Abstract
The adipose tissue inflammation accompanying obesity has important consequences for adipocyte lipid metabolism, and increased adipose tissue TNFα plays an important role for mediating the effect of inflammation on adipocyte function. Recent studies have shown that apolipoprotein E (apoE) is highly expressed in adipose tissue where it plays an important role in modulating adipocyte triglyceride metabolism, triglyceride mass, and adipocyte size. We have previously reported that TNFα reduces adipocyte apoE, and the current studies were undertaken to evaluate the molecular mechanism for this regulation. TNFα repression of adipocyte apoE gene expression required an intact nuclear factor (NF)-κB binding site at −43 in the apoE promoter. Site-directed mutagenesis at this site completely eliminated TNFα regulation of an apoE gene reporter. TNFα treatment activated binding of NFκB p50, isolated from adipocyte nuclei, to the apoE promoter. Two structurally distinct inhibitors of NFκB complex activation or translocation abrogated the TNFα effect on the apoE gene. Using chromatin immunoprecipitation assays, we demonstrated that treatment of adipocytes with TNFα led to increased binding of NFκB p50, and decreased binding of p65 and Sp1, to this region of the apoE promoter in living cells. The key role played by increased p50 binding was confirmed by p50 knockdown experiments. Reduction of p50 expression using small interference RNA completely eliminated TNFα-mediated reduction of endogenous adipocyte apoE gene expression. These results establish the molecular link between adipose tissue inflammation and apoE gene expression in adipocytes. The suppression of adipocyte apoE by the proinflammatory adipose tissue milieu associated with obesity will have important downstream effects on adipocyte triglyceride turnover and content.
ADIPOSE TISSUE IS now recognized as an important regulator of organismal metabolism (1,2). The adipose tissue organ, which can average from 20–30% of total body weight in adult humans, modulates metabolism by regulating systemic substrate flux and by secreting factors that modulate gene expression and metabolism in distant organs. Obesity is accompanied by important changes in adipocyte function and in systemic metabolism that have been associated with increased risk for both diabetes and cardiovascular disease (2,3,4,5). Adipocyte dysfunction in obesity has been linked to an inflammatory response in adipose tissue that is characterized by an influx of inflammatory cells. Cross talk between adipocytes and adipose tissue inflammatory cells, primarily macrophages, importantly contributes to adipocyte dysfunction in obesity. One important product of adipose tissue macrophages in the obese state is TNFα (6,7,8,9,10,11). TNFα regulates the expression of numerous adipocyte genes and has been implicated as a significant factor that contributes to adipocyte dysfunction with adverse systemic metabolic and cardiovascular consequences.
apoE production by adipocytes has been known for many years, but only very recently has additional information related to its physiological function and regulation become available (12,13,14,15,16). With respect to physiological function, we have shown that adipocytes from apoE−/− mice are smaller than those from wild-type mice and contain less triglyceride (16). Adipocytes isolated from apoE−/− mice also accumulate significantly less triglyceride after stimulation with peroxisome proliferator-activated receptor-γ (PPAR-γ) agonists, consistent with an important role for adipocyte apoE in mediating the effect of PPARγ agonists on adipocyte lipid metabolism (16). Differences in triglyceride metabolism and content between apoE-expressing and nonexpressing adipocytes and adipose tissue are maintained after incubation in apoE-containing serum or with apoE-rich very-low-density lipoprotein (16). These latter observations underscore the importance of endogenously expressed adipocyte apoE for modulating adipocyte lipid metabolism. Physiologically relevant regulation of adipocyte apoE expression provides further support for the notion that it plays an important role in adipocyte lipid metabolism. Adipocyte apoE expression responds to the in vivo administration of PPARγ agonists or angiotensin II (13,15) and to organismal nutritional perturbation (14). Diet-induced obesity and leptin-deficient obesity substantially reduce adipocyte apoE expression. Obesity leads to inflammatory cell infiltration with increased adipose tissue TNFα, and we have shown that TNFα markedly suppresses adipocyte apoE (13).
The 5′ proximal promoter of the apoE gene has been shown to contain multiple potential positive and negative control elements, including an important Sp1 binding site located at −54 that is required for maximal transcriptional activity (17). Two duplicated 620-bp enhancer elements located 3.3 and 15.9 kb downstream of the apoE gene (designated ME1 and ME2) direct its in vivo expression in macrophages and adipocytes (18,19). These ME elements contain an liver X receptor binding site that mediates the response of the apoE gene to cellular sterol (18). Beyond the importance of these downstream enhancer elements, little is known regarding the molecular mechanism for regulating apoE gene expression in adipocytes, and there is no information in any cell type regarding the molecular mechanism for TNFα regulation of the apoE gene. In the current series of experiments, we investigated the molecular pathway responsible for TNFα regulation of the adipocyte apoE gene.
Materials and Methods
Materials
Cell culture media, supplements, and antibiotics were purchased from Invitrogen (Carlsbad, CA). Recombinant murine TNFα was from R&D Systems (Minneapolis, MN). SN50 and 6-amino-4-(4-phenoxyphenylethylamino) quinazoline (QNZ) were purchased from Biomol (Plymouth Meeting, PA). All other materials were from previously identified sources (20,21).
