesis, persons with Bloom’s syndrome do indeed develop colon cancer at young age (40, 41).
Aneuploidy, a Common Cause of Cancer and Genetic Instability?
In view of these new and many previous results (42, 43), we propose aneuploidy as a common cause of cancer and genetic instability. This aneuploidy–cancer hypothesis makes, in fact, fewer assumptions than the hypothesis that cancer is caused by gene mutations (36, 44). However, in view of the popularity of gene mutation as a cause of phenotype alteration, it is currently hardly known that aneuploidy is, by itself, sufficient to change normal, eukaryotic cellular phenotypes into one of the many cancer-specific phenotypes listed above. Therefore, the evidence for this is briefly recorded.
Because cellular phenotypes are determined by molecules that are assembled by thousands of kinetically linked enzymes (45, 46), aneuploidy can dominantly alter normal phenotypes because it multiplies or divides the diploid, biosynthetic assembly lines of normal cells (32, 33, 47–49, 62). In light of this hypothesis, normal cells are the equivalent of car factories with concerted assembly lines producing appropriate numbers of engines, bodies and wheels, but cancer cells would be equivalent to factories with disconcerted assembly lines that produce two or more bodies per engine, or more or less than four wheels per body. Qualitative changes would be achieved by multiplying or dividing assembly lines that produce regulatory molecules.
Thus, cellular phenotypes can be changed in two ways: (i) by canonical mutation of specific enzymes involved in the synthesis of a molecule that determines a phenotype or (ii) by increasing or decreasing the number of normally diploid assembly lines of kinetically linked enzymes whose end product determines a phenotype. According to metabolic control analysis the effects of specific gene mutations are very limited (45, 46, 50, 62). Positive or activating mutations would be strongly buffered because the remaining enzymes of an assembly line will continue to work at their native rate. Negative mutations are buffered by the fact that enzymes work in vivo far below saturation, i.e. at only a few percent of their capacity. As a result, biosynthetic assembly lines resist very efficiently any changes in the activities of their enzymes, although this aspect of the evolutionary design of metabolism continues to be ignored in attempts to modify phenotypes and yields by genetic engineering of bacteria and animals (50, 62). Thus typically only homozygous null mutations of essential genes, generating a metabolic block, would significantly alter the phenotype (45). However, most of these would be lethal mutations that are not relevant to cancer. In light of this, it is not surprising that mutated cellular oncogenes and tumor-suppression genes fail to perform dominant transforming functions (51–55), are typically not more active than unmutated counterparts (37, 43, 54–56), and are not consistently present in otherwise identical cancers (13, 15, 37, 53, 55, 57, 58).
However, the presence of low levels of aneuploidy in rare, noncancerous cells of normal persons (4, 59–61) and in noncancerous, congenital diseases like Down’s syndrome (32, 33) indicates that there must be a threshold of aneuploidy for cancer. Likewise, the two flat but near-diploid aneuploid CHE clones, M8-1 and M8-3, described above may be examples of aneuploidy below the threshold of morphological transformation. This threshold of aneuploidy for carcinogenesis would depend either on aneuploidy of chromosomes that encode the proper molecules to change the phenotype of a given differentiated cell to a cancer cell or on aneuploidy of chromosomes that generate genetic instability, which would then change the phenotype via further aneuploidy.
As an explanation for genetic instability, aneuploidy also makes fewer assumptions than the mutation hypothesis. For example, the mutation hypothesis has to assume a “mutator” phenotype (14) to explain the “confusing plethora” (9) of diverse phenotypes and karyotypes among cells from the same cancer (1, 11, 13), but a mutator phenotype has not been experimentally confirmed (13, 15, 18–21). In view of this, even a hit and run mutator has recently been postulated (63). According to the aneuploidy hypothesis, genetic instability is just an inherent consequence of aneuploidy.
It follows that aneuploidy offers a consistent explanation for cancer-specific phenotypes and genetic instability that is independent of gene mutation. This hypothesis also resolves the apparent paradox that cancers are clonal yet heterogeneous: cancers are clonal with regard to aneuploidy but not with regard to the resulting heterogeneity. Nevertheless, the mechanism and kinetics of how carcinogens generate aneuploidy and what levels of aneuploidy are necessary for carcinogenesis have yet to be studied.
