Abstract
Cytochrome c release from mitochondria promotes apoptosome formation and caspase activation. The question as to whether mitochondrial permeabilization kills cells via a caspase-independent pathway when caspase activation is prevented is still open. Here we report that proneural cells of embryonic origin, when induced to die but rescued by apoptosome inactivation are deprived of cytosolic cytochrome c through proteasomal degradation. We also show that, in this context, those cells keep generating ATP by glycolysis for a long period of time and that they keep their mitochondria in a depolarized state that can be reverted. Moreover, under these conditions, such apoptosome-deficient cells activate a Beclin 1–dependent autophagy pathway to sustain glycolytic-dependent ATP production. Our findings contribute to elucidating what the point-of-no-return in apoptosis is. They also help in clarifying the issue of survival of apoptosome-deficient proneural cells under stress conditions. Unraveling this issue could be highly relevant for pharmacological intervention and for therapies based on neural stem cell transfer in the treatment of neurological disorders.
INTRODUCTION
Apoptosome formation is the most common pathway involved in apoptosis. This complex is formed when, upon mitochondrial outer membrane permeabilization (MOMP) caused by apoptotic inducers, cytochrome c is released from the mitochondrial intermembrane space where it normally resides (Liu et al., 1996; Kluck et al., 1997). In the cytoplasm, cytochrome c binds to Apaf1 and induces its oligomerization. Procaspase-9 is recruited to the complex and activated. Active caspase-9, in turn, promotes activation of executioner caspases that are responsible for the cleavage of cellular substrates, resulting in the biochemical and morphological changes typical of apoptosis (Li et al., 1997; Srinivasula et al., 1998; Earnshaw et al., 1999; Saleh et al., 1999; Zou et al., 1999).
During apoptosis, MOMP allows the release of cytochrome c and other proapoptotic proteins. Mitochondrial permeabilization is regulated by proteins of the Bcl-2 family, although its exact mechanism has not been clarified (Susin et al., 1996; Kluck et al., 1997; Yang et al., 1997; Li et al., 1998; Luo et al., 1998).
The absence of the apoptosome inhibits the completion of the apoptotic process in many systems. In these conditions, cells that have been induced to die cannot complete the apoptotic program, even though they show some early biochemical modifications that are typical of programmed cell death and that might affect cell survival. One of them is the release of the cytochrome c from mitochondria. Are these modifications irreversible and do they lead invariably to cell death, or can cells with apoptosome impairment triggered to death still be metabolically active and therefore able to recover? What is the point-of-no-return in apoptosis? These are frequently addressed questions that remain to be clarified (Martinou et al., 1999; Cozzolino et al., 2004).
The release of cytochrome c has often been considered the point-of-no return in cell death because cytochrome c, as a part of the mitochondrial respiratory chain, is responsible for the generation of the mitochondrial membrane potential (ΔΨm). ΔΨm is needed for various functions including the production of ATP via oxidative phosphorylation. Permeabilization of the mitochondrial outer membrane and release of cytochrome c should therefore, in principle, have a strong impact on mitochondrial function because the proton gradient generated by the electron transport chain may be impaired.
However, this point is still controversial. Some reports show that release of cytochrome c and disruption of ΔΨm will kill the cell in a caspase-independent way if downstream apoptotic effects are blocked. In fact, in cells lacking caspase-9 or Apaf1, death is delayed but still inevitable, and such cells are considered essentially “zombies,” i.e., severely impaired cells that cannot be defined as living normal cells (Ekert et al., 2004). By contrast, other studies find that cytochrome c released and diffused in the cytoplasm is able to maintain ΔΨm and ATP generation in caspase-inhibited cells and in Apaf1−/− cells. This suggests that mitochondrial recovery and cell survival are possible after the release of cytochrome c, and therefore, that this is not the point-of-no-return (Goldstein et al., 2000; Waterhouse et al., 2001). However, it has been reported by the same authors that the ability to recover the ΔΨm and to produce ATP in apoptosome-deficient cells is possible only soon after toxic treatment, whereas over time cells show a decrease of ΔΨm (which is considered the point-of-no-return) as well as ATP production (Waterhouse et al., 2001).
