Abstract
Cation/proton exchange has been recognized for decades in mammalian mitochondria, but the exchanger proteins have eluded identification. In this study, a cDNA from a human brain library, previously designated NHA2 in the genome, was cloned and characterized. The NHA2 transcript bears more similarity to prokaryotic than known eukaryotic sodium/proton exchangers, but it was found to be expressed in multiple mammalian organs and cultured cells. A mAb to NHA2 was generated and found to label an approximately 55-kD native protein in multiple tissues and cell lines. The specificity of this antibody was confirmed by demonstrating the loss of the native NHA2 band on immunoblots when cultured cells were treated with NHA2-specific small interfering RNA. Although NHA2 protein was detected in multiple organs, within each, its expression was restricted to specific cell types. In the kidney, co-localization with calbindin 28k and reverse transcription–PCR of microdissected tubules revealed that NHA2 is limited to the distal convoluted tubule. In cell lines, native NHA2 was localized both to the plasma membrane and to the intracellular compartment; immunogold electron microscopy of rat distal convoluted tubule demonstrated NHA2 predominantly but not exclusively on the inner mitochondrial membrane. Furthermore, co-sedimentation of NHA2 antigen and mitochondrial membranes was observed with differential centrifugation, and two mitochondrial markers co-localized with NHA2 in cultured cells. Regarding function, human NHA2 reversed the sodium/hydrogen exchanger–null phenotype when expressed in sodium/hydrogen exchanger–deficient yeast and restored the ability to defend high salinity in the presence of acidic extracellular pH. In summary, NHA2 is a ubiquitous mammalian sodium proton/exchanger that is restricted to the distal convoluted tubule in the kidney.
Sodium/hydrogen exchangers (NHE) are ubiquitous ion transporters present in lipid bilayers in simple prokaryotes and eukaryotes, including plants, fungi, and animals, that harness the electrochemical gradient of one ion to energize the uphill transport of the other.1 More than 40 yr ago, Mitchell and Moyle2 first described a cation/proton antiport system allowing mitochondria to extrude sodium and potassium against a highly unfavorable inward cationic electrochemical gradient. Ten NHE isoforms have since been cloned in mammals, which include plasmalemmal NHE1 through 5 and intracellular NHE6 through 9. In addition, a sperm-specific plasmalemmal NHE was cloned.3 Although it was first described functionally, the molecular identity of the mitochondrial cation/proton antiport system remains elusive.
By controlling cell volume and maintaining cellular and organellar pH, NHE have been proposed to play a pivotal role in diverse physiologic processes, including control of cell cycle and cell proliferation, cell migration, transepithelial proton, bicarbonate and sodium transport, salt tolerance, and vesicle trafficking and biogenesis.1,4 At the whole-organism level, NHE are key players in extremely diverse areas such as cancer biology, central nervous system integrity, and cardiovascular pathophysiology.4,5
The superfamily of monovalent cation/proton antiporters (CPA) as proposed by Saier6 has two large clades, named CPA1 and CPA2. CPA1 includes the currently known mammalian NHE with the exception of the recently cloned sperm NHE. In a comprehensive phylogenetic analysis of CPA families, Brett et al.7 proposed two novel, previously unrecognized, putative NHE in the mammalian CPA2 clade. These putative NHE have higher homology to fungal and bacterial NHE, especially Escherichia coli NhaA, than the currently known mammalian NHE. On the basis of this homology, they have been named NHA1 and NHA2. NHA1, also referred to as NHEDC1, has been cloned and proposed to be expressed exclusively in testis.8 No functional characterization has been reported so far. One recent article described the ubiquitous distribution of NHA2,9 but another reported a highly specific distribution in osteoclasts only.10,11 Although there was evidence for cell surface expression in one study,9 an exclusively intracellular distribution was described in the other.11
In this study, we describe tissue distribution, intracellular localization, and functional characterization of NHA2. NHA2 is a new intracellular and plasmalemmal NHE with wide tissue distribution, and, in the kidney, it is specifically localized to the distal convoluted tubules (DCT).
RESULTS
We have cloned the human NHA1 and NHA2 isoforms (HsNHA1 and HsNHA2, respectively) from a human brain cDNA library. The human NHA2 open reading frame (ORF) spans 1614 bp, giving rise to a protein of 537 amino acids with a predicted molecular weight of approximately 57 kD. All completely sequenced metazoan genomes contain orthologs of NHA2 with high conservation on the amino acid level (Figure 1).7 By hydropathy-based analysis and depending on the algorithm used, the protein is predicted to contain 10 to 12 transmembrane domains with short intracellular N- and C-termini (www.expasy.org/tools). The E. coli Nha crystal structure shows 12 transmembrane spans.12 The predicted structure of the two termini are in contrast to the currently known mammalian NHE, which have long intracellular C-termini important in the functional regulation.
Figure 1.
Sequence comparison of NHA2. Amino acid alignment of human (Hs), mouse (Mm), and rat (Rn) NHA2. Sequence identity is 81% between human and mouse or human and rat NHA2. Epitope of MAB45D12D is marked in yellow highlight.
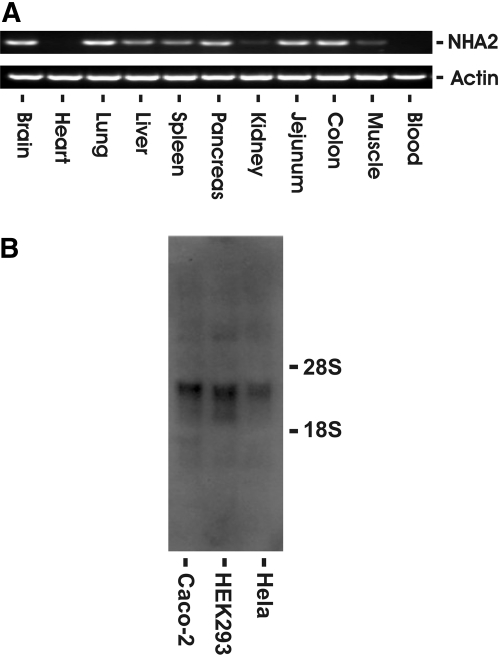
Human NHA2 is expressed in all human cell lines tested. Figure 2A shows a representative RNA blot in HeLa, HEK293, and Caco-2 cells. The corresponding mRNA is approximately 2.4 kb without evidence of alternate splice variants with differential mobility in these cell lines. Presence of NHA2 in these cell lines was also confirmed with reverse transcription–PCR (RT-PCR) of NHA2 ORF followed by sequencing of the PCR product (data not shown). We next performed RT-PCR using mouse-specific NHA2 primers from different mouse tissues. With the exception of heart and blood, NHA2 transcript was detected in all organs, including the kidney (Figure 2B).