Cell culture
3T3-L1 cells were purchased from American Type Culture Collection (Rockville, MD) and maintained as fibroblasts. Cells were differentiated into adipocytes, as previously described in detail (13,16). After differentiation, cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum. All experiments were performed on postdifferentiation d 8–10.
Quantitative RT-PCR and immunoblot
Total RNA was isolated from adipocytes after the treatments described in the figure legends. First-strand cDNA was synthesized from 1 μg total RNA using the ThermoScript RT-PCR system according to the manufacturer’s instructions (Life Technologies, Inc., Rockville, MD). cDNA (1.5 μl) was amplified by real-time PCR using Taq SYBR Green Supermix with ROX (Bio-Rad, Foster City, CA). For normalization of RNA loading, samples were standardized to β-actin message abundance. The apoE and β-actin primer pairs have been described previously (13,14). The quantitative PCR was performed in a total of 25 μl. Mixtures were preincubated at 95 C for 3 min, followed by 35 cycles of PCR at 95 C for 30 sec, 59 C for 50 sec, and 72 C for 50 sec. In every case, cycle threshold (CT) taken for quantitation were in the linear portion of the amplification range. The relative abundance of β-actin compared with apoE was higher and was therefore always detected at an earlier cycle that remained relatively stable from experiment to experiment. Each individual sample was analyzed in triplicate using the Mx3000P quantitative PCR system (Stratagene, La Jolla, CA). RNA levels are reported as fold change compared with control using the comparative quantitation analysis software available with the Mx3000P. Change in expression (fold) for apoE is calculated as 2−Δ(ΔCT) where ΔCT = CT (target) − CT (housekeeping), and Δ(ΔCT) = ΔCT (treated) − ΔCT(control). Reaction product purity was confirmed by examination of melting curves for a single peak. Immunoblots were performed and quantitated as previously described (13,16).
Transient transfection and luciferase activity
The 620-bp downstream enhancer (ME1) of the human apoE gene was subcloned into the pGL3 basic luciferase reporter using PCR amplification to create BamH1/Sal1 sites to generate pGLen. Human apoE promoter fragments −623 to +86, −447 to +86, and −300 to +86 were generated by restriction enzyme digestion, and fragment −503 to +86 was generated by PCR amplification. apoE promoter sequences were subcloned into the Kpn1/HindIII site of pGLen or pGL3. The day before transfection, differentiated 3T3-L1 cells (d 8–10) were trypsinized and diluted with fresh growth medium without antibiotics (1.5 × 105 cells/ml) and transferred to 24-well plates (1 ml/well). In each well, 0.75 μg of the apoE reporter construct and 0.25 μg of the pSV-β-galactosidase reporter were added with 2 μl Lipofectamine according to the manufacturer’s instructions for a 3-h incubation. Cells were then incubated overnight before starting the incubations described in the figure legends. After the treatments indicated in the figure legends, cells were washed twice with cold PBS and lysed in 60 μl cell culture lysis reagent. Luciferase assays were performed using the Luciferase Assay System (Promega, Madison, WI). Ten microliters of the cell lysate were mixed with 50 μl luciferase assay solution, and the luminescence was determined using a Turner Design (Sunnyvale, CA) luminometer. Measurements on each sample were performed in triplicate and normalized to β-galactosidase activity expressed by cotransfected plasmid. For transfection of siRNA, differentiated 3T3-L1 cells (d 8–10) were trypsinized and diluted with fresh growth medium without antibiotics (3 × 105 cells/ml) and transferred to 12-well plates (1 ml/well). In each well, 100 pm NFκB, p50, or control silencing small interference RNA (siRNA) complexed with 2 μl Lipofectamine were added. After an overnight incubation, the medium was replaced with growth medium. Forty-eight hours after transfection, cells were incubated with or without TNFα and processed as indicated in the figure legends.
Site-directed mutagenesis
Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene). The oligonucleotide GGGGG CGGGA CAGac cGAGa gCTAT AATTG GACAA GTCTG GG, containing mutant binding site (in lowercase letters), and its complementary sequences were used for mutagenesis of the −300/+86 apoE promoter construct. The nucleotide sequence of the mutant promoter was confirmed by DNA sequencing.
RNA interference
The siRNA targeted to down-regulate NFκB p50 mRNA (ON-TARGETplus SMARTpool siRNA catalog item L-047764-00-0005; Dharmacon, Lafayette, CO) was transfected into 3T3-L1 cells. Nontargeting siRNA provided by the manufacturer was used as a negative control. Cell lysates were collected at 72 h after transfection. Immunoblots to measure knockdown of the p50 protein were performed using an antibody from Santa Cruz Biotechnologies, Santa Cruz, CA (catalog item SC-11904).
