Abstract
The highly developed endoplasmic reticulum (ER) structure of pancreatic β-cells is a key factor in β-cell function. Here we examined whether ER stress-induced activation of activating transcription factor (ATF)-6 impairs insulin gene expression via up-regulation of the orphan nuclear receptor small heterodimer partner (SHP; NR0B2), which has been shown to play a role in β-cell dysfunction. We examined whether ER stress decreases insulin gene expression, and this process is mediated by ATF6. A small interfering RNA that targeted SHP was used to determine whether the effect of ATF6 on insulin gene expression is mediated by SHP. We also measured the expression level of ATF6 in pancreatic islets in Otsuka Long Evans Tokushima Fatty rats, a rodent model of type 2 diabetes. High glucose concentration (30 mmol/liter glucose) increased ER stress in INS-1 cells. ER stress induced by tunicamycin, thapsigargin, or dithiotreitol decreased insulin gene transcription. ATF6 inhibited insulin promoter activity, whereas X-box binding protein-1 and ATF4 did not. Adenovirus-mediated overexpression of active form of ATF6 in INS-1 cells impaired insulin gene expression and secretion. ATF6 also down-regulated pancreatic duodenal homeobox factor-1 and RIPE3b1/MafA gene expression and repressed the cooperative action of pancreatic duodenal homeobox factor-1, RIPE3b1/MafA, and β-cell E box transactivator 2 in stimulating insulin transcription. The ATF6-induced suppression of insulin gene expression was associated with up-regulation of SHP gene expression. Finally, we found that expression of ATF6 was increased in the pancreatic islets of diabetic Otsuka Long Evans Tokushima Fatty rats, compared with their lean, nondiabetic counterparts, Long-Evans Tokushima Otsuka rats. Collectively, this study shows that ER stress-induced activation of ATF6 plays an important role in the development of β-cell dysfunction.
THE HIGHLY DEVELOPED structure of the endoplasmic reticulum (ER) in pancreatic β-cells is related to its heavy engagement in insulin secretion. Thus, proper function of the ER is a key factor in β-cell survival, and any perturbation of ER function inevitably impacts insulin secretion (1,2). There are many physiological and pathological conditions involving an increased demand for insulin biosynthesis that can disturb ER function, resulting in what is termed ER stress (1,2). The ER stress response involves the function of three molecular components (1,2): pancreatic ER kinase (PERK), interferon response element (IRE)-1/X-box binding protein (XBP)-1, and activating transcription factor (ATF)-6. Accumulating evidence indicates that PERK- and IRE1-mediated signaling pathways that are activated in response to ER stress play an important role in the pathogenesis of β-cell dysfunction (3,4). Mutations that affect PERK activation in response to ER stress and the downstream effector of PERK, the translation initiation complex eukaryotic initiation factor 2 (eIF2), have a profound impact on islet cell function and survival (3). Studies of the Akita mouse, which carries a missense mutation in Insulin2 (resulting in a Cys96Tyr substitution) that results in a conformational change in the protein, have shown that ER stress in β-cells leads to apoptosis via CCAAT/enhancer-binding protein homologous protein (CHOP) induction (4). It has also been shown that chemical-induced ER stress causes the degradation of mRNA transcripts of the insulin gene through activation of the IRE1-XBP1 pathway, without affecting the activity of the insulin promoter (5). However, little is known on the role of ATF6 in β-cell function, particularly in high glucose-induced ER stress and insulin gene transcription.
ATF6 is an ER membrane-bound transcription factor (6). It is a member of the ATF/cAMP response element-binding protein basic-leucine zipper family of DNA-binding proteins (7). Upon the induction of ER stress, ATF6 translocates from the ER to the Golgi (8,9), in which it is cleaved by site 1 and 2 proteases (10). Proteolytic cleavage of ATF6 directly induces transcriptional activation of chaperone molecules and other enzymes that are essential for protein folding (8,10,11,12). In addition to posttranslational modification of ATF6, accumulating evidence suggests that ER stressors, including hypoxia, tunicamycin, and thapsigargin, up-regulate ATF6 mRNA expression, which suggests that an increase in the expression of ATF6 is also important for the ER stress response (13,14,15). In a previous study (16), we demonstrated that glucotoxicity in cultured INS-1 cells is mediated by atypical orphan nuclear receptor, small heterodimer partner (SHP), which functions as an inhibitor of nuclear receptor signaling in a variety of metabolic pathway (17,18,19,20,21,22). We found that ATF6 increases SHP gene expression, which suggested that ATF6 may have a role in the development of β-cell dysfunction, similar to IRE1 and PERK.
In the current study, we examined whether high glucose conditions induces ER stress in cultured INS-1 cells and whether ER stress impairs insulin gene transcription. We investigated whether ER stress-impaired insulin gene transcription is mediated by ATF6 and examined whether ATF6 expression is up-regulated in the pancreatic islets of Otsuka Long Evans Tokushima Fatty (OLETF) rats, an animal model of type 2 diabetes.
Materials and Methods
Cell culture
The INS-1 rat insulinoma cell line was cultured in 5% CO2-95% air at 37 C in RPMI 1640 (Life Technologies, Inc., Grand Island, NY) containing 11.2 mmol/liter glucose and 2 mmol/liter l-glutamine. The medium was supplemented with 10% fetal bovine serum (FBS), 1 mmol/liter pyruvate, 10 mmol/liter HEPES, 50 μmol/liter 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin (INS-1 medium). All experiments were performed using INS-1 cells between their 20th and 30th passages.
Animals
Male 32-wk-old OLETF rats and their lean nondiabetic counterparts, Long-Evans Tokushima Otsuka (LETO) rats, were supplied by the Otsuka Pharmaceutical Co. (Tokushima, Japan). All animal procedures were carried out in accordance with institutional guidelines for animal research.