Figure 2.
NHA2 mRNA expression in cells and tissue. (A) RNA blot of three human cell lines using a 32P-labeled human NHA2 cDNA probe. Ten micrograms of total RNA was loaded per lane. Human NHA2 transcript size is approximately 2.4 kb. (B) RT-PCR of mouse NHA2 from various tissues. One microgram of total RNA was subjected to oligo-dT–primed reverse transcription followed by PCR using mouse NHA2- or β-actin–specific primers for 35 cycles.
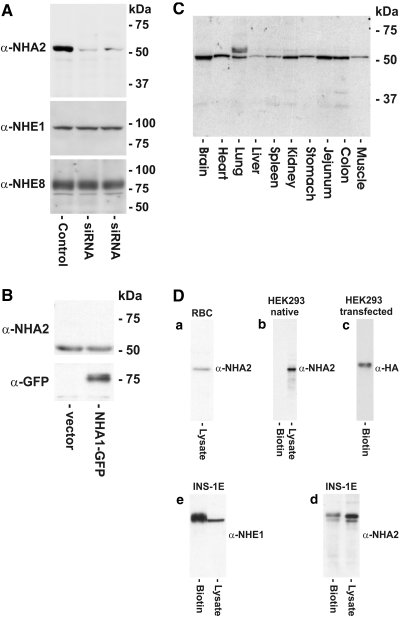
To determine NHA2 expression at the protein level, we raised a mAb to a conserved epitope (DTARSHGEKQLEDYG; Figure 1, yellow highlight). We obtained a highly specific mAb, designated MAB45D12D of the IgG2a subclass. On immunoblot, this antibody recognizes a single band of approximately 55 kD, which is compatible with the predicted ORF (Figure 3A). For verification of the specificity of our antibody, endogenous NHA2 in NRK cells was knocked down using RNA interference. Compared with control cells, transfection of cells with small interference RNA (siRNA) targeting rat NHA2 led to almost complete disappearance of the approximately 55 kD band on immunoblot (Figure 3A). Global silencing was ruled out by the fact that protein levels of NHE1 or NHE8 in these cells were not affected (Figure 3A). To rule out cross-reactivity with the highly homologous NHA1 isoform, we transiently transfected plasmalemmal NHE-deficient AP-1 cells with a C-terminally GFP-tagged human NHA1 construct. Although the construct was expressed as detected by a monoclonal anti-GFP antibody, MAB45D12D failed to label NHA1 (Figure 3B).
Figure 3.
Immunoblot of native and overexpressed human NHA2. (A) NRK cells were transfected with two different concentrations of a mix of two rat NHA2-specific siRNA (middle lane 50 pg, right lane 30 pg) and compared with mock-transfected cells. Twenty-four hours after transfection, 50 μg of total lysate protein was separated by SDS-PAGE and endogenous NHA2, NHE1, or NHE8 protein levels were detected by immunoblotting using MAB45D12D, monoclonal anti-NHE1, or anti-NHE8 antibodies. (B) MAB45D12D does not react with NHA1. Human C-terminally GFP-tagged NHA1 or empty vector was transiently expressed in AP-1 cells. Fifty micrograms of total protein was separated by SDS-PAGE and blotted with MAB45D12D or monoclonal anti-GFP antibody. (C) Expression of NHA2 in various mouse tissues. Fifty micrograms of total lysate protein were separated by SDS-PAGE followed by immunoblotting with MAB45D12D. (D) Surface expression of native and overexpressed NHA2 in different cells assessed by surface biotinylation. Native NHA2 is present on the surface membrane of RBC (a), a rat pancreatic β cell line (d), and HEK293 cells transiently transfected with human C-terminally HA-tagged NHA2 (c), but native NHA2 is absent from the surface membrane in HEK293 cells (b). Endogenous NHE1 was used as biotinylation control (e).
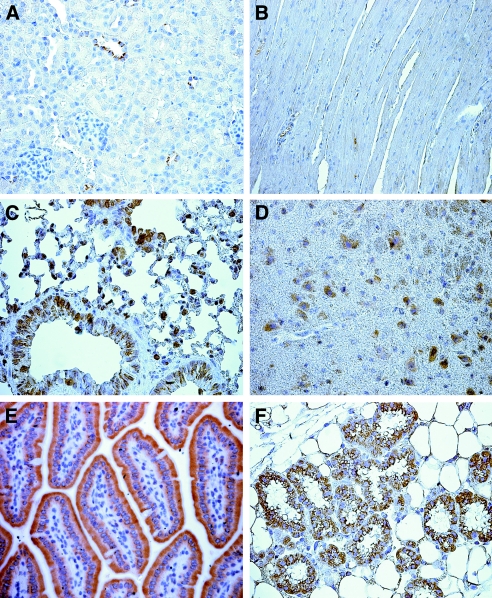
We next analyzed the tissue distribution of NHA2 at the protein level in the mouse using MAB45D12D. The protein expression pattern mirrors our RT-PCR findings with the exception of the heart (Figure 3C). NHA2 transcript in heart was also absent when two different PCR primer pairs were used and in another mouse strain (C57 Black/6 strain; data not shown). This discrepancy can be explained by either the presence of alternate splice variants in heart that failed to anneal to our primers or that, although the tissues were perfused with PBS before homogenization, residual red blood cells (RBC) in the capillary bed could be responsible for the signal. Also, the possibility of cross-reactivity of MAB45D12D in RBC and heart cannot be completely ruled out. Indeed, we and others have detected NHA2 in RBC (Figure 3D),9 despite the absence of mRNA detected by RT-PCR of whole blood (Figure 2B). The negative RT-PCR findings are supported by the absence of a signal in heart by immunohistochemistry (Figure 4).
Figure 4.