To shed light on the ability of cells devoid of the apoptosomal function to survive under prolonged apoptotic insults, we analyzed cell metabolism in Apaf1-deprived neural precursors (ETNA−/− cells). We decided to use this cell system for the following reasons: 1) The issue of proneural cell survival is of great importance in biomedicine, because therapies based on neural stem cell transfer are promising in treating several neurodegenerations and one of the main concerns regards the viability of such cells after transplantation; 2) ETNA proneural cells are very sensitive to a series of proapoptotic stimuli; and 3) we have demonstrated that their genetic manipulation may interfere with the death pathway without affecting their capability to grow and differentiate in vitro (Cozzolino et al., 2004). Because it has been suggested that the requirement and coupling of caspase-9 to Apaf1 are both context-dependent, which means that Apaf1 and caspase-9 deficiency can have different effects on the ability of cells to resist to apoptosis (Hakem et al., 1998; Ho et al., 2004), we also set out to establish another apoptosome-deficient system, i.e., a cell line overexpressing a dominant negative form of caspase-9 (C9DNETNA cells).
We found that apoptosome-deficient proneurons, escaping apoptosis induced by tunicamycin or actinomycin D, switch to autophagy and survive through a glycolytic metabolism for a long period of time before dying. In this context, autophagy is essential to support glycolysis, ATP production and therefore to allow cell survival. Moreover, we found that, after its release from mitochondria, cytochrome c is degraded by a proteasome-dependent mechanism. These findings may be relevant to devise novel strategies to improve or to reduce cell survival under stressful conditions.
MATERIALS AND METHODS
Reagents and Plasmid Constructs
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. MG132 and lactacystin were purchased from Calbiochem (La Jolla, CA) and hygromycin B from GIBCO (Rockville, MD).
Wild type caspase-9 was amplified from a mouse cDNA library using the sense primer 5′-GCAATCTCGAGGCCATGGACGAGGCGGACCGGCAG CTCCTGC and the antisense primer 5′-AACAGCTCGAGGCCTCATGAAGTTTTAAAAAACAGCTTTTTC. The PCR product was cloned into pDrive (Qiagen, Chatsworth, CA), and from there it was cut with EcoRI and ligated into pcDNA3.1 vector (Invitrogen, Carlsbad, CA) at EcoRI site to obtain the pcDNA3.1-C9WT expression vector. Myc-tagged cytochrome c (pcDNA3.1Cytc-myc) was created by amplifying the cytochrome c from a murine cDNA library using the following primers: 5′-CTATAAGCTTACCATGGGTGATGTTGAAAA and 5′-CTTTGATATCCTCATTAGTAGCCTT and by cloning the PCR product into pcDNA/3.1myc-His (Invitrogen) between the HindIII and EcoRV sites. The pcDNA3.1 plasmid coding mutant dominant negative caspase-9 (pcDNA3.1-C9DN) was created by performing a site-directed mutagenesis on pcDNA3.1-C9WT following the manufacturer's instructions (Stratagene, La Jolla, CA) through which the cysteine 325 (C325S) of the caspase-9 active site (QACGG) was changed in serine (QASGG). For this purpose the following primers were used: 5′-CTCTTCTTCATCCAAGCTTCCGGTGGTGAGCAGAAG-3′ and 5′-CTTTCTGCTCACCACCGGAAGCTTGGATGAAGAAGAG-3′. To obtain the four mutants of myc-tagged murine holo-cytochrome c (pcDNA3.1Cytc-mycK6R, K8R, K9R, and K8,9R), cytochrome c cDNA was amplified from a mouse cDNA library using four different sense primers. For pcDNA3.1Cytc-mycK6R construct: 5′-CTATAAGCTTACCATGGGTGATGTTGAAAGAGGCAAGAAGATTTTTGTTCAG. For pcDNA3.1Cytc-mycK8R construct: 5′-CTATAAGCTTACCATGGGTGATGTTGAAAAAGGCAGGAAGATTTTTGTTCAG. For pcDNA3. 1Cytc-mycK9R construct: 5′- CTATAAGCTTACCATGGGTGATGTTGAAAAAGGCAAGAGGATTTTTGTTCAG. For pcDNA3.1cytc-mycK8,9R construct: 5′- CTATAAGCTTACCATGGGTGATGTTGAAAAAGGCAGGAG GATTTTTGTTCAG and for all of them the antisense primer 5′-CTTTGATATCCTCATTAGTAGCCTT. The PCR products were cloned into pcDNA/3.1myc-His between the HindIII and EcoRV sites.