Intraperitoneal glucose tolerance test
For the ip glucose tolerance test (IPGTT), rats were fasted for 14 h, and then glucose was injected ip (2 g/kg body weight). Blood was drawn from the tail vein every 30 min after glucose injection. Blood glucose levels were measured using glucose reagent strips and a glucometer (Abbott, Bedford, MA).
Islet isolation
Pancreatic islets were isolated from OLETF and LETO rats using a collagenase digestion technique. Briefly, animals were anesthetized by Nembutal and then euthanized by exsanguination. A solution of collagenase type V (1 mg/ml; Sigma, St. Louis, MO) in Hanks’ balanced salt solution was transfused into the pancreatic ducts via the common bile duct. The dissected pancreas was incubated for 10–15 min at 37 C in a water bath with shaking every 3 min. The islets were removed under a stereoscope (×1.5 magnification) and placed in Krebs Ringer bicarbonate (KRB) buffer containing 1% BSA, penicillin (100 U/ml), and streptomycin (0.1 mg/ml). Between 400 and 500 islets per rat were retrieved.
Generation of recombinant adenovirus
The cDNA encoding a constitutively active form of ATF6 (amino acids 1–373, 50 kDa, cytosolic N-terminal portion of ATF6) was inserted into the EcoRI/XhoI sites of the shuttle vector pAdTrack-CMV. The vector was then electrophoresed into BJ5183 cells that contained the adenoviral vector Adeasy to generate a recombinant adenoviral plasmid. Recombinants were amplified in HEK-293 cells and purified by CsCl (Sigma) gradient centrifugation. Viral preparations were collected and desalted, and titers were determined using Adeno-X rapid titer (BD Bioscience, San Jose, CA), according to the manufacturer’s instructions. The efficiency of adenoviral infection was assessed using a recombinant adenovirus encoding ATF6 fused to green fluorescent protein (GFP; data not shown).
Construction of small interfering RNA (siRNA) for ATF6 and SHP
The ATF6 and SHP siRNA was chemically synthesized by Samchully Pharm and Bioneer, respectively (Seoul, Korea), deprotected, and annealed. Transfections were carried out according to the manufacturer’s instructions. Briefly, INS-1 cells were plated at a density of 3 × 106 cells per 60-mm dish and cultured for 2 d in INS-1 medium. Cells were transfected with 100 nmol/liter of ATF6 or SHP siRNA oligonucleotide using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). Twenty-four hours after transfection, total RNA was isolated for Northern blot analysis. The sequences of ATF6 and SHP siRNAs and the nonspecific control siRNA were as follows: rat ATF6 siRNA, CCA UUG UGU UAC CAG CAA U tt (sense); rat SHP siRNA, AAA GAU CUU GCU AGA GGA ACC tt (sense); nonspecific control siRNA, GGA GUA CGC AUA CCU GAA AGG tt (sense). The effect of ATF6 siRNA and SHP siRNA on the expression of endogenous SHP mRNA and ATF6 mRNA were measured by RT-PCR.
XTT assay
INS-1 cells were placed in a 96-well plate to obtain 1 × 103 per well. Cells were then cultured for various times in media containing the indicated concentrations of tunicamycin, thapsigargin, dithiotreitol (DTT), or adenovirus expressing ATF6 or SHP. The survival rate after each treatment was measured by the 2,3-bis(2-methoxy-4-nitro-5-sulfonphenyl)-2H-tetrazolium-5-carboxanilide inner salt (XTT) assay kit (WEL GENE, Seoul, Korea) according to the manufacturer’s instructions. The data were compared and analyzed by using the OD of the control group as a reference. After repeating each measurement three times, the average of the results of the three measurements was used.
Northern blot analysis
INS-1 cells were plated at a density of 3 × 106 per 60-mm dish and cultured for 2 d in INS-1 medium. Cells were then cultured for various times in media containing the indicated concentrations of glucose, with or without adenovirus. The media were changed every 24 h. An XTT assay for cell viability was performed before RNA was isolated (supplemental Fig. 1 published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org). Total RNA was isolated from the cells using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, and 20 μg of total RNA from each sample were used. The probes for CHOP, ATF6, SHP, insulin, pancreatic duodenal homeobox factor-1 (PDX-1), RIPE3b1/MafA, and β-cell E box transactivator 2 (BETA2) were labeled with [α-32P]dCTP using a random-primer DNA-labeling system (Amersham Biosciences, Little Chalfont, UK).
Western blot analysis
Cells were plated and cultured as described above for Northern blot analysis. Cell lysates were prepared using IPH lysis buffer [50 mmol/liter Tris (pH 8.0), 150 mmol/liter NaCl, 5 mmol/liter EDTA, 0.1 mmol/liter phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40] containing proteinase inhibitors and DTT. The proteins were resolved by SDS-PAGE and then transferred electrophoretically to a polyvinyl difluoride membrane (Millipore, Bedford, MA). The membrane was blocked by incubation in blocking buffer, incubated with anti-PDX-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-RIPE3b1/MafA antibody (Bethyl Laboratories Inc., Montgomery, TX), anti-BETA2 antibody (Santa Cruz), antiphosphorylated (p)-eIF2α antibody (Cell Signaling, Beverly, MA), anti-ATF4 antibody (Santa Cruz), or anti-ATF6 antibody (Labprontier, Seoul, Korea), and then washed and incubated with horseradish peroxidase-conjugated secondary antibody. Immunoreactive proteins were visualized using chemiluminescence, according to the manufacturer’s instructions (Amersham). The membrane was reblotted with antiactin antibody to verify equal loading of protein in each lane. Densitometry was used to quantitate the results, using the digitalized scientific software program UN-SCAN-IT (Silk Scientific Corp., Orem, UT).