Tissue distribution of NHA2 in the mouse assessed by immunohistochemistry. Tissue was harvested and prepared as described in the Concise Methods section. MAB45D12D was used for staining. Tissues are as follows: Kidney cortex (A), heart muscle (B), lung (C), brain cortex (D), jejunum (E), and mammary gland (F).
To study plasmalemmal expression of endogenous NHA2, we used a surface biotinylation assay. Figure 3D shows absence of endogenous NHA2 from the plasma membrane in HEK293 cells as determined by this method. Simultaneous presence of NHA2 in lysate suggests an intracellular localization in these cells. Identical results were obtained from other cell lines (HeLa, NRK; results not shown). In contrast, plasmalemmal NHA2 could be detected in INS-1E cells, a rat insulinoma cell line,13 and in HEK293 cells transiently transfected with a C-terminally HA-tagged human NHA2 construct and in RBC. NHE1 was used as a biotinylation control (Figure 3D) to show that absence of biotinylatable NHA2 was not due to inaccessibility of the reagent in these cells.
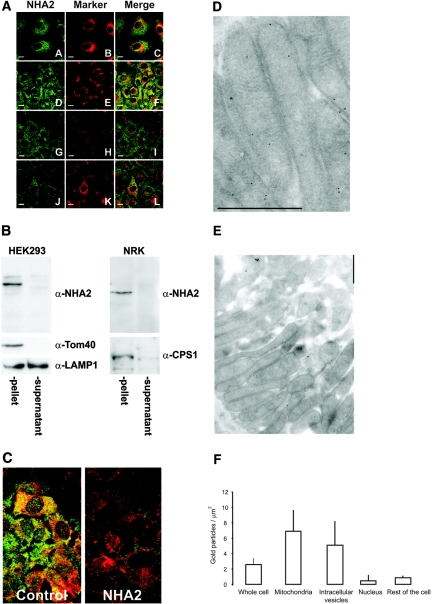
To study further native NHA2 expression, we performed confocal microscopy studies in NRK cells as shown in Figure 5A. By fluorescence immunocytochemistry, native NHA2 was not found on the plasma membrane of NRK cells. Identical staining patterns were observed in other cell lines tested for native NHA2 localization (HeLa, HEK293, MDCK; data not shown). To delineate further the intracellular localization of NHA2, we performed co-staining with the mitochondrial marker Mitotracker. There is partial co-localization of endogenous NHA2 with mitochondria in NRK cells. Although there is considerable variance from cell to cell, 39 ± 17% (mean ± SD; n = 40 cells) of the intracellular signal co-localizes with mitochondria (Figure 5A). Because there are some nonoverlapping signals of NHA2 and mitochondria, we performed additional co-localization of NHA2 with anti-CPS1 as a second mitochondrial marker, anti-GOLPH4 antibody for Golgi, and anti-PDI for endoplasmic reticulum (ER). The overlap of NHA2 with CPS1 was 30 ± 19% (mean ± SD; n = 52 cells; Figure 5A). Although there is no overlap with Golgi, there is some co-localization with ER (Figure 5A). We next isolated mitochondria from HEK293 and NRK cells by differential centrifugation. Native NHA2 is enriched in the mitochondrial fraction (3000 × g pellets; Figure 5B) as are the mitochondrial proteins Tom40 and CPS1. Although there is some co-purification of lysosomes as evidenced by the presence of the lysosomal membrane protein LAMP1 in the pellet, the majority of LAMP1 is present in the 3000 × g supernatant (Figure 5B). Knockdown of endogenous NHA2 leads to disappearance of the co-staining in NRK cells (Figure 5C). The intracellular localization as determined by imaging thus fits the biochemical evidence obtained with fractionation studies.
Figure 5.
Intracellular localization of endogenous NHA2. (A) Fluorescence immunocytochemistry. Bar = 10 μm. NRK cells were treated with the specific markers (mitochondria: Mitotracker [B] and α-CPS1 [E]; Golgi: α-GOLPH4 [H]; ER: α-PDI [K]) and then with MAB45D12D (α-NHA2 [A, D, G, and J]). Merged images (C, F, I, and L) show overlap of the organellar markers with NHA2. Note the absence of plasmalemmal staining for NHA2. (B) Mitochondrial isolation of HEK293 and NRK cells. Native NHA2 is present in the 3000 × g pellets as the mitochondrial proteins Tom40 and CPS1. LAMP1, a lysosomal membrane protein, partially co-purifies with the mitochondrial pellet but is also present in the 3000 × g supernatant. (C) Fluorescence immunocytochemical images of NRK cells treated with Mitotracker and stained with MAB45D12D. Disappearance of the MAB45D12D signal after siRNA transfection. (D and E) Immunogold electron microscopy of rat DCT cells using MAB45D12D. NHA2 is present on the inner mitochondrial membrane. Bar = 1 μm. (F) Quantitative gold particle density of rat DCT. Bars and error bars are means ± SD (n = 56). Background particle density (gold-conjugated secondary antibody only) was <0.5/μm2.
To study intracellular localization of NHA2 in native tissue, we performed immunogold electron microscopy of rat DCT cells in the kidney using MAB45D12D, which demonstrates presence of NHA2 on the inner membrane of mitochondria (Figure 5, D and E). In addition to mitochondria, NHA2 is seen in other intracellular organelles. Most of these are vesicular structures. A small amount of NHA2 is also seen in other locations. Figure 5F shows quantitative gold particle density of NHA2 in the DCT. The most abundant sites were mitochondria and intracellular vesicles. A few or no gold particles were seen in the proximal tubule, which was very variable and much lower than that of the DCT (data not shown).