Cell Cultures and Transfection
ETNA wild-type and ETNA−/− cells were obtained as described elsewhere (Cozzolino et al., 2004). They were routinely grown in DMEM (GIBCO) + 10% FCS, at 33°C in an atmosphere of 5% CO2 in air.
To induce apoptosis, cells were treated with 20 μM actinomycin D, 5 μM staurosporine or 3 μg/ml tunicamycin. To inhibit the proteasome, cells were treated with 3 μg/ml tunicamycin or 20 μM actinomycin D for 24 h and then MG132 (5 μM) or lactacystin (10 μM) were added for an additional 40 h.
Transient expression of each vector (1.5 μg DNA/3–5 × 105 cells) was obtained with LipofectAMINE Plus reagent (Invitrogen) according to the manufacturer's instructions. ETNA cells stably expressing C9DNETNA were obtained by transfecting pCDNA3.1-C9DN (1.5 μg DNA/3–5 × 105 cells) concomitantly with a vector conferring hygromycin B resistance (0.15 μg DNA/3–5 × 105 cells), and clones were selected by treatment with hygromycin B. To discard the possible selection of clones highly resistant to caspase-independent cell death, we performed most of the experiments using two ETNA−/− cell clones (ETNA−/− and ETNA−/−C2) and two C9DNETNA clones (C9DNETNA and C9DNETNA-C6). For preparation of mouse embryonic fibroblasts (MEFs), embryonic day 13.5 (e13.5) embryos from heterozygous Apaf1 pregnant females were killed; the heads, viscera, and limbs were removed, and the rest of the bodies was treated as previously reported (Cecconi et al., 1998).
RNA Interference
Small interfering RNA (siRNA) oligonucleotides corresponding to the murine Beclin 1 sequence were purchased from Dharmacon (siGENOME SMARTpool M-055895, Mouse BECN1; Research, Boulder, CO). Cells (2 × 105 well) were transfected with 100 pmol siRNA in 35-mm Petri dishes by Lipofectamine 2000 (Invitrogen) as indicated by the manufacturer. Forty-eight hours after transfection cells were treated with tunicamycin for 48 h or left untreated. Beclin 1 down-regulation was assessed by Western blot analysis.
Immunoprecipitation and Western Blotting
After rinsing the cultures with ice-cold phosphate-buffered saline (PBS), cell lysis was performed in lysis buffer (20 mM Tris-HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2) containing 1 mM PMSF and a protease inhibitor cocktail (Sigma-Aldrich). A clear supernatant was obtained by centrifugation of lysates at 13,000 × g for 10 min. Protein content was determined using Bradford protein assay (Bio-Rad, Richmond, CA).
For the detection of ubiquitinated cytochrome c, 100 μg of protein was immunoprecipitated with anti-cytochrome c mAb (6H2.B4, PharMingen, San Diego, CA) coupled to protein G-Sepharose beads (Roche, Indianapolis, IN).