Analysis of splicing of XBP-1 mRNA
Total RNA was obtained from INS-1 cells using Trizol reagent (Invitrogen). The cDNA was synthesized using a first-strand cDNA synthesis kit (Fermentas, Hanover, MD) and 2 μg of total RNA, according to the manufacturer’s instructions. PCR was carried out under the following conditions using Taq polymerase (Takara, Tokyo, Japan): 92 C for 3 min for 40 cycles; 92 C for 45 sec; 52 C for 45 sec; and 72 C for 45 sec. The following primers were used: XBP-1, 5′-AAA CAG AGT AGC AGC GCA GAC TGC-3′ (forward) and 5′-GGA TCT CTA AAA CTA GAG GCT TGG TG-3′ (reverse); β-actin, 5′-GGC ATC GTC ACC AAC TGG GAC-3′ (forward), and 5′-CGA TTT CCC GCT CCG TGG-3′ (reverse). The purified PCR products were digested by Pst1 for 5 h at 37 C and then separated by 2% agarose gel electrophoresis.
In vitro transient transfection and gene reporter assays
INS-1 cells were plated at a density of 3 × 105 cell/well in a 12-well plate and cultured for 2 d in INS-1 medium. COS-1 cells were plated at a density of 1 × 105 in a 12-well plate and cultured for 1 d in DMEM (Life Technologies) containing 10% FBS. Cells were transiently transfected with the indicated promoter constructs (200 ng/well) and other cDNAs using Lipofectamine 2000 transfection reagent (Invitrogen). Cells were cotransfected with a plasmid encoding β-galactosidase as an internal control. Cells were transfected for 4 h, washed to remove plasmids, and then cultured in media containing the indicated concentrations of glucose (INS-1) or in culture medium alone (COS-1). Cells were harvested approximately 24 h after transfection and assayed for luciferase and β-galactosidase activity. Twenty microliters of cell lysate containing 15 μg of protein were analyzed using the luciferase assay system, according to the manufacturer’s instructions (Promega, Madison, WI). Luciferase activity was measured using a SIRUS luminometer (Berthold, Pforzheim, Germany), and luciferase activity was normalized to β-galactosidase activity. The human insulin promoter construct phINS-362Luc (−362 to +27) was generously provided by Dr. K. Yamagata (Osaka University, Osaka, Japan) (23).
Measurement of insulin secretion
INS-1 cells were plated and cultured as described above for the reporter assay. To examine the effect of expression of ATF6 on glucose-stimulated insulin secretion (GSIS), INS-1 cells were infected with adenovirus encoding ATF6 (Ad-ATF6) or GFP (Ad-GFP) at a multiplicity of infection (MOI) of 50 for 2 h. The culture medium was then changed to fresh RPMI 1640 containing 10% FBS, and the cells were cultured for an additional 24 h. The cells were then starved in medium containing 3 mmol/liter glucose and 2% FBS for 5 h and then incubated for 1 h at 37 C in modified KRB solution [114 mmol/liter NaCl, 4.4 mmol/liter KCl, 1.28 mmol/liter CaCl2, 1 mmol/liter MgSO4, 29.5 mmol/liter NaHCO3, 10 mmol/liter HEPES, 5 mmol/liter glucose, 0.1% BSA (pH 7.4) (adjusted with NaOH)] with or without 16.7 mmol/liter glucose. The supernatant (200 μl) was carefully collected and subjected to a rat insulin RIA (Linco Research, St. Charles, MO).
Immunofluorescence analysis
Cells were plated on cover glasses before incubating them for 2 d in INS-1 medium. After cultivation for various times in media containing 30 mmol/liter glucose, the cells were fixed in 2% paraformaldehyde for 15 min and then permeabilized with 0.2% Triton X-100 for 15 min at room temperature. After incubation with ATF6 antibody and then with Alexa fluor 488 secondary antibody (Invitrogen, Eugene, OR), the cells were observed through an inverted MRc5 Carl Zeiss fluorescence microscopy (Thornwood, NY).
Immunohistochemical analysis
The entire pancreas was isolated from OLETF and LETO rats, fixed in 4% paraformaldehyde, and then processed for paraffin embedding. Immunohistochemical staining was performed using anti-ATF6 primary antibody (Labprontier) and horseradish peroxidase-conjugated antirabbit IgG secondary antibody (Dako, Glostrup, Denmark), according to manufacturer’s instructions.
Statistical analysis
Data represent the means ± se of at least three independent experiments. Statistical comparisons were made with the Student’s unpaired t test. P < 0.05 was considered statistically significant.
Results
High glucose induces ER stress in INS-1 cells
To determine whether exposure to high concentrations of glucose induces ER stress, INS-1 cells were cultured in the presence of elevated glucose, and insulin mRNA was analyzed by Northern blot. Under basal level glucose conditions (5 mmol/liter glucose), insulin mRNA expression was abundant in INS-1 cells. However, in the presence of 30 mmol/liter glucose, insulin mRNA expression decreased gradually in a time-dependent manner and was faintly detectable after 96 h of exposure. In contrast, CHOP mRNA was expressed at low levels under basal glucose conditions, and exposure to 30 mmol/liter glucose increased its expression (Fig. 1A). In the presence of high glucose, phosphorylation of eIF2α gradually increased from 24 to 96 h. The level of ATF4 increased slightly during the first 24–48 h of exposure but then decreased on longer exposure (72–96 h) to high glucose. The activated form of ATF6 (50 kDa, cytosolic N-terminal portion of ATF6) gradually increased during 24–96 h of exposure to high glucose (Fig. 1B). Splicing of XBP-1 was induced after 24 h of high glucose exposure. Although initially the unspliced form of XBP-1 decreased, it subsequently increased at the 48-h time point, which suggested that prolonged elevation of ER stress by chronic exposure to high glucose induces XBP-1 transcription (Fig. 1C).
Figure 1.