Acknowledgments
We thank Athel Cornish-Bowden (Marseille), Alwin Kraemer (Mannheim), and Felix Mitelman (Lund) for critical reviews of the manuscript, discussions, and encouragement. Most of this work was carried out and supported by the Forschungsfond der Fakultaet fuer Klinische Medizin Mannheim, and by the III Medizinische Klinik in Mannheim of the University of Heidelberg. P.D. was working at the III Medizinische Klinik during a sabbatical leave from University of California, Berkeley. All work in Berkeley was supported by Robert Leppo (philanthropist, San Francisco), the Abraham J. and Phyllis Katz Foundation (New York), and the Nathan Cummings Foundation (San Francisco), with additional donations from Carol J. Wilhelmy (San Mateo) and other private sources.
ABBREVIATIONS
- CHE
Chinese hamster embryo
- mn
modal chromosome number
- pf
ploidy factor
References
- 1.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Cancer: Science and Society. San Francisco: Freeman; 1978. [Google Scholar]
- 3.Heim S, Mandahl N, Mitelman F. Cancer Res. 1988;48:5911–5916. [PubMed] [Google Scholar]
- 4.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. 2nd Ed. New York: Elsevier; 1990. [Google Scholar]
- 5.Mitelman F. Catalogue of Chromosome Aberrations in Cancer. 4th Ed. New York: Wiley–Liss; 1994. [DOI] [PubMed] [Google Scholar]
- 6.Johansson B, Mertens F, Mitelman F. Genes Chromosomes Cancer. 1996;16:155–163. doi: 10.1002/(SICI)1098-2264(199607)16:3<155::AID-GCC1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Mitelman, F., Mertens, F. & Johansson, B. (1997) Nat. Genet. 15, Suppl., 417–474. [DOI] [PubMed]
- 8.Shapiro J, Yung W-K, Shapiro W R. Cancer Res. 1981;41:2349–2359. [PubMed] [Google Scholar]
- 9.Heim S, Mitelman F. Cancer Cytogenetics. New York: Wiley–Liss; 1995. [Google Scholar]
- 10.Hauschka T S. Cancer Res. 1961;21:957–981. [PubMed] [Google Scholar]
- 11.Rubin H. Cancer Res. 1985;45:2935–2942. [PubMed] [Google Scholar]
- 12.Holliday R. Trends Genet. 1989;5:42–45. doi: 10.1016/0168-9525(89)90020-6. [DOI] [PubMed] [Google Scholar]
- 13.Heppner G, Miller F R. Int Rev Cytol. 1998;177:1–56. doi: 10.1016/s0074-7696(08)62230-5. [DOI] [PubMed] [Google Scholar]
- 14.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 15.Strauss B S. Cancer Res. 1992;52:249–253. [PubMed] [Google Scholar]
- 16.Orr-Weaver T L, Weinberg R A. Nature (London) 1998;392:223–224. doi: 10.1038/32520. [DOI] [PubMed] [Google Scholar]
- 17.Lengnauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 18.Kinzler K, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 19.Harris C C. Cancer Res. 1991;51:5023s–5044s. [PubMed] [Google Scholar]
- 20.Barrett J C, Tsutsui T, Tsly T, Oshimura M. In: Genetic Mechanisms in Carcinogenesis and Tumor Progression. Harris C C, Liotta L A, editors. New York: Wiley–Liss; 1990. pp. 97–114. [Google Scholar]
- 21.Jakubczak R J, Merlino G, French J E, Muller W J, Paul B, Adhya S, Garges S. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heim S, Johansson B, Mertens F. Mutat Res. 1989;21:39–51. doi: 10.1016/0165-1110(89)90044-4. [DOI] [PubMed] [Google Scholar]
- 23.Burke D, Gasdaska P, Hartwell L. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futcher B, Carbon J. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer V W, Aguilera A. Mutat Res. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Yerganian G, Duesberg P, Kraemer A, Willer A, Rausch C, Hehlmann R. Proc Natl Acad Sci USA. 1997;94:14506–14511. doi: 10.1073/pnas.94.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cram L S, Bartholdi M F, Ray F A, Travis G L, Kraemer P M. Cancer Res. 1983;43:4828–4837. [PubMed] [Google Scholar]
- 28.Benedict W F. J Natl Cancer Inst. 1972;49:585–590. [PubMed] [Google Scholar]
- 29.Matsuoka A, Ozaki M, Takeshita K, Sakamoto H, Glatt H, Hayashi M, Sofuni T. Mutagenesis. 1997;12:365–372. doi: 10.1093/mutage/12.5.365. [DOI] [PubMed] [Google Scholar]
- 30.American Type Culture Collection. Catalogue of Cell Lines & Hybridomas. Rockville, MD: American Type Culture Collection; 1992. [Google Scholar]