Western blots were performed on polyvinylidene difluoride membranes (Immobilon P, Millipore, Bedford, MA). Equal loading of samples was confirmed by β-tubulin normalization. Mouse anti-ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit polyclonal antibody raised against cytochrome c (Santa Cruz Biotechnology) was used on anti-cytochrome c immunoprecipitates for the detection of ubiquitinated cytochrome c. Cleaved caspase-3 was detected with a polyclonal antibody against the large fragment of activated caspase-3 (Cell Signaling, Beverly, MA). For the detection of both full-length and cleaved active form of PARP, an anti-PARP rabbit polyclonal antibody (Cell Signaling) was used. For the analysis of cytochrome c expression the mouse mAb anti-cytochrome c 7H8.2C12 (BD PharMingen) was used. Polyclonal anti-caspase-9 was purchased from Cell Signaling, polyclonal antibody anti-MnSOD from Stressgen (San Diego, CA), and rabbit polyclonal anti-LC3 from Molecular Biology Laboratory (Heidelberg, Germany) and mouse antibody anti-Beclin 1 from BD Biosciences (San Jose, CA).
Subcellular Fractionation
Cells were harvested in hypotonic buffer (2 mM MgCl2, 10 mM KCl, 10 mM Tris-HCl, pH 7.6) supplemented with protease inhibitor cocktail (Sigma) and incubated for 20 min on ice. One volume of 2× Mitobuffer (450 mM sucrose, 10 mM Tris-HCl, pH 7.4, 400 μM EGTA, pH 7.4, 2 μM DTT) was added, and the cell suspension was homogenized for 40 strokes with a Dounce homogenizer. Samples were centrifuged twice at 900 × g for 5 min at 4°C to eliminate cell nuclei and unbroken cells. The resulting supernatant was centrifuged at 10,000 × g for 30 min at 4°C to recover the heavy membrane pellet enriched for mitochondria, and the resulting supernatant was stored as the cytosolic fraction.
Immunocytochemistry
Cells cultured in 35-mm Petri dishes were washed in PBS and fixed with 4% paraformaldehyde in PBS for 15 min. After permeabilization with 0.2% Triton X-100 in PBS for 5 min, cells were blocked in 2% horse serum in PBS and incubated for 1 h at 37°C with primary antibodies. We used an anti-cytochrome c mouse mAb (clone 6H2.B4, BD PharMingen), an anti-caspase-9 polyclonal antibody (Cell Signaling), and a polyclonal antibody anti-MnSOD (Stressgen). Cells were then washed in blocking buffer and incubated for 1 h with labeled anti-mouse (Alexa Fluor 488 or Alexa Fluor 555, Molecular Probes, Eugene, OR) or anti-rabbit (FITC or Cy3, Jackson ImmunoResearch, West Grove, PA) secondary antibodies. After rinsing in blocking buffer, cell nuclei were stained with 1 μg/ml DAPI and examined under a Leica TCS SP5 confocal microscope equipped with a 40× (NA 1.25) or 63× (NA 1.4) oil-immersion objective (Deerfield, IL). Fluorescence images were adjusted for brightness, contrast and color balance by using Adobe Photoshop CS (San Jose, CA).
Measurement of Cellular ATP
Measurement of cellular ATP was performed using the ATPlite assay (Perkin Elmer-Cetus, Norwalk, CY) according to the manufacturer's instructions. In brief, cells seeded in 96-well microplates were resuspended in 50 μl lysis buffer, and after 5 min, 40 μl of substrate solution (Luciferase/Luciferin) was added to each sample. The luminescence was measured using a luminescence plate reader (Victor2 1420 multilabel counter; Wallac Oy, Turku, Finland). The ATP concentration was standardized using the total cellular protein concentration estimated by using Bradford protein assay (Bio-Rad). To confirm results, the experiments have been repeated by normalizing the ATP content to the number of cells.
MTS Assay
MTS Cell Titer 96AQueous One solution cell proliferation assay was performed according to the supplier's instructions (Promega, Madison, WI). The MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] tetrazolium compound is bioreduced into a colored formazan product by NADPH or NADH produced by dehydrogenase enzymes in metabolically active cells. Cells were plated in triplicate in 96-well plates at 3–7 × 103 cells per well. After treatment, Cell Titer 96AQueous One solution was added to each well for 90 min, and the light absorbance at 490 nm was recorded using a 96-well plate reader (Wallac Oy; Victor2 1420 multilabel counter). Values were normalized by protein content or by number of cells.