High glucose concentrations induce ER stress in INS-1 cells. A, Representative Northern blot analysis of insulin and CHOP mRNA expression in the presence of high glucose. INS-1 cells were incubated with 30 mmol/liter glucose for increasing lengths of time, and mRNAs of the indicated genes were examined. 18S rRNA levels were analyzed as an internal control. B, Representative Western blot analysis of p-eIF2α, ATF4, and the 50-kDa activated form of ATF6 in the presence of high glucose. INS-1 cells were incubated with 30 mmol/liter glucose for the indicated times. C, Representative RT-PCR analysis of XBP-1 splicing in the presence of high glucose. INS-1 cells were incubated with 30 mmol/liter glucose for the indicated time or 1 μmol/liter thapsigargin (Tg) for 5 h, and mRNA levels at each time point were assessed. β-Actin mRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.05; **, P < 0.01; and #, P < 0.001, compared with 0 h (5 mmol/liter glucose).
High glucose induces nuclear localization of ATF6
Because the induction of ER stress results in the translocation of ATF6 from the ER to the nucleus, we examined whether high glucose increases the nuclear localization of ATF6 by using immunofluorescence. As shown in Fig. 2, at basal levels of glucose, ATF6 localized to the cytoplasm in INS-1 cells. As expected, treatment with thapsigargin-induced nuclear localization of ATF6 in INS-1 cells. Moreover, exposure to 30 mmol/liter glucose for 48 h induced the nuclear localization of ATF6 and this nuclear localization was sustained for 96 h.
Figure 2.
High glucose induces nuclear localization of ATF6. Representative immunofluorescence images of ATF6 in presence of high glucose are shown. INS-1 cells were incubated with 30 mmol/liter glucose for 48 and 96 h or 1 μmol/liter thapsigargin (Tg; positive control) for 1 h. ATF6 is visualized in red, 4′,6′-diamino-2-phenylindole (DAPI) nuclear staining in blue.
ER stress in INS-1 cells impairs insulin gene expression
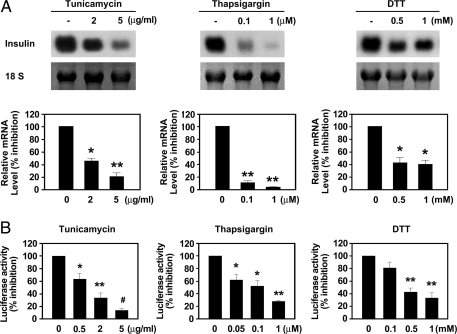
To determine whether increased ER stress impairs insulin gene expression, we examined the effect of tunicamycin, thapsigargin, or DTT on insulin mRNA expression by Northern blot analysis. Tunicamycin, thapsigargin, and DTT markedly decreased insulin mRNA expression in INS-1 cells in a dose-dependent manner (Fig. 3A). Cell viability after treatment with ER stress inducers was assessed by an XTT assay. ER stress inducers had no influence on cell viability (supplemental Fig. 1, A–C). To determine whether ER stress-induced suppression of insulin gene expression is mediated at the transcriptional level, we examined human insulin promoter activity in the presence of tunicamycin, thapsigargin, and DTT. As shown in Fig. 3B, tunicamycin, thapsigargin, and DTT significantly inhibited insulin promoter activity in a dose-dependent manner. These results suggested that ER stress in pancreatic β-cells impairs insulin gene expression at the transcriptional level.
Figure 3.
ER stress inhibits insulin gene transcription in INS-1 cells. A, Representative Northern blot analysis of insulin mRNA expression in cells treated with tunicamycin, thapsigargin, or DTT. INS-1 cells were incubated with indicated concentrations of tunicamycin for 24 h, thapsigargin for 5 h, or DTT for 5 h. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01; **, P < 0.001, compared with untreated cells. B, Effect of tunicamycin, thapsigargin, or DTT on insulin promoter activity. INS-1 cells were transfected with a human insulin promoter-luciferase reporter gene construct (insulin promoter, 200 ng/well) and then stimulated with the indicated concentrations of tunicamycin for 24 h, thapsigargin for 5 h, or DTT for 5 h. Data represent the means ± se of three independent measurements. *, P < 0.05; **, P < 0.01; #, P < 0.001, compared with untreated cells.
ER stress-induced impairment of insulin gene transcription is mediated by ATF6
Next, we examined which of the three different signaling arms of the ER stress response (ATF6, IRE1, and PERK) mediated the impairment of insulin gene expression. As shown in Fig. 4A (upper panel), transient expression of active form of ATF6 in INS-1 cells significantly inhibited insulin promoter activity in a dose-dependent manner. In contrast, expression of the spliced form of XBP1, which functions downstream of IRE1, tended to decrease insulin promoter activity, but this effect was not significant (Fig. 4A, middle panel). Quite unexpectedly, ATF4, a key factor in the PERK pathway, increased insulin promoter activity (Fig. 4A, lower panel). To further confirm that the suppression of insulin gene expression by high glucose concentrations is mediated by ATF6, we down-regulated endogenous ATF6 expression by transfecting INS-1 cells with siRNA-ATF6 before incubating them in medium containing 5 mmol/liter or 30 mmol/liter glucose for 48 h. siRNA-ATF6 significantly inhibited the up-regulation of ATF6 expression at the high glucose concentration (Fig. 4B). Moreover, it significantly blocked high glucose-induced suppression of insulin gene expression (Fig. 4C).
Figure 4.
ER stress-induced suppression of insulin gene transcription is mediated by ATF6. A, Upper panel, Effect of ATF6 on insulin promoter activity. INS-1 cells were cotransfected with the insulin promoter (200 ng/well) and the indicated amounts of an expression vector for active form of ATF6 for 24 h. Middle panel, Effect of XBP-1 on insulin promoter activity. INS-1 cells were cotransfected with the insulin promoter (200 ng/well) and the indicated amounts of an expression vector for the spliced form of XBP-1 for 24 h. Lower panel, Effect of ATF4 on insulin promoter activity. INS-1 cells were cotransfected with the insulin promoter (200 ng/well) and the indicated amounts of an expression vector for ATF4 for 24 h. Data represent the means ± se of three independent measurements. *, P < 0.05; **, P < 0.01; #, P < 0.001, compared with untreated cells. B, INS-1 cells were transfected with 100 nmol/liter of siRNA-ATF6 or control siRNA (Con) and then incubated with the indicated concentrations of glucose for 48 h. The RNA levels at the various time points were normalized by using the β-actin levels. C, Representative Northern blot analysis of the effect of siRNA-ATF6 on the high glucose concentration-induced decrease in insulin gene expression. INS-1 cells were transfected with 100 nmol/liter of siRNA-ATF6 or control siRNA and then incubated with the indicated concentrations of glucose for 48 h. 18S rRNA levels were analyzed as an internal control. Data in the bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01, compared with 30 mmol/liter glucose with or without control siRNA (Con).