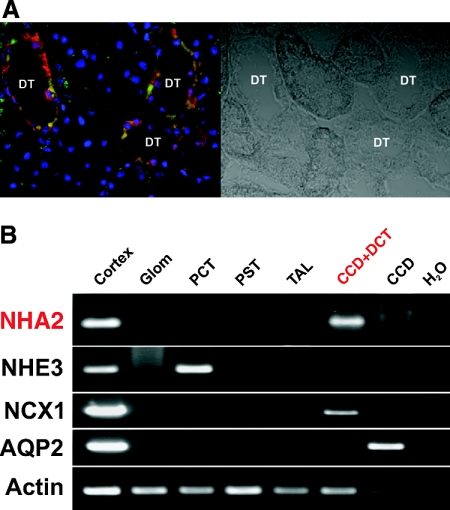
The quasi-ubiquitous expression of NHA2 is striking, and the question arises in which cell types NHA2 expression is present in different tissues. By immunohistochemistry analysis of mouse tissues, NHA2 is present in epithelial cells of the small intestine, lung, mammary gland, and renal tubules. In addition, NHA2 is strongly expressed in neurons of the brain (Figure 4). Although quite ubiquitous in organ distribution, within each organ NHA2 is not universally expressed in all cells. In the kidney, NHA2 seems to be exclusively expressed in distal tubular cells (Figure 4) by light microscopy. This is further supported by co-labeling of NHA2 with calbindin 28k, a DCT marker (Figure 6A), and exclusive presence of the transcript in DCT by RT-PCR of microdissected rat tubules (Figure 6B). This is comparable to the immunogold electron microscopy described in Figure 5.
Figure 6.
NHA2 localizes to DCT. (A) Immunofluorescence confocal microscopy of rat kidney cortex showing co-staining of calbindin 28k (red) and NHA2 (green) in distal tubules. (B) RT-PCR of NHA2 with appropriate control transcripts from microdissected rat tubules. Microdissection and RT-PCR were performed as described in the Concise Methods section. NHA2 could be amplified from DCT–cortical collecting duct (CCD) section. Note that separation of DCT from CCD was not possible because of technical limitations.
To study function of NHA2, we took advantage of the fact that overexpressed NHA2 can be detected on the plasma membrane of mammalian cells. For this purpose, we used cells devoid of plasmalemmal cation/proton exchange activity to express our constructs. This included AP-1 cells that have been derived from CHO cells by means of random chemical mutagenesis and skin fibroblasts derived from NHE1 knockout mice.14 First, we used a well-documented NHE transport assay using the intracellularly trapped pH-sensitive dye BCECF15 and were unable to detected cation/proton exchange (data not shown). We then resorted to a recently developed electrophysiologic approach that allows determination with high sensitivity of simultaneously ion fluxes and currents elicited by NHE16; however, even with the latter highly sensitive method combined with using highly favorable pH and cation gradients, we were unable to demonstrate NHA2-induced cation (Na+, K+, or H+) fluxes or currents in a mammalian system using untagged expression constructs and NHA2 from different species (human and mouse; data not shown).
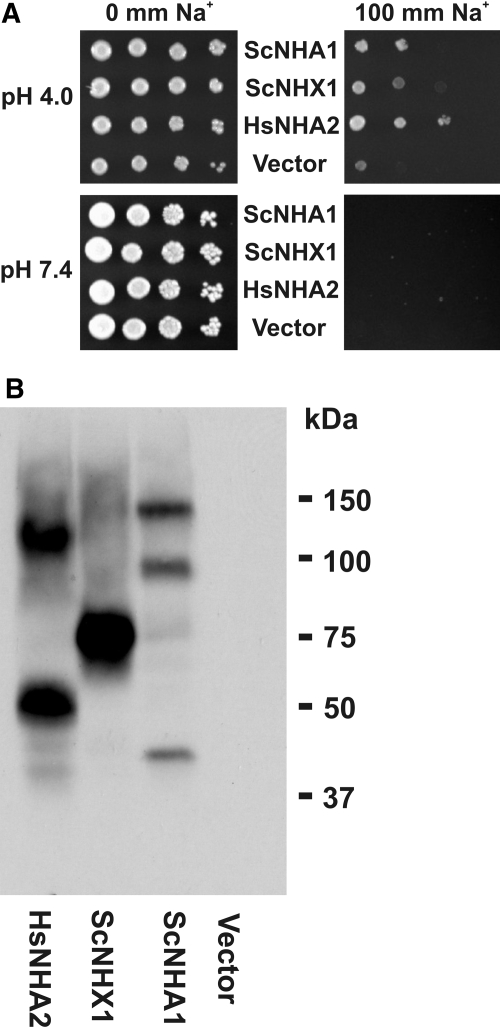
Because NHA2 has high similarity to bacterial and fungal NHE, we examined the ability of heterologously expressed human NHA2 to rescue the salt-sensitive growth phenotype of Saccharomyces cerevisiae strains lacking two B31 2/6c (ena1–4Δnha1Δ) or three AXT3 (ena1–4Δnha1Δnhx1Δ) major salt-handling ion transporters.17–19 As expected, the host strain AXT3 transformed with the empty vector is unable to grow at 100 mM extracellular Na+, at both extracellular pH of 4.0 and 7.4 (Figure 7A). Expression of prevacuolar (Nhx1) and plasmalemmal (Nha1) S. cerevisiae NHE and human NHA2 all rescue the salt-sensitive phenotype at pH 4 but not at pH 7.4. This finding is compatible with NHA2-induced Na+/H+ exchange activity. Expression of C-terminally HA-tagged NHA2 constructs in yeast are shown in Figure 7B. Findings were identical for the yeast strain B31 2/6c (data not shown). At mildly acidic extracellular pH of 6.0, no rescue of the salt-sensitive phenotype could be observed. Also, no rescue could be observed when Na+ was replaced by K+ (data not shown). We also determined K+ transport in a K+ uptake–deficient S. cerevisiae strain WΔ3 (trk1Δtrk2Δ) that grows only on high external K+ medium.20 At different pH, no rescue could be observed with heterologous expression of NHA2 (data not shown). All of these observations are in agreement with the functional characterization by Xiang et al.9 and suggest that NHA2 indeed is a functional Na+-selective NHE.
Figure 7.
Functional complementation of a salt-sensitive S. cerevisiae strain by heterologous expression of human NHA2. (A) Salt-sensitive yeast strain AXT3 was transformed with S. cerevisiae NHE (ScNha1 and ScNhx1), human NHA2, or empty vector p426TEF. Cells (5 × 104) were plated with serial 10-fold dilutions (left to right) on AP medium plates with indicated Na+ concentrations and pH. Growth was assessed after 4 d. (B) S. cerevisiae AXT3 strain expressing C-terminally HA-tagged expression constructs. Equal amounts of yeast lysates from 10 OD units were loaded, separated by SDS-PAGE, and immunoblotted with a polyclonal anti-HA antibody.