Assessment of ΔΨm
ΔΨm was measured using tetramethylrhodamine ethyl ester (TMRE, Molecular Probes). Cells were incubated at 33°C for 15 min in media containing TMRE (50 nM). As a control for ΔΨm dissipation, cells were treated with 10 μM carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP). Cells were then rinsed in fresh medium and detached from the dish. TMRE fluorescence was detected by flow cytometry on a FACScan (Becton Dickinson, Franklin Lakes, NJ) in FL-2.
Measurement of Reactive Oxygen Species Production
The reactive oxygen species (ROS)-sensitive probe DCF-DA (2′,7′-dichlorodihydrofluorescein-diacetate; 30 μM) was added directly to the culture medium for 1 h. Cells were washed with PBS and analyzed immediately by flow cytometry using the FITC channel on a FACScan (Becton Dickinson).
Oxygen Consumption Measurement in Intact Cells
The respiration rate was measured by a Clark oxygen electrode (Gilson Medical Electronics, Middleton, WI; 3000 W Mod 5/6 H). Cells were injected in a thermojacketed chamber containing 1650 μl TD buffer (25 mM Tris-HCl, pH 7.4, 137 mM NaCl, 10 mM KCl, 0.7 mM Na2HPO4) prewarmed at 33°C. Respiration was driven by endogenous substrates. After obtaining a stable rate, oligomycin and then bongkrekic acid were added. After blocking complex III with antimycin A, the cytochrome c oxidase–dependent oxygen consumption rate (obtained by addition of N,N,N′,N′-tetramethyl-p-phenylenediamine, TMPD + ascorbate) was inhibited by KCN or NaN3 (Duan et al., 2003). Rates of oxygen consumption are expressed as nanograms atoms of oxygen per minute per milligram proteins.
Electron Microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 45 min at 4°C, rinsed in cacodylate buffer 0.1 M, pH 7.4, postfixed in 1% OsO4 in 0.1 M cacodylate buffer, pH 7.4, dehydrated, and embedded in Epon. Ultrathin sections were briefly contrasted with uranyl acetate and photographed in a Zeiss EM 900 electron microscope (Thornwood, NY).
Statistical Analysis
Microsoft Excel was used for statistical analysis (Redmond, WA). Statistical significance was determined using the Student's t test; p ≤ 0.05 was considered significant.
RESULTS
Dissection of the Role of the Apoptosome Components: Apaf1-deficient Cells (ETNA−/−) and C9DNETNA Are Resistant to Apoptosis
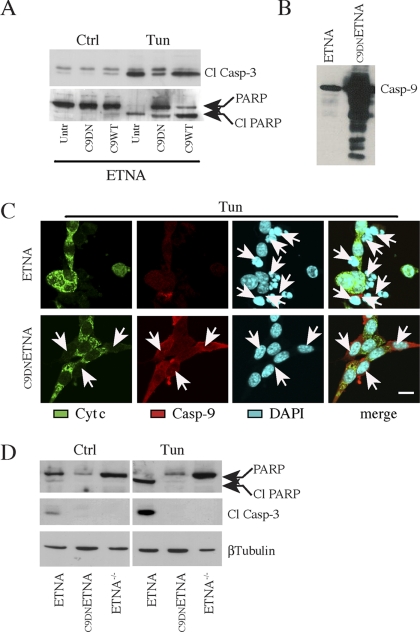
Our purpose was to analyze in detail the behavior of cells devoid of the apoptosome under prolonged apoptotic insults. We had previously established a cell line in which Apaf1 is deleted (ETNA−/−) and that is therefore unable to form the apoptosome. We had already proved the resistance of ETNA−/− cells to several apoptotic stimuli (Cozzolino et al., 2004). For this work, we also produced a cell line expressing a mutant form of murine caspase-9 acting as dominant negative (C9DNETNA). To this aim, we performed a site-directed mutagenesis of the caspase-9 active site (QACGG) in which the cystein 325 was changed in serine (QASGG). This form of mutant caspase-9 is able to bind Apaf1 and to allow the formation of the apoptosome; however, it does exert a dominant negative influence on effector caspase activation and apoptosis (Li et al., 1997). As shown in Figure 1A, transient transfection of an expression vector encoding dominant negative caspase-9 (C9DN), in ETNA cells followed by treatment with tunicamycin (an inhibitor of N-linked glycosylation and of the formation of N-glycosidic protein-carbohydrate linkages), induces decreased activation of caspase-3 as well as a reduced cleavage of PARP, if compared with untransfected ETNA cells (Untr) or with ETNA cells overexpressing wild-type caspase-9 (C9WT). We therefore established a second apoptosome-deficient system, i.e., ETNA cells stably overexpressing C9DN (C9DNETNA), in which the formation of the apoptosome is allowed but caspase-9 is mutated and therefore enzymatically inactive (Figure 1, B and C). We demonstrated that not only ETNA−/− cells but also C9DNETNA cells are protected from apoptosis because treatment with tunicamycin or with actinomycin D (an inhibitor of mRNA transcription) does not produce nuclear fragmentation (Figures 1C and 2A and data not shown), PARP cleavage or caspase-3 activation (Figure 1D and data not shown). The same results were obtained upon treatment with staurosporine, a widespread protein kinase inhibitor (data not shown).