Effect of ATF6 on insulin mRNA expression and GSIS
Based on the results of the transient transfection assay, we were interested in determining the physiological role of ATF6 in β-cell function, specifically insulin expression and GSIS. To determine the effect of ATF6 on the expression of the insulin gene, INS-1 cells were infected with an adenovirus encoding a constitutively active form of ATF6 (Ad-ATF6) at an MOI of 10, 50, or 100 and then cultured in medium containing 5 mmol/liter glucose. Consistent with the effect of ATF6 on insulin promoter activity, Ad-ATF6 down-regulated insulin mRNA expression in a dose-dependent manner (Fig. 5A). Ad-ATF6 also impaired GSIS in INS-1 cells. Static stimulation with 16.7 mmol/liter glucose significantly increased insulin secretion in INS-1 cells, and GSIS was significantly inhibited by Ad-ATF6 but not Ad-GFP (Fig. 5C).
Figure 5.
Effect of overexpression of ATF6 on insulin gene expression; the transcriptional activity of PDX-1, BETA2, and RIPE3b1/MafA; and glucose-stimulated insulin secretion. A, Representative Northern blot analysis of insulin, PDX-1, BETA2, and RIPE3b1/MafA mRNA expression in cells infected with Ad-ATF6. INS-1 cells were infected with the indicated doses of Ad-ATF6 or Ad-GFP for 24 h. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01; **, P < 0.001, compared with Ad-ATF6 (0 MOI) and Ad-GFP (50 and 100 MOI). B, Representative Western blot analysis of PDX-1, BETA2, and RIPE3b1/MafA protein expression in cells infected with Ad-ATF6. INS-1 cells were infected with the indicated does of Ad-ATF6 or Ad-GFP for 24 h. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01; **, P < 0.001, compared with Ad-ATF6 (0 MOI) and Ad-GFP (50 and 100 MOI). C, Effect of Ad-ATF6 on glucose-stimulated insulin secretion. INS-1 cells were infected with Ad-ATF6 or Ad-GFP at an MOI of 50 for 24 h, starved in 3 mmol/liter glucose for 5 h, and then incubated in KRBB with 3 or 16.7 mmol/liter glucose for 1 h. Insulin secretion was measured using a RIA kit. Data represent the means ± se of three independent measurements. *, P < 0.01, compared with 3 mmol/liter glucose. D, Effect of ATF6 expression on PDX-1-, BETA2-, and RIPE3b1/MafA-stimulated insulin promoter activity. COS-1 cells were cotransfected with the insulin promoter (200 ng) and the indicated expression vectors (pcDNA3) for PDX-1 (100 ng), BETA2 (100 ng), and RIPE3b1/MafA (100 ng), with (+) or without (−) an expression vector for activated ATF6 (100 ng) for 24 h. Data represent the means ± se of three independent measurements. *, P < 0.01; **, P < 0.001.
Effects of ATF6 on the expression of β-cell-enriched transcription factors
The transcriptional activity of the insulin gene is primarily regulated by transcription factors that are enriched in expression in β-cells. These include PDX-1, BETA2/NeuroD, and RIPE3b1/MafA (22,24,25,26,27). To determine the mechanism of impairment of insulin gene expression by ATF6, we examined the effect of ATF6 on the expression of transcription factors that are enriched in β-cells. Infection of INS-1 cells with Ad-ATF6 down-regulated PDX-1 and RIPE3b1/MafA mRNA expression in a dose-dependent manner (Fig. 5A). In contrast, Ad-ATF6 had no effect on BETA2 expression (Fig. 5A). The differential pattern of expression of PDX-1, RIPE3b1/MafA, and BETA2 in response to ATF6 correlated with protein expression, as assessed by Western blot analysis (Fig. 5B). Cell viability after infection with Ad-ATF6 was assessed by an XTT assay. Ad-ATF6 had no influence on cell viability (supplemental Fig. 1D).
Effect of ATF6 on cooperative transcriptional activity of PDX-1, BETA2, and RIPE3b1/MafA on insulin promoter
Transcriptional regulation of the insulin gene is a very complex process that requires the cooperation of a number of transcription factors, including PDX-1, BETA2, and RIPE3b1/MafA (28,29). In COS-1 cells, in the absence of exogenous ATF6, expression of PDX-1, BETA2, and RIPE3b1/MafA, either in combinations of two or all three together, significantly activated the insulin promoter. When the cells were also transfected with an expression construct for active form of ATF6, the cooperative action of PDX-1, BETA2, and RIPE3b1/MafA in the stimulation of insulin transcription was repressed (Fig. 5D). Taken together, these results suggested that in addition to suppressing the expression of β-cell-enriched transcription factor genes, ATF6 impairs insulin gene transcription by inhibiting the cooperative action of PDX-1, BETA2, and RIPE3b1/MafA in stimulating the insulin promoter.