DISCUSSION
In this study, we describe the detailed characterization of a novel, almost ubiquitously expressed sodium/hydrogen exchanger NHA2. Mitchell and Moyle2 were the first to propose a mitochondrial cation/proton antiport system. Because of the highly favorable electrochemical gradient, mitochondria represent a sink for cations. High mitochondrial sodium concentrations can lead to mitochondrial swelling and, in parallel with sodium/calcium exchange, can lead to calcium accumulation in the mitochondria.21 The finding of mitochondrial localization of NHA2 may at first glance contrast with the striking cell-specific expression of NHA2 in the organs examined. There is, however, both functional and biochemical evidence for a heterogeneity of mitochondrial cation/proton antiporters.22–27 Also, there is ample evidence for tissue-specific differences in mitochondria, even along the nephron.28,29 NHA2 does not have the classical N-terminal motif for mitochondrial import but may use internal recognition signals for translocation into mitochondria.30 The sodium-selective NHE of mitochondria has been partially purified and reconstituted in proteoliposomes with a predicted molecular weight of approximately 59 kD.23 This is very close to the predicted and detected molecular weight of approximately 57 kD of human and rodent NHA2. In S. cerevisiae, NHE activity can be demonstrated for NHA2 under highly favorable conditions, including a 3-log-unit difference in proton concentrations. The physiologic implications are unclear because this surrogate system is considerably divergent from the native habitat of NHA2. High proton gradients may be found in gastric epithelia, medullary collecting duct of the kidney under maximal urinary acidification, or possibly microdomains between organelles and cytoplasm. It is also conceivable that the overexpression system S. cerevisiae is lacking accessory proteins or the right lipid environment for proper function of NHA2, mandating the requirement of a high pH gradient.
During the preparation of this manuscript, Battaglino and Xiang and co-workers9–11 published initial reports on NHA2. Our data partially support and extend these initial findings. The one major discrepancy between the findings of Battaglino and those of Xiang and ours is that Battaglino found bone-restricted expression of NHA2. The reason for this incongruity is unclear.
Although NHA2 is present on the plasma membrane of certain specialized cells, in many cell types it seems to be a completely intracellular protein. It has been documented for several “intracellular” NHE that they can also reside and function on the plasma membrane under certain circumstances,31–33 and dual mitochondrial and plasmalemmal localization has also been reported for the voltage-dependent anion channel VDAC.34 For the intracellular NHE isoform NHE6, the scaffolding protein RACK1 was recently shown to regulate the balance between plasmalemmal and intracellular localization.31 Clearly, further studies are needed to elucidate the regulatory pathways underlying this dual localization.
One salient finding is that renal NHA2 is specific to the DCT. Because only 1% of mitochondrial proteins are encoded by the mitochondrial genome, protein import from nucleus-originated transcripts is not uncommon and in fact is expected to exhibit cell specificity. One unique feature of the mammalian DCT is that it mediates the highest transcellular flux of calcium. The mitochondria act in concert with many other mechanisms to provide intracellular calcium buffering in many cell types,35 including the DCT.36 Mitochondrial sodium/calcium exchange may be deployed by the DCT to unload the mitochondria of calcium as previously shown in cardiac cells.37 This forward sodium/calcium exchange may burden the mitochondria with increased sodium load. This can be extruded by sodium/proton exchange by expending part of the proton gradient that is usually used for ATP generation. These and other speculations for the DCT-specific expression remain to be proved.
In summary, our data indicate that NHA2 is both an intracellular and a plasmalemmal sodium/proton exchanger expressed in multiple tissues but in a cell type–specific manner. Whether this is indeed the cation/proton exchanger of Mitchell and what the physiologic function for NHA2 is remains to be determined.
CONCISE METHODS
Chemicals
Unless otherwise specified, all chemicals and reagents were obtained from Sigma (St. Louis, MO).
DNA Constructs
Human brain cDNA library in pADSL-Nx vector was a gift from I. Stagljar (University of Toronto, Toronto, ON, Canada). Human NHA2 (accession no. NM_178833) was cloned into pMH vector (Roche, Indianapolis, IN) containing a C-terminal HA-tag. Human NHA1 (accession no. BC132712) was cloned into pEGFP-N3 (Clontech, Mountain View, CA). Monoclonal anti-GFP antibody was obtained from Clontech. S. cerevisiae Nha1 and Nhx1 cDNA were obtained from Open Biosystems (Huntsville, AL) and subcloned into p426TEF expression vector.38 C-terminal HA tags were introduced by PCR. All constructs were verified by sequencing.
Generation of MAb
Mouse mAb were raised against the peptide H2N-CDTARSHGEKQLEDYG-COOH linked to KLH as described previously.39 The anti-NHA2 mAb designated MAB45D12D was determined to belong to the IgG2a subclass as revealed by an ELISA-based mouse typer subisotyping kit (Bio-Rad, Hercules, CA).
Cell Culture, Transfection, and siRNA
NRK and HEK293 cells were obtained from ATCC and cultured in high-glucose (450 mg/dl) DMEM supplemented with 10% (vol:vol) FBS. INS-1E cells were obtained from P. Maechler (University of Geneva, Geneva, Switzerland) and grown in RPMI-1640 medium supplemented with 10% (vol:vol) FBS. NHE1-deficient AP-1 cells, derived from CHO cells (gift from S. Grinstein, Hospital for Sick Children, Toronto, ON, Canada), were maintained in α-modified minimal essential medium. Transient transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and cells were used 24 to 36 h after transfection for further experiments.
Two independent stealth siRNA targeting rat NHA2 (1, forward 5′-AGGAACGUCCCAGUCAUCAGUGAUA-3′; 2, 5′-CAUCCCUCUUAUCCGCUCUUCAUAU-3′) were obtained from Invitrogen and transfected into NRK cells according to the manufacturer's instructions.