NFκB DNA binding assay
Transcription factor assay kits (TransAM Flexi NFκB; Active Motif, Carlsbad, CA) were used to detect TNFα-induced NFκB p50 binding to DNA. Briefly, an NFκB consensus oligonucleotide or an apoE oligonucleotide (from −70 to −24 bp) containing the putative NFκB binding site (at −43) was end-labeled with biotin. Nuclear extracts were prepared (Nuclear Extract Kit; Active Motif) from untreated cells or cells treated with TNFα, and 10 μg of nuclear extract was incubated with biotin-labeled oligos alone or with a 10-fold molar excess of unlabeled competitors. The reaction mixtures were added to streptavidin-coated plates and incubated for 1 h. The p50 bound to DNA was detected colorimetrically after sequential incubation with an antibody directed against p50 followed by a horseradish peroxidase-conjugated secondary antibody.
Chromatin immunoprecipitation (ChIP) assay
The ChIP-IT Express kit (Active Motif) was used to perform ChIP assays. 3T3-L1 adipocytes were treated with TNFα for the time points indicated in the figure legends. Cells were treated with 1% formaldehyde, followed by glycine stop solution. Cells were then collected and centrifuged at 4 C and resuspended in lysis buffer. After an additional 30 min of lysis and 10 strokes in a Dounce homogenizer, nuclei were collected by 10 min of centrifugation at 2400 × g and resuspended in shearing buffer. Enzymatic shearing cocktail was added to digest DNA for preparing DNA fragments ranging in size from 200–500 bp. After centrifugation, 10 μl of the supernatant (containing sheared chromatin) was used as assay input. Immunoprecipitation was carried out overnight at 4 C using an incubation mixture containing 10 μl sheared chromatin, protein G magnetic beads, and 3 μg antibodies (NFκB p65, sc-109X; NFκB p50, sc-1190X; or Sp1, sc-59X; Santa Cruz Biotechnology). After centrifugation, washing, and elution, the samples were reverse cross-linked and treated with ribonuclease A and proteinase K. DNA was then purified with the DNA minicolumns provided with the kit, and real-time PCR was performed with primers 5′-GGG AGA GAA CAA CCC GCC TCG TGA CAG-3′ and 5′-TCC GAA ACA AGT CCT TAG CCT CCA GGG TCT-3′, designed to amplify a 142-bp product of the mouse apoE promoter. ChIP assay for polymerase II binding to the GAPDH promoter was used as the input control.
Statistical analysis
Statistical differences between experimental groups were evaluated using ANOVA (SPSS, Chicago, IL). P < 0.05 was considered significant. Results are expressed as mean ± sd of the number of replicates indicated in the figure legends. The experiments shown are representative of two to three experiments with similar results.
Results
Identification of a proximal apoE gene promoter site necessary for TNFα regulation
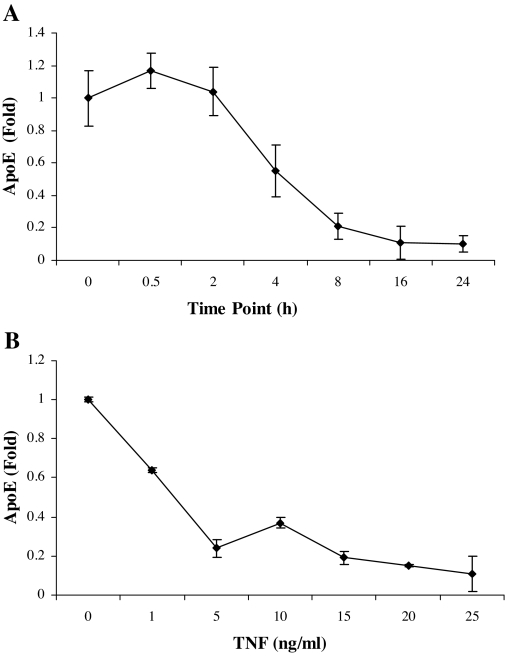
The results in Fig. 1 show the results of experiments to evaluate the time course and dose-response effects of TNFα on adipocyte apoE gene expression. In Fig. 1A, cells were harvested at the times shown after addition of TNFα, and apoE mRNA levels were compared with cells harvested at the same time without TNFα. TNFα led to a reduction of apoE mRNA as early as 4 h after its addition. Figure 1B shows that TNFα at a concentration of 5 ng/ml produced maximal suppression of apoE gene promoter activity, and this concentration was used in all subsequent experiments.
Figure 1.
Time- and dose-dependent response of adipocyte apoE to TNFα. A, 3T3-L1 adipocytes were treated with TNFα (5 ng/ml) in complete medium for the times shown. Total RNA was extracted and apoE mRNA was quantitated by RT-PCR as described in Materials and Methods. Abundance of apoE mRNA was compared with untreated control cells harvested at the same time. B, 3T3-L1 cells were transfected to express a luciferase reporter gene under the control of the −623/+86 bp of the apoE promoter and the 620-bp enhancer. After 36 h, the cells were treated with TNFα at the doses indicated for 24 h. Relative luciferase activity was quantitated after normalization for β-galactosidase expression as described in Materials and Methods. All points shown are the mean ± sd from triplicate samples.