SHP mediates ATF6-induced β-cell dysfunction
Previously, we reported that high levels of glucose induce SHP expression and that SHP inhibits insulin gene expression (16). We next examined whether ATF6 up-regulated SHP expression in INS-1 cells and found that Ad-ATF6 increased SHP mRNA expression in a time- and dose-dependent manner (Fig. 6, A and B). To determine whether impaired insulin gene expression was mediated by SHP, we examined the effects of SHP on insulin gene expression and tested whether down-regulation of endogenous SHP expression with SHP-specific siRNA (siRNA-SHP) blocks ATF6-induced suppression of insulin gene expression. Moreover, in cells transfected with siRNA-SHP, Ad-ATF6-induced suppression of insulin gene expression was significantly blocked, whereas the control siRNA had no effect (Fig. 6C). Additionally, we observed that siRNA-SHP significantly blocked Ad-ATF6-induced suppression of PDX-1 gene expression (Fig. 6C). These observations suggested that ER stress-induced β-cell dysfunction is mediated, at least in part, by endogenous SHP.
Figure 6.
Effect of overexpression of ATF6 on SHP expression. A, Representative Northern blot analysis of SHP mRNA expression over time in cells infected with Ad-ATF6. INS-1 cells were infected with Ad-ATF6 or Ad-GFP at an MOI of 100 for the indicated times. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01, compared with Ad-ATF6 (0 h) and Ad-GFP (12 or 24 h). B, Representative Northern blot analysis of SHP mRNA expression in the presence of increasing doses of Ad-ATF6. INS-1 cells were infected with indicated dose (MOI) of Ad-ATF6 or Ad-GFP for 24 h. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01, compared with Ad-ATF6 (0 MOI) and Ad-GFP (50 and 100 MOI). C, Representative Northern blot analysis of the effect of siRNA-SHP on Ad-ATF6-induced decrease of insulin and PDX-1 gene expression. INS-1 cells were transfected with 100 nmol/liter of siRNA-SHP or control siRNA and then incubated with Ad-ATF6 at an MOI of 50 for 24 h. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the means ± se of three independent measurements (bottom panel). *, P < 0.01, compared with Ad-GFP; **, P < 0.01, compared with Ad-ATF6 alone or Ad-ATF6 with control siRNA (Con).
ATF6 expression is up-regulated in the pancreatic islets of OLETF rats
Using immunohistochemical staining, we examined ATF6 expression in the pancreatic islets of two animal models of diabetes. Islets of 32-wk-old LETO rats appeared normal and exhibited minimal positive reactivity, which indicated that ATF6 is expressed at low levels under normal glucose conditions. In 32-wk-old OLETF rats, the islets were enlarged and disorganized, and there was a marked increase in ATF6 expression in the pancreatic islets (Fig. 7A). To assess the ER stress response in the pancreatic islets of two animal models of diabetes, we examined the expression of p-eIF2α and the activated ATF6 by Western blot. The expression of p-eIF2α and activated ATF6 was higher in the pancreatic islets of 32-wk-old OLETF rats, compared with LETO rats. In contrast, the islets of OLETF rats exhibited a lower level of insulin gene expression, compared with LETO rats (Fig. 7, B and C). Next, we examined the expression of SHP in the pancreatic islets of the two rat types by real-time RT-PCR. Consistent with a previous report (16), the pancreatic islets of 32-wk-old OLETF rats showed significantly increased SHP expression, compared with the islets of LETO rats (Fig. 7D). The development of diabetes in 32-wk-old OLETF rats was confirmed by IPGTT (Fig. 7E).
Figure 7.
Expression of ATF6 in the pancreatic islets of OLETF rats. A, Representative immunohistochemical staining of ATF6. Micrographs of pancreatic islets of 32-wk-old OLETF and LETO rats subjected to immunohistochemical staining using anti-ATF6 antibodies. Brown-colored areas represent ATF6-positive cells. B, Representative Northern blot analysis of insulin mRNA expression and Western blot analysis of p-eIF2α and active form ATF6 protein expression in pancreatic islets of OLETF and LETO rats. Pancreatic islets were isolated from 32-wk-old OLETF and LETO rats. C, Quantification of insulin mRNA and active form ATF6 protein expression. Data represent the means ± se of five independent measurements (n = 5 for each group). *, P < 0.01, compared with LETO rats. D, SHP mRNA expressions were measured by real-time RT-PCR. The results are expressed as fold increases of the ratios of mRNA expression of SHP/actin relative to the ratio in LETO rats. Data represent the means ± se of five independent measurements (n = 5 for each group). *, P < 0.01, compared with the equivalent LETO rat values. E, IPGTT. Changes in blood glucose concentration during an IPGTT in 32-wk-old OLETF and LETO rats. Data represent the means ± se of five independent measurements (n = 5 for each group). *, P < 0.01, compared with LETO rats.
Discussion
In this study, we demonstrated that high glucose induces ER stress in INS-1 cells and that ER stress-induced activation of ATF6 impairs insulin gene expression and GSIS. Expression of ATF6 down-regulated PDX-1 and RIPE3b1/MafA gene expression and repressed the cooperative action of PDX-1, BETA2, and RIPE3b1/MafA in stimulating insulin transcription. ATF6-induced β-cell dysfunction was associated with up-regulation of the expression of the orphan nuclear receptor, SHP. Moreover, we showed that the expression of ATF6 protein is higher in the pancreatic islets of OLETF rats, compared with LETO rats. Collectively, these results suggest that ATF6 plays an important role in β-cell dysfunction induced by high glucose conditions.