Yeast Transformation and Growth Selection
Expression constructs were transformed into S. cerevisiae strains AXT3 (MATα, ade2, his3, leu2, trp1, ura3, ena1::HIS3::ena4, nha1::LEU2, nhx1::TRP1), WΔ3 (MATα, ade2, his3, leu2, trp1, ura3, trk1::LEU2, trk2::HIS3), or B31 2/6c (MATα, ade2, can1, his3, leu2, trp1, ura3, mal10 ena1::HIS3::ena4, nha1::LEU2). Transformation of yeast was performed as described previously.40 Yeast were grown at 30°C on AP medium agar plates containing 10 mM arginine, 8 mM phosphoric acid, 2% glucose, 2 mM MgSO4, 1 mM KCl, 0.2 mM CaCl2, trace minerals, and vitamins.41 pH was adjusted with phosphoric acid or Tris base. Salt-sensitive growth was assayed after 4 d.
For immunoblot, 10 OD units of yeast were washed once with 1 mM EDTA/H2O and lysed in 2 M NaOH for 10 min on ice. Proteins were isolated by TCA/acetone precipitation, resuspended in SDS sample buffer (25 mM Tris-Cl [pH 6.8], 9 M urea, 1 mM EDTA, 1% [wt:vol] SDS, 0.7 M β-mercaptoethanol, 10% [vol:vol] glycerol), separated by SDS-PAGE, and analyzed by immunoblotting.
Mitochondrial Isolation
Mitochondria were isolated using the Pierce (Rockford, IL) mitochondrial isolation kit based on differential centrifugation. As mitochondrial fraction, the 3000 × g pellet was used. Polyclonal anti-Tom20 and anti-Tom40 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); polyclonal anti-Lamp1 antibody was obtained from Abcam (Cambridge, MA).
RBC Isolation
RBC ghosts were isolated from freshly drawn human whole blood as described previously.42 Briefly, sedimented RBC were washed three times with saline, and RBC ghosts were prepared by hypotonic lysis in 5 mM sodium phosphate/1 mM EDTA (pH 8.0) in the presence of proteinase inhibitors (Roche). The lysate was centrifuged at 25,000 × g for 20 min, and the supernatant was removed. The membrane pellets were resuspended and washed five additional times in lysis buffer before further use.
Immunoprecipitation, Cell-Surface Biotinylation, and Immunoblotting
Immunoprecipitation and cell-surface biotinylation were performed as described previously.15 For immunoblotting, undiluted hybridoma cell culture supernatant containing 40 mM Tris-HCl (pH 7.4) and 0.2% Tween 20 or monoclonal or polyclonal anti-HA antibodies (1:1000 dilution) were used followed by secondary horseradish peroxidase–conjugated goat anti-mouse or goat anti-rabbit IgG antibodies (1:10,000 and 1:20,000 dilution, respectively; Bio-Rad). Signals were detected using enhanced chemiluminescence. Monoclonal anti-NHE1 antibody (clone 4E9) was obtained from Calbiochem (Temecula, CA); monoclonal anti-NHE8 antibody has been described before.32
Immunocytochemistry and Immunohistochemistry
Preparation of samples for immunocytochemistry and immunohistochemistry was performed as described previously.15 Confluent quiescent NRK cells were fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized in 0.1% Triton X-100 in PBS for 3 min, and blocked by 1.5% BSA and 10% goat serum in PBS for 1 h. Fixed monolayers were incubated with anti-NHA2 mAb (1:3), followed by Alexa 488–conjugated goat anti-mouse antibody (1:800; Invitrogen), and then incubated with anti-GOLPH4 antibody (1:400; Abcam) or anti-PDI antibody (ER marker, 1:50; Abcam) or anti-CPS1 antibody (mitochondrial marker, 1:5000; Abcam) followed by Alexa 568–conjugated goat anti-rabbit antibody (1:800; Invitrogen). Some samples were stained with Mitotracker Red 580 (Invitrogen). Images were visualized with a Zeiss LSM-510 laser scanning confocal microscope. Degree of overlap between NHA2 and the organellar larkers were quantified by ImageJ43 as fluorescence area yellow/yellow + green integrated over approximately 50 randomly selected cells.
Kidney sections, after blocking with 1.5% BSA/10% goat serum, were incubated with undiluted hybridoma supernatant (MAB45D12D) or rabbit anti-calbindin 28k (Swant, Bellinzona, Switzerland) antibody diluted 1:100 in 1.5% BSA/5% goat serum, followed by Alexa 488–or Alexa 568–conjugated goat anti-mouse or goat anti-rabbit antibody diluted 1:500 in 1.5% BSA/5% goat serum. Confocal images were obtained using a Zeiss LSM-510 laser-scanning confocal microscope.