Figure 1.
ETNA cells overexpressing a dominant negative form of caspase-9 (C9DNETNA) are resistant to apoptotic stimuli. (A) Total cell lysates from untransfected ETNA cells (Untr), ETNA cells transiently transfected with dominant negative caspase-9 (C9DN) or ETNA cells transiently transfected with wild-type caspase-9 (C9WT) untreated (Ctrl) or treated for 40 h with 3 μg/ml tunicamycin (Tun) were subjected to SDS-PAGE and analyzed for caspase-3 processing and for PARP processing. (B) Western blot analysis of caspase-9 expression in untransfected ETNA cells (ETNA) or ETNA cells stably overexpressing caspase-9 (C9DNETNA). (C) Triple-labeling immunofluorescence microscopy of ETNA and C9DNETNA cells treated for 40 h with 3 μg/ml tunicamycin (Tun). Cytochrome c (green), caspase-9 (red), nuclear staining (DAPI, blue), and merge of the three patterns are shown. ETNA cells show release of cytochrome c and extensive nuclear fragmentation (white arrows), whereas C9DNETNA cells exhibit released cytochrome c but unaffected nuclei (white arrows). Notice that in the immunofluorescence assay the used anti-caspase-9 antibody (which can recognize both the procaspase-9 and its cleaved form) enables caspase-9 detection only upon its overexpression (in C9DNETNA cells). Scale bar, 20 μm. (D) Western blot analysis of caspase-3 processing and PARP processing in total lysates of ETNA, C9DNETNA, and ETNA−/− cells untreated (Ctrl) or treated with tunicamycin (Tun) as described before. Immunoblot detection of β-tubulin is used as control of equal protein loading.
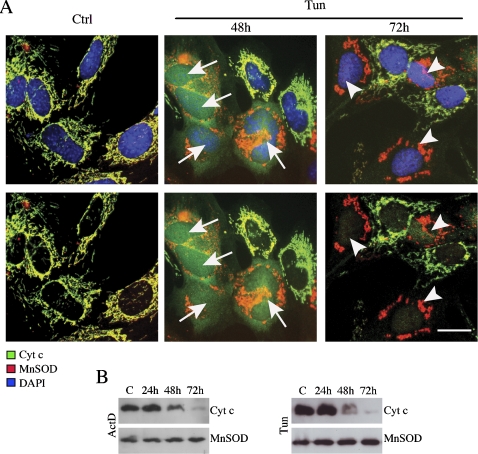
Figure 2.