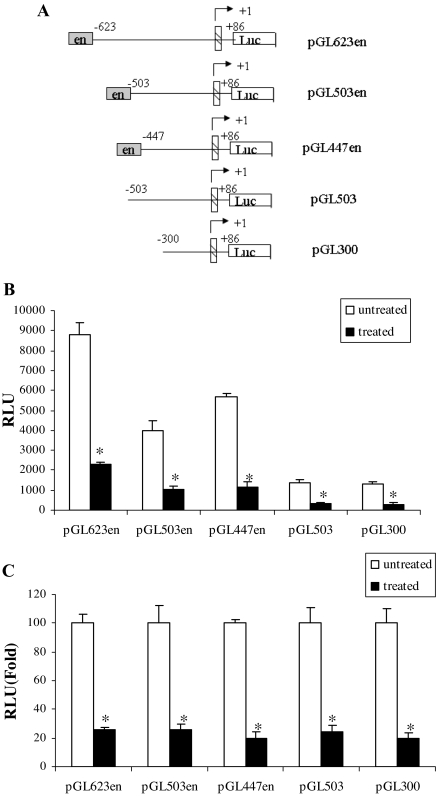
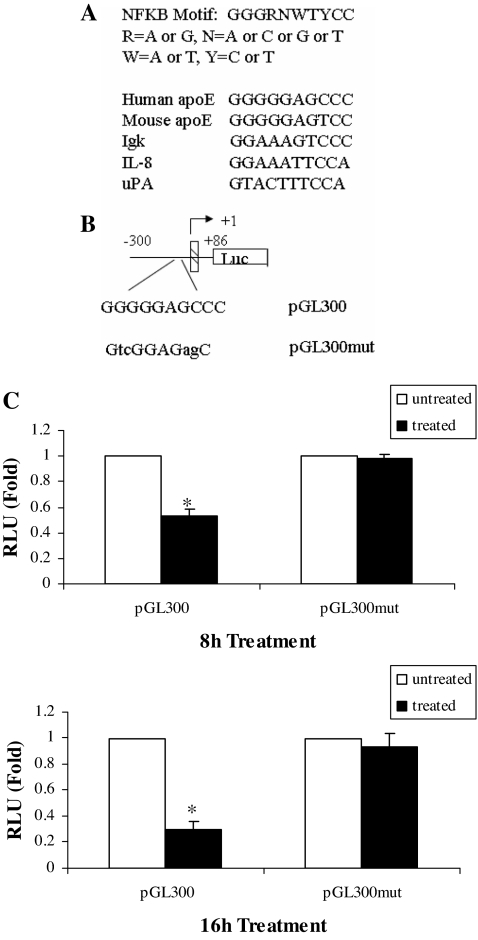
We next constructed the apoE promoter-luciferase reporter constructs shown in Fig. 2A. These constructs contain an apoE 5′ promoter sequence of varying lengths with or without the 620-bp ME1 element (indicated as en in the figure). Total luciferase activity decreased with sequential truncation of the 5′ promoter (Fig. 2B). Luciferase activity remained easily measurable in the smallest construct (pGL300), which demonstrated significantly reduced expression after treatment with TNFα. To compare the magnitude of suppression produced by TNFα in reporter constructs with different baseline expression, we pooled the results of three separate experiments (each done in triplicate) and examined changes in reporter expression relative to untreated control (Fig. 2C). TNFα-mediated suppression was again observed in the smallest promoter construct examined, and furthermore, the percentage of suppression produced by TNFα treatment was approximately the same (80%) for all constructs. There was not a significant difference among the constructs with respect to percentage suppression by TNFα treatment. This result indicated that the apoE gene regulatory elements necessary for response to TNFα were contained in the smallest promoter construct between −300 and +86. This portion of the promoter contains multiple potential binding sites for transcription factors; however, we focused on a potential NFκB binding site because many effects of TNFα in adipocytes are mediated via NFκB (10). Using computer-assisted analysis, we identified a putative NFκB binding site in the apoE promoter located between −43 and −34 (Fig. 3A), and we used site-directed mutagenesis to further evaluate the importance of this site for mediating the TNFα effect. The response of pGL300 or pGL300mut reporter constructs (containing the native or mutant apoE promoter sequences shown in Fig. 3B) is shown in Fig. 3C. In three independent experiments (each conducted in triplicate), treatment of cells with TNFα led to a 50% reduction at 8 h and 75% reduction at 16 h of pGL300 expression. Mutation of the putative NFκB site completely eliminated TNFα-mediated suppression.
Figure 2.
Evaluation of apoE gene promoter sequences responsive to TNFα in adipocytes. A, The reporter constructs used to evaluate apoE gene elements responsive to TNFα are shown. Base pairs are numbered relative to the transcription start site, and en represents inclusion of the 620-bp ME1. The hatched box represents the first apoE exon. B, 3T3-L1 adipocytes were transfected with the indicated apoE promoter-luciferase constructs. TNFα (5 ng/ml) was added for 24 h in complete growth medium before harvesting cells for measurement of luciferase and β-galactosidase activities. *, P < 0.01 for comparing TNFα-treated to untreated control. Results shown are mean ± sd from triplicate samples. C, Percent change in relative luciferase units (RLU) after treatment with TNFα is shown for each construct after normalization to the untreated control. *, P < 0.01 for comparing TNFα-treated to untreated control. The results shown are mean ± sd from three independent experiments, each performed using triplicate samples.
Figure 3.