Immunogold Electron Microscopy and Gold Particle Quantification
Anesthetized rats were laparatomized, and the aorta was ligated proximal to the renal arteries. Kidneys were fixed in situ by perfusion with 2.5% paraformaldehyde in PBS (pH 7.4), delivered via the distal aorta. Kidneys were then removed and postfixed in 4% paraformaldehyde in PBS at 4°C for 4 h. Immunogold labeling of ultrathin frozen tissue sections was performed according to the method of Tokuyasu.44 Kidney cortex was infiltrated with 2.3 M sucrose overnight and frozen in liquid nitrogen, and 70- to 80-nm sections were cut on a ultramicrotome (Leica ultracut UCT; Leica Mikrosysteme Gmbh, Vienna, Austria) and mounted on Formvar-coated nickel grids. Sections were processed using the following steps: Rehydration with PBS containing 20 mM glycine for 10 min followed by PBS (5 min × 2); blocking with 1.5% BSA in PBS for 15 min; incubation with anti-NHA2 mAb (1:3) for 60 min; washing with 1.5% BSA in PBS for 5 min and PBS (5 min × 3); incubation with gold-conjugated goat anti-mouse IgG antibody (10-nm gold particles, 1:5 dilution) for 60 min; washing with PBS (5 min × 3); fixation with 1% glutaraldehyde for 5 min; washing in water (5 min × 3); staining with a mixture (8:5:1) of 3% methylcellulose-water-3% uranyl acetate. Samples were visualized with a JEOL 1200 EX transmission electron microscope at a magnification of ×30,000. Random sampling was used at every stage (selection of renal cortical tissue blocks, tissue sections, and transmission electron microscope fields) to yield a minimum of least 60 randomly selected electron micrographs for each analysis.45 Label quantification was performed according to the methods of Mayhew and Kaur.45,46 Gold particles were counted on each electron micrograph, and the areas represented by mitochondria, intracellular vesicles, nucleus, and other cellular parts were measured using the ImageJ software.43 Labeling density (number of gold particles/μm2) for each cellular compartment was calculated, and results were expressed as relative labeling index.45
Mouse Tissue Isolation and Rat Single-Tubule Microdissection
Mice were perfused with ice-cold PBS, and individual organs were then harvested for RNA or protein isolation. For protein isolation, organ tissue was homogenized in ice-cold RIPA buffer (in mM: 150 NaCl, 50 Tris-HCl [pH 8.0], 5 EDTA, 1 EGTA, Triton X-100 1% [vol:vol[, deoxycholate 0.5% [wt/vol], SDS 0.1% [wt/vol], and protease inhibitor cocktail from Roche), lysed for 1 h at 4°C, cleared twice at 10,000 × g for 15 min and immediately used for SDS-PAGE. Rat tubules were dissected as described previously.47 Total RNA from microdissected tubules was prepared using the Microprep kit (Stratagene, La Jolla, CA). Total RNA from cells and rat or mouse tissue was prepared as described by Chomczynski et al.48 cDNA was generated by oligo-dT–primed reverse transcription using the Thermoscript method (Invitrogen). The following primers were used for PCR: Mouse NHA2 forward 5′-TTTATCCAGTACTTCCCGAGCAGG-3′, reverse 5′-TTTCTGCTCTGAGAGACACAATGG-3′; mouse β-actin forward 5′-ATTGTTACCAACTGGGACGAC-3′, reverse 5′-CACCATGCTGGTCTCCGTATG-3′. The fidelity of PCR products was confirmed by direct sequencing. Other primers used included rat NHA2 forward 5′-ATTACGGGCCCTGAATGTCTTCCT-3′, reverse 5′-AGATGACGACCACTTGTGCTGGAT-3′; rat NHE3 forward 5′-AATCTCGAGATCGGATCCTG-3′, reverse 5′-CTCTGTTCCAAGGACTGCAT-3′; rat NCX1 forward 5′-CCAGCACTGCCACCATAACCATTT-3′, reverse 5′-AATGCGCCTCTCCTCCTCTTCTTT-3′; rat AQP2 forward 5′-TTGTCTTCTTTGGCCTTGGCTCAG-3′, reverse 5′-ACAGAGAAACCAATGGAGAGGGCA-3′; and rat β-actin forward 5′-TGACCCAGATCATGTTTGAGACC-3′, reverse 5′-CCATACCCAAGAAGGAAGGC-3′. For RNA blot, 10 μg of total RNA was size-fractionated by formaldehyde gel electrophoresis, transferred to nitrocellulose membranes, and probed with a random-primed 32P-labeled human NHA2 cDNA probe under high stringency conditions.
DISCLOSURES
None.
Acknowledgments
This work was supported by the Swiss National Science Foundation (grant 3100A0-117732), by a research fellowship of the European Union (Marie Curie Fellowship) and a research fellowship from the Pak Center of Mineral Metabolism to D.G.F.; and by the National Institutes of Health (R01-DK-48482 and P01-DK-54392), the Nephrology O'Brien Center (P30-DK079328), and the Simmon Family Foundation to O.W.M.
Some of the data were presented in abstract form at the annual meeting of the American Society of Nephrology, October 31 through November 5, 2007; San Francisco, CA.
We thank Drs. Lydie Mareschova, Rajini Rao, and José Pardo for yeast strains; Profs. Pierre Maechler and Claes Wollheim for INS-1E cells; and Dr. Ming-Chang Hu for helpful discussions. We acknowledge the technical expertise of Mr. Carlos Wotzkow.
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1.Orlowski J, Grinstein S: Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch 447: 549–565, 2004 [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P, Moyle J: Stoichiometry of proton translocation through the respiratory chain and adenosine triphosphatase systems of rat liver mitochondria. Nature 208: 147–151, 1965 [DOI] [PubMed] [Google Scholar]
- 3.Wang D, King SM, Quill TA, Doolittle LK, Garbers DL: A new sperm-specific Na+/H+ exchanger required for sperm motility and fertility. Nat Cell Biol 5: 1117–1122, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Putney LK, Denker SP, Barber DL: The changing face of the Na+/H+ exchanger, NHE1: Structure, regulation, and cellular actions. Annu Rev Pharmacol Toxicol 42: 527–552, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Bobulescu IA, Di Sole F, Moe OW: Na+/H+ exchangers: Physiology and link to hypertension and organ ischemia. Curr Opin Nephrol Hypertens 14: 485–494, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saier MH: Transport Classification Database. Available at: http://www.tcdb.org/tcdb/index.php?tc=2.A.36. Accessed May 5, 2008
- 7.Brett CL, Donowitz M, Rao R: Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol 288: C223–C239, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Ye G, Chen C, Han D, Xiong X, Kong Y, Wan B, Yu L: Cloning of a novel human NHEDC1 (Na+/H+ exchanger like domain containing 1) gene expressed specifically in testis. Mol Biol Rep 33: 175–180, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Xiang M, Feng M, Muend S, Rao R: A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc Natl Acad Sci U S A 105: 18677–18681, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham L, Purcell P, Morse L, Stashenko P, Battaglino RA: Expression analysis of nha-oc/NHA2: A novel gene selectively expressed in osteoclasts. Gene Expr Patterns 7: 846–851, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battaglino RA, Pham L, Morse LR, Vokes M, Sharma A, Odgren PR, Yang M, Sasaki H, Stashenko P: NHA-oc/NHA2: A mitochondrial cation-proton antiporter selectively expressed in osteoclasts. Bone 42: 180–192, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H: Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435: 1197–1202, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, Maechler P: Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology 145: 667–678, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Rotin D, Grinstein S: Impaired cell volume regulation in Na(+)-H+ exchange-deficient mutants. Am J Physiol 257: C1158–C1165, 1989 [DOI] [PubMed] [Google Scholar]