Because glucose is the major activator of insulin biosynthesis in β-cells, it can be expected that prolonged activation of insulin synthesis by high glucose concentrations may increase ER stress in pancreatic β-cells. Moreover, recent studies indicated cross talk between ER stress and oxidative stress (30,31), although the mechanistic link is still not fully understood. Because β-cells are sensitive to oxidative stress, high glucose-induced oxidative stress in pancreatic β-cells could be one possible mechanism through which high glucose concentrations induce ER stress. In the current study, we demonstrated that prolonged exposure of INS-1 cells to high glucose concentration increases ER stress in INS-1 cells and that the chemical ER stressors tunicamycin, thapsigargin, and DTT-induced ER stress impaired insulin gene expression. We showed that ER stress-induced suppression of insulin gene expression was mediated by activation of ATF6 and that other ER stress response pathways, such as those involving IRE1-XBP1 and PERK-eIF2α-ATF4, were less likely to be involved. We demonstrated that adenovirus-mediated overexpression of ATF6 in INS-1 cells impairs insulin gene expression and GSIS but not basal insulin secretion. Although it is not clear why ATF6 affected GSIS without affecting basal insulin secretion, ATF6 might influence the signaling pathway of GSIS. In addition, the expression of ATF6 was higher in the pancreatic islets of diabetic OLETF rats than in the islets of their nondiabetic counterparts. These results suggest that ER stress-induced β-cell dysfunction is mediated by ATF6 and that this mechanism may play an important role in the pathogenesis of type 2 diabetes. However, thapsigargin (especially at 0.1 μmol/liter) had a more severe inhibitory effect on insulin mRNA expression than insulin promoter activity. Moreover, insulin mRNA expression is very stable, and inhibition of insulin promoter activity should lead to decreased insulin mRNA expression only after 12–24 h and not after 5 h, as presently observed. This raises the possibility that ER stress, besides inhibiting insulin transcription, also induces insulin mRNA degradation, as shown by Pirot et al. (5). Therefore, a remaining issue is to determine which of the component(s) of decreased insulin gene transcription or insulin mRNA degradation contribute the most to ER stress-induced suppression of insulin mRNA expression.
Previously we reported that glucotoxicity in INS-1 cells is mediated by the orphan nuclear receptor SHP (16). High concentrations of glucose increased SHP expression, and this was followed by decreases in insulin enhancer-driven promoter activation, insulin gene expression, and insulin secretion. The mechanism of β-cell dysfunction by SHP involved down-regulation of PDX-1 and RIPE3b1/MafA expression. In this study, we found that SHP gene expression is induced by adenovirus-mediated overexpression of ATF6, suggesting SHP mediates β-cell dysfunction induced by ATF6. Moreover, when endogenous SHP gene expression in INS-1 cells was inhibited by an SHP-specific siRNA, ATF6-induced suppression of insulin gene expression was partly, but significantly, reversed. These results indicate that β-cell dysfunction induced by ER stress is mediated, at least in part, by ATF6-induced transcriptional activation of SHP. However, it remains to be elucidated whether ATF6 directly regulates the expression of the insulin, PDX-1, and RIPE3b1/MafA genes without SHP as mediator because siRNA-SHP did not fully block the effect of Ad-ATF6 on insulin mRNA expression.
The results of the current study showed that exposure of INS-1 cells to high glucose leads to an increase in the expression or activity of factors involved in the ER stress response, including ATF6, XBP1, and CHOP. On the other hand, the increase in ATF4 protein levels in response to high glucose was slight and transient. Moreover, we found unexpectedly that expression of exogenous ATF4 increases insulin promoter activity. The precise mechanism or physiological role of ATF4 in the regulation of insulin promoter activity remains to be elucidated. Recently Yusta et al. (32) showed that the potentiation of ATF4 induction by glucagon-like peptide-1 receptor agonists accelerates recovery from ER stress-mediated translational repression in INS-1 cells. Thus, it is possible that activation of ARF4 plays a role in recovering from β-cell dysfunction induced by high glucose concentration.
In conclusion, we have shown that ER stress-induced activation of ATF6 results in β-cell dysfunction and that ATF6-induced β-cell dysfunction is mediated, at least in part, by the up-regulation of SHP gene expression. However, our data disagree with recent studies showing that cytokines induce ER stress and β-cell death without ATF6 activation (33) and that other pathways of ER stress (such as PERK-eIF2α and IRE1-XBP1) are more relevant to β-cell dysfunction than to ATF6 (34,35). Thus, to more clearly elucidate the role of ATF6 in the development of β-cell dysfunction, it would be useful to generate animals in which ATF6 expression in the pancreatic β-cells is conditionally knocked out.
Supplementary Material
Acknowledgments
We thank Professor Prywer (Department of Biological Sciences, Columbia University, New York, NY) for the gift of ATF6 plasmids.
Footnotes
This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MOST) (R01-2007-000-10057-0 and M10642140004-06N4214-00410) and the NRL program (Grant M106 00000271-06J000-27110). H.-Y.S. and K.-M.L. were supported by the Brain Korea 21 project in 2007.
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 1, 2008
Abbreviations: ATF, Activating transcription factor; BETA2, β-cell E box transactivator 2; CHOP, CCAAT/enhancer-binding protein homologous protein; DTT, dithiotreitol; eIF2, eukaryotic initiation factor 2; ER, endoplasmic reticulum; FBS, fetal bovine serum; GFP, green fluorescent protein; GSIS, glucose-stimulated insulin secretion; IPGTT, ip glucose tolerance test; IRE, interferon response element; KRB, Krebs Ringer bicarbonate; LETO, Long-Evans Tokushima Otsuka; MOI, multiplicity of infection; OLETF, Otsuka Long Evans Tokushima Fatty; p, phosphorylated; PDX-1, pancreatic duodenal homeobox factor-1; PERK, pancreatic ER kinase; SHP, small heterodimer partner; siRNA, small interfering RNA; XBP, X-box binding protein; XXT, 2,3-bis(2-methoxy-4-nitro-5-sulfonphenyl)-2H-tetrazolium-5-carboxanilide inner salt.