Importance of −43 to −34 apoE gene promoter sequence for mediating the response to TNFα in adipocytes. A, NFκB binding sites at −43 bp in the 5′-flanking region of the human and mouse apoE genes are compared with the sequences in other NFκB responsive genes. B, The presumptive sequence for NFκB binding site at −43 of the apoE gene was mutated as described in Materials and Methods. Lowercase letters indicate mutated nucleotides. C, pGL300 and the pGL300mut were transfected into 3T3-L1 cells, and cells were treated with 5 ng/ml TNFα for 8 or 16 h. The results shown are the mean ± sd of three independent experiments, each done in triplicate. *, P < 0.001 for comparison with untreated control. RLU, Relative luciferase units.
Role of NFκB pathway protein elements for TNFα suppression of adipocyte apoE gene expression
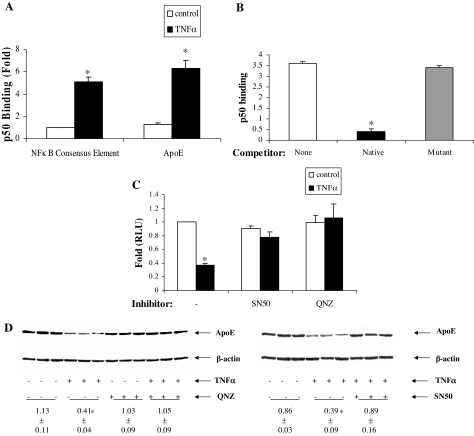
We further probed the importance of NFκB binding site to the proximal apoE promoter binding site by evaluating the involvement of NFκB protein elements. We first evaluated the effect of TNFα treatment on NFκB p50 subunit binding to the apoE promoter. 3T3-L1 cells were treated with TNFα and nuclear extracts were prepared. The binding of p50 isolated from adipocyte nuclei to an NFκB consensus element, and to the native apoE promoter, was markedly stimulated by TNFα (Fig. 4A). The need for the intact apoE gene binding site at −43 for enhanced p50 binding was evaluated by adding the intact site or the mutated site (from Fig. 3B) as competitors for p50 binding to the NFκB consensus sequence. The addition of a 10-fold molar excess of the unlabeled native apoE promoter significantly competed for binding of p50 to the NFκB consensus sequence; however, no competition was observed with a 10-fold molar excess of the apoE promoter containing the mutated apoE binding site (Fig. 4B).
Figure 4.
Involvement of NFκB protein complex for the apoE gene response to TNFα. A, 3T3-L1 adipocytes were treated with TNFα (5 ng/ml) for 24 h, and then 10 μg nuclear protein was isolated and tested for binding to a consensus NFκB binding sequence or to the putative NFκB binding site on the apoE gene proximal promoter. The results shown are mean ± sd from three independent experiments, each done in triplicate. B, Binding of nuclear extract to the NFκB consensus sequence was evaluated with no competitor or in the presence of a 10-fold molar excess of the native apoE promoter sequence or of the apoE promoter mutant shown in Fig. 3B. Results shown are from three independent experiments, each done in triplicate. C, 3T3-L1 cells were pretreated with 50 μg/ml SN50 (an inhibitor of NFκB complex translocation to the nucleus) or with 20 nm QNZ (an inhibitor of NFκB complex activation) for 1 h, and then TNFα (5 ng/ml) was added for 8 h to cells previously transfected to express pGL623en. The results shown are mean ± sd from triplicate samples and are representative of three independent experiments, each done in triplicate. D, Representative Western blot for the effect of NFκB pathway inhibitors on TNFα (5 ng/ml for 16 h) suppression of apoE protein. The apoE signal densities corrected for β-actin (mean ± sd) are shown. *, P < 0.01 for comparison with untreated control (A, C, and D) or for comparison with binding in the absence of the competitor oligonucleotide (B). RLU, Relative luciferase units.
We next examined the effect of two structurally distinct inhibitors of NFκB protein complex translocation or activation. Treatment of cells with SN50, an inhibitor of NFκB complex translocation to the nucleus, completely abrogated the suppressive effect of TNFα on the apoE gene (Fig. 4C). Treatment of cells with QNZ, an inhibitor of NFκB complex activation, similarly eliminated the apoE gene response to TNFα (Fig. 4C). Treatment of adipocytes with TNFα reduced apoE protein expression (measured by Western blot) by approximately 50%. Both SN50 and QNZ completely eliminated TNFα-mediated suppression of adipocyte apoE protein (Fig. 4D).