Cytochrome c detection, in ETNA−/− cells, is abolished after long treatment with tunicamycin or actinomycin D. (A) Double-labeling confocal immunofluorescence microscopy of ETNA−/− cells untreated (Ctrl) or treated for 48 or 72 h with 3 μg/ml tunicamycin (Tun). Merged patterns of cytochrome c (green), MnSOD (red), and nuclei (DAPI, blue; top panels), and merged patterns of cytochrome c (green) and MnSOD (red) without nuclear staining of the same fields are shown (bottom panels). White arrows point to cells with released cytochrome c. White arrowheads point to cells with degraded cytochrome c. Scale bar, 20 μm. (B) Time-course analysis of cytochrome c expression in total cell lysates of ETNA−/− cells treated with 3 μg/ml tunicamycin (Tun) or 20 μM actinomycin D (ActD). MnSOD was also assayed as a control of equal protein loading.
Cytochrome c Is Degraded by a Proteasome-dependent Mechanism Once It Is Released from Mitochondria in Both ETNA−/− and C9DNETNA Cells
By using ETNA−/− and C9DNETNA cells we wanted to evaluate the possibility and the modality of recovery of cells receiving apoptotic stimuli in the absence of the apoptosome. To answer this question, we analyzed some metabolic parameters of ETNA−/− and C9DNETNA cells under apoptotic induction.
The first event in the apoptotic process we wanted to check was the release of cytochrome c. This molecule is extremely relevant both for cell metabolism and for the apoptotic process. When ETNA−/−cells are treated for 48 h with tunicamycin or actinomycin D, cytochrome c is released from mitochondria, and it is found diffused in the cytosol and in the nucleus (Figure 2A and not shown). However, cytochrome c staining disappears upon longer treatment with apoptotic inducers (72 h), whereas mitochondria staining, as revealed by detection of two mitochondrial resident proteins (MnSOD and Hsp60), is maintained (Figure 2A and data not shown). This observation is confirmed by a time-course evaluation of the amount of cytochrome c performed by Western blot (Figure 2B): after 72 h of both treatments, cytochrome c completely disappears.
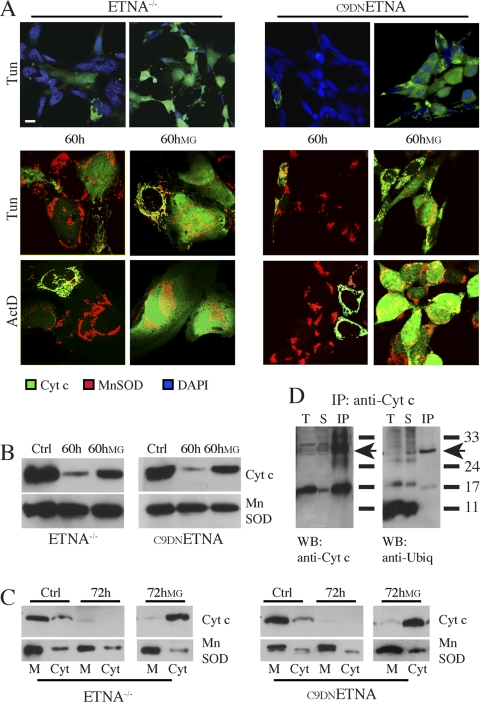
In ETNA−/− cells, cytochrome c released in the cytosol cannot bind the apoptosome because its partner Apaf1 is absent. To elucidate whether cytochrome c degradation takes place in the presence of Apaf1, we checked the amount of cytochrome c in C9DNETNA cells upon apoptotic induction. Again, after a prolonged treatment, a considerable number of cells display a complete loss of cytochrome c staining (Figure 3A, 60 h); this evidence is confirmed by Western blot analysis on total extracts and on mitochondrial/cytosolic fractions (Figure 3, B, 60 h, and C, 72 h).
Figure 3.