References
- Oyadomari S, Araki E, Mori M 2002 Endoplasmic reticulum stress mediated apoptosis in pancreatic β-cell. Apoptosis 7:335–345 [DOI] [PubMed] [Google Scholar]
- Harding HP, Ron D 2002 Endoplasmic reticulum stress and the development of diabetes. Diabetes 51:S455–S461 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D 2001 Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7:1153–1163 [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M 2002 Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest 109:525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirot P, Naamane N, Libert F, Magnusson NE, Ørntoft TF, Cardozo AK, Eizirik DL 2007 Global profiling of genes modified by endoplasmic reticulum stress in pancreatic β cells reveals the early degradation of insulin mRNAs. Diabetologia 50:1006–1014 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanaqi H, Yura T, Mori K 1998 Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. J Biol Chem 273:33741–33749 [DOI] [PubMed] [Google Scholar]
- Hai TW, Liu F, Coukos WJ, Green MR 1989 Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev 3:2083–2090 [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K 1999 Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10:3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen J, Prywes R 2002 The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem 277:13045–13052 [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL 2000 ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6:1355–1364 [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R 2000 Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem 275:27013–27020 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K 2000 ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20:6755–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH 2003 XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H 2005 AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol 25:9554–9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Ishihara T, Tanaka K, Hoshino T, Mizushima T 2007 Transcriptional activation of ATF6 by endoplasmic reticulum stressors. Biochem Biophys Res Commun 355:543–548 [DOI] [PubMed] [Google Scholar]
- Park KG, Lee KM, Seo HY, Suh JH, Kim HS, Wang L, Won KC, Lee HW, Park JY, Lee KU, Kim JG, Kim BW, Choi HS, Lee IK 2007 Glucotoxicity in the INS-1 rat insulinoma cell line is mediated by the orphan nuclear receptor small heterodimer partner. Diabetes 56:431–437 [DOI] [PubMed] [Google Scholar]
- Seol W, Chung M, Moore DD 1997 Novel receptor interaction and repression domains in the orphan receptor SHP. Mol Cell Biol 17:7126–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgius LJ, Steffensen KR, Gustafsson JA, Treuter E 2002 Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J Biol Chem 277:49761–49766 [DOI] [PubMed] [Google Scholar]
- Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J 2002 The small heterodimer partner interacts with the liver X receptor α and represses its transcriptional activity. Mol Endocrinol 16:2065–2076 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A 2004 Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem 279:23158–23165 [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim HJ, Kim KT, Park YY, Seong HA, Park KC, Lee IK, Ha H, Shong M, Park SC, Choi HS 2004 Orphan nuclear receptor small heterodimer partner represses hepatocyte nuclear factor 3/Foxa transactivation via inhibition of its DNA binding. Mol Endocrinol 18:2880–2894 [DOI] [PubMed] [Google Scholar]
- Kim JY, Chu K, Kim HJ, Seong HA, Park KC, Sanyal S, Takeda J, Ha H, Shong M, Tsai MJ, Choi HS 2004 Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix-loop-helix transcription factor BETA2/neuroD. Mol Endocrinol 18:776–790 [DOI] [PubMed] [Google Scholar]
- Okita K, Yang Q, Yamagata K, Hangenfeldt KA, Miyagawa J, Kajimoto Y, Nakajima H, Namba M, Wollheim CB, Hanafusa T, Matsuzawa Y 1999 Human insulin gene is a target gene of hepatocyte nuclear factor-1α (HNF-1α) and HNF-1β,. Biochem Biophys Res Commun 24:566–569 [DOI] [PubMed] [Google Scholar]
- Poitout V, Olson LK, Robertson RP 1996 Chronic exposure of βTC-6 cells to supraphysiologic concentrations of glucose decreases binding of the RIPE3b1 insulin gene transcription activator. J Clin Invest 97:1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson RP, Zhang HJ, Pyzdrowski KL, Walseth TF 1992 Preservation of insulin mRNA levels and insulin secretion in HIT cells by avoidance of chronic exposure to high glucose concentrations. J Clin Invest 90:320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen HV, Serup P, Leonard J, Michelsen BK, Madsen OD 1994 Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc Natl Acad Sci USA 91:10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrot M, Rud J, Moss LG, Sharma A 2002 Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian RIPE3b1/MafA. Proc Natl Acad Sci USA 99:6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Guo M, Huang S, Stein R 2002 Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 22:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R 2005 The islet β cell-enriched RIPE3b1/MafA activator is a key regulator of insulin gene transcription. J Biol Chem 280:11887–11894 [DOI] [PubMed] [Google Scholar]
- Xue X, Piao JH, Nakajima A, Sakon-Komazawa S, Kojima Y, Mori K, Yagita H, Okumura K, Harding H, Nakano H 2005 Tumor necrosis factor α (TNFα) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFα. J Biol Chem 280:33917–33925 [DOI] [PubMed] [Google Scholar]
- Hsieh YH, Su IJ, Lei HY, Lai MD, Chang WW, Huang W 2007 Differential endoplasmic reticulum stress signaling pathways mediated by iNOS. Biochem Biophys Res Commun 359:643–648 [DOI] [PubMed] [Google Scholar]
- Yusta B, Baggio LL, Estall JL, Koehler JA, Holland DP, Li H, Pipeleers D, Ling Z, Drucker DJ 2006 GLP-1 receptor activation improves β cell function and survival following induction of endoplasmic reticulum stress. Cell Metab 4:391–406 [DOI] [PubMed] [Google Scholar]
- Cardozo AK, Ortis F, Storling J, Feng YM, Rasschaert J, Tonnesen M, Van Eylen F, Mandrup-Poulsen T, Herchuelz A, Eizirik DL 2005 Cytokines down-regulate the sarcoendoplasmic reticulum pump Ca2+ ATPase 2b and deplete endoplasmic reticulum Ca2+, leading to induction of endoplasmic reticulum stress in pancreatic β-cells. Diabetes 54:452–461 [DOI] [PubMed] [Google Scholar]
- Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, Urano F 2006 Regulation of insulin biosynthesis in pancreatic β cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab 4:245–254 [DOI] [PubMed] [Google Scholar]
- Cnop M, Ladriere L, Hekerman P, Ortis F, Cardozo AK, Dogusan Z, Flamez D, Boyce M, Yuan J, Eizirik DL 2007 Selective inhibition of eukaryotic translation initiation factor 2α dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic β-cell dysfunction and apoptosis. J Biol Chem 282:3989–3997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.