Role of NFκB p50 for the TNFα response
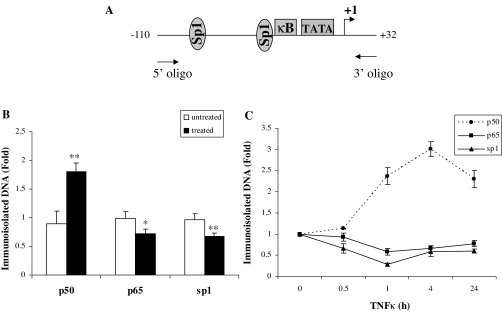
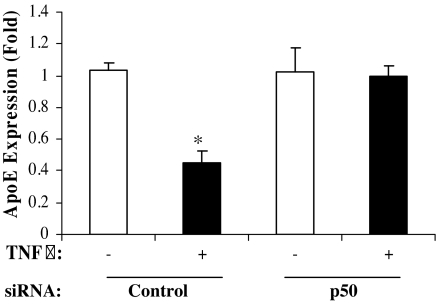
The data thus far indicated that apoE gene transcriptional repression by TNFα required intact apoE gene promoter sequences between −43 and −34 with homology to an NFκB binding site, that binding of NFκB p50 to this apoE promoter region was stimulated after TNFα treatment, and that two different inhibitors of the NFκB pathway prevented TNFα-mediated suppression. We next used a ChIP assay to assess the time course for the binding of NFκB p50 and NFκB p65 to the apoE promoter in living cells after treatment with TNFα. The binding of Sp1, previously shown to be required for maximal apoE gene expression and has a binding site immediately upstream of the NFκB site, was also assessed (Fig. 5A). One hour of TNFα treatment (Fig. 5B) increased binding of p50 to the apoE gene promoter by over 2-fold but reduced binding of both p65 and Sp1. A time course for the effect of TNFα treatment on binding of p50, p65, and Sp1 is shown in Fig. 5C. The increased binding of p50 is sustained for 24 h after treatment with TNFα. This result is consistent with recent reports in other cell types, suggesting that increased binding of p50 to gene regulatory elements is an important mechanism mediating transcriptional repression by the NFκB pathway (22,23). We therefore focused on the need for p50 in TNFα-mediated transcriptional repression of the apoE gene by using a silencing RNA that produced a 50% reduction of p50 protein expression (not shown). In cells treated with a control siRNA, treatment with TNFα for 8 h produced a 60% reduction of apoE mRNA levels in adipocytes (Fig. 6). Knockdown of p50, however, completely eliminated the reduction of apoE mRNA produced by treatment with TNFα (Fig. 6). This result identifies p50 as the key mediator of the TNFα effect on the endogenous adipocyte apoE gene.
Figure 5.
ChIP assay. A, The location of the primers used and the target sequence of the apoE promoter are shown. 3T3-L1 cells were treated with or without TNFα (5 ng/ml) for 1 h (B) or times shown (C) in complete medium. Real-time PCR quantitation of fragments precipitated in the ChIP assay using antibodies specific for Sp1, p50, or p65 were analyzed in triplicate as described in Materials and Methods and data expressed as fold change relative to untreated control samples. Comparing treated and untreated samples: *, P < 0.05; **, P < 0.001. Values shown are mean ± sd of three independent experiments each done in triplicate.
Figure 6.
Effect of silencing NFκB p50 on TNFα-mediated suppression of the apoE gene in adipocytes. 3T3-L1 cells were transfected with 100 nm p50 siRNA or a nontarget control siRNA for 24 h as described in Materials and Methods. Forty-eight hours after transfection of the siRNA, cells were incubated with or without TNFα (5 ng/ml) for 8 h. Total RNA was extracted for quantitation of apoE mRNA by RT-PCR as described in Materials and Methods. Results shown are mean ± sd from three independent experiments, each done in triplicate. A representative agarose gel is shown. *, P < 0.01 for the effect of TNFα on apoE mRNA level.
Discussion
The results in this manuscript establish the molecular mechanism by which TNFα suppresses apoE gene transcription in adipocytes. We have shown that reduced apoE gene transcription after TNFα treatment is eliminated by two structurally distinct inhibitors of the NFκB pathway. Furthermore, the apoE gene transcriptional response to TNFα requires intact apoE gene promoter sequences between −43 and −34 with a high degree of homology to the NFκB binding consensus sequence. Treatment of adipocytes with TNFα stimulates binding of NFκB p50 to this region of the apoE promoter, and mutation of this site eliminates the ability of the apoE promoter to compete with the NFκB consensus sequence for binding of NFκB p50. ChIP analysis demonstrates that TNFα treatment of living cells recruits p50 binding to this portion of the apoE promoter with concomitant reduction of p65 and Sp1 binding. Finally, knockdown of NFκB p50 completely eliminates TNFα suppression of native apoE gene expression by TNFα.
Transcriptional mechanisms accounting for stimulation of gene transcription by NFκB have been extensively characterized. The molecular mechanisms accounting for NFκB suppression of gene transcription, however, are less clear, particularly in adipocytes. In other cell types and for other genes, it has been suggested that transcriptional repression by NFκB is produced by the binding of p50 homodimers to the NFκB binding site (22,23,24,25,26). The prototypical complex for NFκB-mediated transcriptional activation is a p65/p50 heterodimer, and increased binding of p50 homodimers may displace activating p65/p50 heterodimers leading to transcriptional repression. Alternatively, functional interactions have been observed between elements of the NFκB transcription complex and other transactivating factors, including Sp1, with negative effects on gene transcription (27,28,29). Many of these interactions have involved the NFκB p65 subunit. The results of the p50 knockdown experiments (Fig. 6) establish that p50 is the essential element required for transcriptional repression of the adipocyte apoE gene by TNFα. Furthermore, the total elimination of TNFα suppression of the native adipocyte apoE gene by p50 knockdown confirms the NFκB pathway as the important molecular pathway necessary for this suppression.
apoE is highly expressed in macrophages (30), and common regulatory enhancers are important for macrophage and adipocyte expression of apoE. The response of the apoE gene to TNFα, however, appears to differ between these two cell types. In early monocyte-macrophages, TNFα treatment increases apoE expression. In fully mature macrophages, treatment with TNFα for times up to 48 h leads to a small 8–10% nonsignificant reduction in apoE mRNA level as measured by Northern hybridization (31). The divergent apoE response to TNFα in adipocytes and macrophages suggests that the role subserved by endogenous apoE for modulating cellular lipid flux is different in these two cell types, particularly in response to a proinflammatory milieu.