- 31.Heim S. In: Genetic Instability in Cancer. Lindahl T, Tooze J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. , Cancer Surv., vol. 28, pp. 247–260. [Google Scholar]
- 32.Shapiro B L. Am J Med Genet. 1983;14:241–269. doi: 10.1002/ajmg.1320140206. [DOI] [PubMed] [Google Scholar]
- 33.Epstein C. The Consequences of Chromosome Imbalance: Principles, Mechanisms, and Models. Cambridge, U. K.: Cambridge Univ. Press; 1986. [Google Scholar]
- 34.Cahill D P, Lengnauer C, Yu J, Riggins G J, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 35.Holliday R. In: Genetic Instability in Cancer. Lindahl T, Tooze J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. , Cancer Surv. 28, pp. 103–115. [Google Scholar]
- 36.Lewin B. Genes V. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 37.Duesberg P H, Schwartz J R. Prog Nucleic Acid Res Mol Biol. 1992;43:135–204. doi: 10.1016/s0079-6603(08)61047-8. [DOI] [PubMed] [Google Scholar]
- 38.Koshland D. Science. 1994;266:1925. doi: 10.1126/science.7801114. [DOI] [PubMed] [Google Scholar]
- 39.German J. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. pp. 601–617. [Google Scholar]
- 40.German J. Cancer Genet Cytogenet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 41.German J. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 42.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. In: What is Cancer? Theories on Carcinogenesis. Root-Bernstein R, Den Otter W, editors. Kapandriti, Greece: Anticancer Research; 1998. [PubMed] [Google Scholar]
- 43.Bialy H. Nat Biotechnol. 1998;16:137–138. doi: 10.1038/nbt0298-137. [DOI] [PubMed] [Google Scholar]
- 44.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. New York: Freeman; 1995. [Google Scholar]
- 45.Kacser H, Burns J A. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fell D. Understanding the Control of Metabolism. London: Portland Press; 1997. [Google Scholar]
- 47.Lindsley D L, Sandler L, Baker B S, Carpenter A T C, Denell R E, Hall J C, Jacobs P A, Gabor Miklos G L, Davis B K, Gethmann R C, et al. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandler L, Hecht F. Am J Hum Genet. 1973;25:332–339. [PMC free article] [PubMed] [Google Scholar]
- 49.Papp I, Iglesias V A, Moscone E A, Michalowski S, Spiker S, Park Y-D, Matzke M A, Matzke A J M. Plant J. 1996;10:469–478. doi: 10.1046/j.1365-313x.1996.10030469.x. [DOI] [PubMed] [Google Scholar]
- 50.Cornish-Bowden A, Homeyr J-H S, Cardenas M L. Bioorg Chem. 1995;23:439–449. [Google Scholar]
- 51.Lijinsky W. Environ Mol Mutagen. 1989;14:78–84. doi: 10.1002/em.2850140615. [DOI] [PubMed] [Google Scholar]
- 52.Stanbridge E J. Annu Rev Genet. 1990;24:615–657. doi: 10.1146/annurev.ge.24.120190.003151. [DOI] [PubMed] [Google Scholar]
- 53.Augenlicht L H, Wahrman M Z, Halsey H, Anderson L, Taylor J, Lipkin M. Cancer Res. 1987;47:6017–6021. [PubMed] [Google Scholar]
- 54.Duesberg P. Science. 1995;267:1407–1408. doi: 10.1126/science.7794335. [DOI] [PubMed] [Google Scholar]
- 55.Hua V Y, Wang W K, Duesberg P H. Proc Natl Acad Sci USA. 1997;94:9614–9619. doi: 10.1073/pnas.94.18.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 57.Bos J L, Fearon E R, Hamilton S R, Verlaan-de Vries M, van Boom J H, van der Eb A J, Vogelstein B. Nature (London) 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 58.Cooper G M. Oncogenes. Boston: Jones and Bartlett; 1990. [Google Scholar]
- 59.Jensen J C, Thilly W G. Mutat Res. 1986;160:95–112. doi: 10.1016/0027-5107(86)90033-3. [DOI] [PubMed] [Google Scholar]
- 60.Galloway S M, Buckton K E. Cytogenet Cell Genet. 1978;20:78–96. doi: 10.1159/000130842. [DOI] [PubMed] [Google Scholar]
- 61.Cimino M, Tice R R, Liang J C. Mutat Res. 1986;167:107–122. doi: 10.1016/0165-1110(86)90012-6. [DOI] [PubMed] [Google Scholar]
- 62.Cornish-Bowden A. In: Biotechnology. Rehm H-J, Reed G, editors. New York: VCH; 1995. pp. 121–136. [Google Scholar]
- 63.Loeb L A. In: Cancer Surveys, vol. 28. Lindahl T, Tooze J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 329–342. [Google Scholar]