- 15.Fuster DG, Bobulescu IA, Zhang J, Wade J, Moe OW: Characterization of the regulation of renal Na+/H+ exchanger NHE3 by insulin. Am J Physiol Renal Physiol 292: F577–F585, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuster D, Moe OW, Hilgemann DW: Lipid- and mechanosensitivities of sodium/hydrogen exchangers analyzed by electrical methods. Proc Natl Acad Sci U S A 101: 10482–10487, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintero FJ, Blatt MR, Pardo JM: Functional conservation between yeast and plant endosomal Na(+)/H(+) antiporters. FEBS Lett 471: 224–228, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Maresova L, Sychrova H: Arabidopsis thaliana CHX17 gene complements the kha1 deletion phenotypes in Saccharomyces cerevisiae. Yeast 23: 1167–1171, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Maresova L, Sychrova H: Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol Microbiol 55: 588–600, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Madrid R, Gomez MJ, Ramos J, Rodriguez-Navarro A: Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem 273: 14838–14844, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Li W, Shariat-Madar Z, Powers M, Sun X, Lane RD, Garlid KD: Reconstitution, identification, purification, and immunological characterization of the 110-kDa Na+/Ca2+ antiporter from beef heart mitochondria. J Biol Chem 267: 17983–17989, 1992 [PubMed] [Google Scholar]
- 22.Garlid KD: Sodium/proton antiporters in the mitochondrial inner membrane. Adv Exp Med Biol 232: 37–46, 1988 [DOI] [PubMed] [Google Scholar]
- 23.Garlid KD, Shariat-Madar Z, Nath S, Jezek P: Reconstitution and partial purification of the Na(+)-selective Na+/H+ antiporter of beef heart mitochondria. J Biol Chem 266: 6518–6523, 1991 [PubMed] [Google Scholar]
- 24.Li XQ, Hegazy MG, Mahdi F, Jezek P, Lane RD, Garlid KD: Purification of a reconstitutively active K+/H+ antiporter from rat liver mitochondria. J Biol Chem 265: 15316–15322, 1990 [PubMed] [Google Scholar]
- 25.Kakar SS, Mahdi F, Li XQ, Garlid KD: Reconstitution of the mitochondrial non-selective Na+/H+ (K+/H+) antiporter into proteoliposomes. J Biol Chem 264: 5846–5851, 1989 [PubMed] [Google Scholar]
- 26.Martin WH, Beavis AD, Garlid KD: Identification of an 82,000-dalton protein responsible for K+/H+ antiport in rat liver mitochondria. J Biol Chem 259: 2062–2065, 1984 [PubMed] [Google Scholar]
- 27.Nakashima RA, Garlid KD: Quinine inhibition of Na+ and K+ transport provides evidence for two cation/H+ exchangers in rat liver mitochondria. J Biol Chem 257: 9252–9254, 1982 [PubMed] [Google Scholar]
- 28.Kendall MD, Warley A, Nicholson JK, Appleton TC: X-ray microanalysis of proximal and distal tubule cells in the mouse kidney, and the influence of cadmium on the concentration of natural intracellular elements. J Cell Sci 62: 319–338, 1983 [DOI] [PubMed] [Google Scholar]
- 29.Palmieri F: The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflugers Arch 447: 689–709, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N: Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep 9: 42–49, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohgaki R, Fukura N, Matsushita M, Mitsui K, Kanazawa H: Cell surface levels of organellar Na+/H+ exchanger isoform 6 are regulated by interaction with the receptor for activated C-kinase 1. J Biol Chem 283: 4417–4429, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Bobulescu IA, Goyal S, Aronson PS, Baum MG, Moe OW: Characterization of Na+/H+ exchanger NHE8 in cultured renal epithelial cells. Am J Physiol Renal Physiol 293: F761–F766, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill JK, Brett CL, Chyou A, Kallay LM, Sakaguchi M, Rao R, Gillespie PG: Vestibular hair bundles control pH with (Na+, K+)/H+ exchangers NHE6 and NHE9. J Neurosci 26: 9944–9955, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bathori G, Parolini I, Tombola F, Szabo I, Messina A, Oliva M, De Pinto V, Lisanti M, Sargiacomo M, Zoratti M: Porin is present in the plasma membrane where it is concentrated in caveolae and caveolae-related domains. J Biol Chem 274: 29607–29612, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Berridge MJ, Lipp P, Bootman MD: The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1: 11–21, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Fowler MR, Hunter M: Mitochondrial Ca2+ transport in frog early distal tubule. Exp Physiol 90: 195–201, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Kim B, Matsuoka S: Mitochondrial Ca2+ flux through Na+/Ca2+ exchange. Ann N Y Acad Sci 1099: 507–511, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Mumberg D, Muller R, Funk M: Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122, 1995 [DOI] [PubMed] [Google Scholar]
- 39.Andersson S, Jornvall H: Sex differences in cytochrome P-450-dependent 25-hydroxylation of C27-steroids and vitamin D3 in rat liver microsomes. J Biol Chem 261: 16932–16936, 1986 [PubMed] [Google Scholar]
- 40.Schiestl RH, Gietz RD: High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346, 1989 [DOI] [PubMed] [Google Scholar]
- 41.Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM: Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci U S A 99: 9061–9066, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett V: Proteins involved in membrane-cytoskeleton association in human erythrocytes: Spectrin, ankyrin, and band 3. Methods Enzymol 96: 313–324, 1983 [DOI] [PubMed] [Google Scholar]
- 43.Rasband WS: Image J, Bethesda, National Institutes of Health, 1997 to 2007. Available at: http://rsb.info.nih.gov/ij/
- 44.Tokuyasu KT: Immunochemistry on ultrathin frozen sections. Histochem J 12: 381–403, 1980 [DOI] [PubMed] [Google Scholar]
- 45.Mayhew TM: How to count your gold: A tutorial on TEM immunogold label quantification. Microsc Anal 19: 9–12, 2005 [Google Scholar]
- 46.Kaur R, Raje M: A solid-phase method for evaluation of gold conjugate used in quantitative detection of antigen by immunogold-labeling electron microscopy. J Immunol Methods 279: 33–40, 2003 [DOI] [PubMed] [Google Scholar]
- 47.Baum M, Moe OW, Zhang J, Dwarakanath V, Quigley R: Phosphatonin washout in Hyp mice proximal tubules: Evidence for posttranscriptional regulation. Am J Physiol Renal Physiol 288: F363–F370, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159, 1987 [DOI] [PubMed] [Google Scholar]