Adipocyte apoE expression is reduced in both diet-induced and leptin-deficient obesity (14). The onset of obesity is accompanied by an inflammatory response in adipose tissue reflected by macrophage infiltration into its stromovascular compartment and a proinflammatory phenotype in adipose tissue macrophages, including increased secretion of TNFα (5,6,7,8,9,10,11,32,33,34). Based on our previously reported observations (16), lower adipocyte apoE expression will reduce adipocyte triglyceride synthesis and reduce the uptake of lipid from triglyceride-rich lipoproteins (TGRL) such as very-low-density lipoprotein. This would tend to limit further expansion of adipocyte lipid content and size. In vivo, this reduced delivery of lipid from TGRL to adipose tissue could favor its delivery to other tissues such as liver or muscle. The partitioning of TGRL lipid away from adipose tissue to liver and muscle has been implicated in producing tissue-specific and systemic insulin resistance (35,36,37,38). Our findings provide a molecular mechanism by which adipose tissue inflammation accompanying obesity suppresses adipocyte apoE gene expression. Suppression of endogenous adipocyte apoE can have significant impact on adipocyte triglyceride metabolism, content, and adipocyte size (16), with potentially important downstream consequences for metabolic and vascular health (2).
Acknowledgments
We thank Stephanie Thompson for assistance with manuscript preparation.
Footnotes
This work was supported by Grant DK71711 from the National Institutes of Health (to T.M.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 8, 2008
Abbreviations: apoE, Apolipoprotein E; ChIP, chromatin immunoprecipitation; CT, cycle threshold; NF, nuclear factor; PPARγ, peroxisome proliferator-activated receptor-γ; QNZ, 6-amino-4-(4-phenoxyphenylethylamino) quinazoline; siRNA, small interference RNA; TGRL, triglyceride-rich lipoproteins.
References
- Scherer PE 2006 Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 55:1537–1545 [DOI] [PubMed] [Google Scholar]
- Fantuzzi G, Mazzone T 2007 Adipose tissue and atherosclerosis: exploring the connection. Arterioscler Thromb Vasc Biol 27:996–1003 [DOI] [PubMed] [Google Scholar]
- Berg AH, Scherer PE 2005 Adipose tissue, inflammation, and cardiovascular disease. Circ Res 96:939–949 [DOI] [PubMed] [Google Scholar]
- Bluher M, Wilson-Fritch L, Lezyki J, Lausten PG, Corvera S, Kahn R 2004 Role of insulin action and cell size on protein expression patterns in adipocytes. J Biol Chem 279:31902–31909 [DOI] [PubMed] [Google Scholar]
- Kershaw EE, Flier JS 2004 Adipose tissue as an endocrine organ. J Clin Endocrinol Metab 89:2548–2556 [DOI] [PubMed] [Google Scholar]
- Suganami T, Nishida J, Ogawa Y 2005 A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: role of free fatty acids and tumor necrosis factor α. Arterioscler Thromb Vasc Biol 25:2062–2068 [DOI] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H 2003 Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, DeYoung SM, Bodzin JL, Saltiel AR 2007 Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 56:16–23 [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, Saltiel AR 2007 Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 117:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Hacohen N, Golub TR, Parijs LV, Lodish HF 2002 Tumor necrosis factor-α suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-κB activation by TNF-α is obligatory. Diabetes 51:1319–1336 [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr 2003 Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Moser R, Newman TC, Fried SK, Breslow JL 1991 Apolipoprotein E gene expression in mouse 3T3-L1 adipocytes and human adipose tissue and its regulation by differentiation and lipid content. J Biol Chem 266:10583–10588 [PubMed] [Google Scholar]
- Yue L, Rassouli N, Ranganathan G, Kern PA, Mazzone T 2004 Divergent effects of PPAR agonists and TNF on adipocyte apoE expression. J Biol Chem 279:47626–47632 [DOI] [PubMed] [Google Scholar]
- Huang ZH, Luque RM, Kineman RD, Mazzone T 2007 Nutritional regulation of adipose tissue apolipoprotein E expression. Am J Physiol Endocrinol Metab 293:E203–E209 [DOI] [PubMed] [Google Scholar]
- Rao P, Huang ZH, Mazzone T 2007 Angiotensin II regulates adipocyte apolipoprotein E expression. J Clin Endocrinol Metab 92:4366–4372 [DOI] [PubMed] [Google Scholar]
- Huang ZH, Reardon CA, Mazzone T 2006 Endogenous apoE expression modulates adipocyte triglyceride content and turnover. Diabetes 55:3394–3402 [DOI] [PubMed] [Google Scholar]
- Chang DJ, Paik YK, Leren TP, Walker DW, Howlett GJ, Taylor JM 1990 Characterization of a human apolipoprotein E gene enhancer element and its associated protein factors. J Biol Chem 265:9496–9504 [PubMed] [Google Scholar]
- Lafitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kas HR, Mangeldorf DJ, Tontonoz P 2001 LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci USA 98:507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S-J, Allan C, Grehan S, Tse E, Moran C, Taylor JM 2000 Duplicated downstream enhancers control expression of the human apolipoprotein E gene in macrophages and adipose tissue. J Biol Chem 275:31567–31572 [DOI] [PubMed] [Google Scholar]
- Basheeruddin K, Rechtoris C, Mazzone T 1992 Transcriptional and post-transcriptional control of apolipoprotein E gene expression in differentiating human monocytes. J Biol Chem 267:1219–1224 [PubMed] [Google Scholar]
- Zhao Y, Mazzone T 2000 Transport and processing of endogenously synthesized ApoE on the macrophage cell surface. J Biol Chem 275:4759–4765 [DOI] [PubMed] [Google Scholar]
- Kostadinova RM, Nawrocki AR, Frey FJ, Frey BM 2005 Tumor necrosis factor α and phorbol 12-myristate-13-acetate down-regulates human 11β-hydroxysteroid dehydrogenase type 2 through p50/p50 NFκB homodimers and Egr-1. FASEB J 19:650–652 [DOI] [PubMed] [Google Scholar]
- Tong X, Yin L, Washington R, Rosenberg DW, Giardina C 2004 The p50–p50 NF-κB complex as a stimulus-specific repressor of gene activation. Mol Cell Biochem 265:171–183 [DOI] [PubMed] [Google Scholar]
- Driessler F, Venstrom K, Sabat R, Asadullah K, Schottelius AJ 2004 Molecular mechanisms of interleukin-10-mediated inhibition of NF-κB activity: a role for p50. Clin Exp Immunol 135:64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundstrom S, Anderson P, Scheipers P, Sundstedt A 2004 Bc1–3 and NFκB p50–p50 homodimers act as transcriptional repressors in tolerant CD4+ T cells. J Biol Chem 279:8460–8468 [DOI] [PubMed] [Google Scholar]
- Guan H, Hou S, Ricciardi RP 2005 DNA binding of repressor nuclear factor-κB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J Biol Chem 280:9957–9962 [DOI] [PubMed] [Google Scholar]
- Hirano F, Tanaka H, Hirano Y, Hiramoto M, Handa H, Makino I, Scheidereit C 1998 Functional interference of Sp1 and NF-κB through the same DNA binding site. Mol Cell Biol 18:1266–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CY, Park JH, Seo KH, Kim JM, Im SY, Lee JW, Choi HS, Lee K 2003 Expression of MIS in the testis is downregulated by tumor necrosis factor α through the negative regulation of SF-1 transactivation by NF-κB. Mol Cell Biol 23:6000–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Farmer P, Rubin J, Nanes MS 2004 Integration of the NFκB p65 subunit into the vitamin D receptor transcriptional complex: identification of p65 domains that inhibit 1,25-dihydroxyvitamin D3-stimulated transcription. J Cell Biochem 92:833–848 [DOI] [PubMed] [Google Scholar]
- Mazzone T 1996 ApoE secretion by macrophages: its potential physiological function(s). Curr Opin Lipidol 7:303–307 [DOI] [PubMed] [Google Scholar]
- Duan H, Li Z, Mazzone T 1995 Tumor necrosis factor-α modulates monocyte/macrophage apoprotein E gene expression. J Clin Invest 96:915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Eagon JC, Trujillo ME, Scherer PE, Klein S 2007 Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes 56:1010–1013 [DOI] [PubMed] [Google Scholar]
- Gustafson B, Hammarstedt A, Andersson CX, Smith U 2007 Inflamed adipose tissue: a culprit underlying the metabolic syndrome and atherosclerosis. Arterioscler Thromb Vasc Biol 27:2276–2283 [DOI] [PubMed] [Google Scholar]
- Neels JG, Olefsky JM 2006 Inflamed fat: what starts the fire? J Clin Invest 116:33–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Fillmore JJ, Chen Y, Yu C, Moore IK, Pypaert M, Lutz EP, Kako Y, Velez-Carrasco W, Goldberg IJ, Breslow JL, Shulman GI 2001 Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc Natl Acad Sci USA 98:7522–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, van de WE, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, Jelicks LA, Mehler MF, Hui DY, Deshaies Y, Shulman GI, Schwartz GJ, Scherer PE 2007 Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest 117:2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, She P, Mozzoli M, Cheung P, Gumireddy K, Reddy P, Xiang X, Luo Z, Ruderman N 2005 Free fatty acids produce insulin resistance and activate the proinflammatory nuclear factor-κB pathway in rat liver. Diabetes 54:3458–3465 [DOI] [PubMed] [Google Scholar]
- Lewis GF, Carpentier A, Adeli K, Giacca A 2002 Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev 23:201–229 [DOI] [PubMed] [Google Scholar]