Cytochrome c is ubiquitinated and degraded by a proteasome-dependent mechanism after long apoptotic treatment in both ETNA−/− and C9DNETNA cells. (A) Double-labeling confocal immunofluorescence microscopy of ETNA−/− cells and C9DNETNA cells treated with 3 μg/ml tunicamycin (Tun) or 20 μM actinomycin D (ActD) for 60 h and cells under the same treatments plus 2 μM MG132 (60 hMG) added after 24 h of apoptotic inducer treatment. Merged patterns of cytochrome c (green) and nuclei (DAPI, blue) or merged patterns of cytochrome c (green) and MnSOD (red) are shown. Scale bar, 20 μm. (B) Western blot analysis of cytochrome c expression in total cell lysates of ETNA−/− cells and C9DNETNA cells untreated (Ctrl) or treated with tunicamycin (60 h) or with tunicamycin plus MG132 (60hMG). MnSOD was also assayed as a control of equal protein loading. (C) Cytosolic (Cyt) and membrane (M) fractions of ETNA−/− cells and C9DNETNA cells untreated (Ctrl) or treated with tunicamycin (72 h) or with tunicamycin and MG132 (72hMG) were analyzed by Western blot for the presence of cytochrome c. MnSOD was used as a control of equal protein loading in the membrane fractions. (D) Cytochrome c was immunoprecipitated from C9DNETNA cell extracts (100 μg) with a mAb (IP:anti-Cyt c). Total extracts (T), supernatants from the immunoprecipitates (S), and immunoprecipitates (IP) were then probed with an anti-ubiquitin antibody (WB:anti-Ubiq) or with a rabbit antibody raised against cytochrome c (WB:anti-Cyt c). The arrows indicate the position of ubiquitinated cytochrome c.
These observations led us to hypothesize a role for the ubiquitin-proteasome machinery in the disappearance of cytochrome c. To assess this possibility, we treated the cells with proteasome inhibitors, namely lactacystin or MG132, during the apoptotic stimulation. In particular, we exposed the cells to the apoptotic inducers (tunicamycin or actinomycin D), and after 24 h, we added the proteasome inhibitor. By immunofluorescence, we observed a significant amount of cytochrome c in the cytosol of cells induced to die and treated with the proteasome inhibitor MG132 (Figure 3A, 60 hMG). Western blot analysis also revealed a considerable increase in the steady-state level of cytochrome c in the presence of MG132 (Figure 3, B and C). Taken together these results suggest that, after its release from mitochondria, cytochrome c is degraded by the proteasome. The fact that we obtained the same regulation of degradation both in ETNA−/− and C9DNETNA indicates that cytochrome c degradation in the cytosol does not depend on its binding to Apaf1.
Previous evidence indicates that both forms of Saccharomyces cerevisiae cytochrome c (iso-1 and -2) can be ubiquitinated and that the major and minor ubiquitination sites are, in both proteins, Lys10 and Lys5, respectively (Sokolik and Cohen, 1991). Ubiquitination of apo-iso-1 (but not apo-iso-2) causes its rapid degradation through the proteasome pathway in the cytosol, when its import into the mitochondria and its maturation are blocked. Furthermore, apo-iso-1 was found to be ubiquitinated by overexpressing HA-tagged ubiquitin (Pearce and Sherman, 1997; Sherman, 2005).
To check whether ubiquitination of holo-cytochrome c occurs in mammals, we performed immunoprecipitation of C9DNETNA cell extracts using a monoclonal anti-cytochrome c antibody. Western blot on the cytochrome c immunoprecipitates (IPs) was then performed by using a polyclonal antibody anti-cytochrome c or a polyclonal antibody anti-ubiquitin (Figure 3D). We observed a band around 27 kDa (arrows) that is a putative ubiquitinated cytochrome c, because it is recognized both by anti-cytochrome c and by anti-ubiquitin antibodies. However, Lys6, Lys8, and Lys9 (corresponding to Lys10 and the Lys5 in yeast) are not involved in the ubiquitination of mammalian cytochrome c (Figure S1).
Under Proapoptotic Treatment, in the Absence of Cytochrome c, Apoptosome-Null Cells Are Able to Produce ATP Although They Have Depolarized Mitochondria
Our next goal was to detect the modalities of surviving of ETNA−/− and C9DNETNA cells without cytochrome c. First of all we wanted to check if such cells survive fully or if they are merely cells on the verge of death (“zombies”; Ekert et al., 2004). For this purpose, we analyzed some aspects of their metabolism by measuring the amount of ATP produced in normal conditions, under apoptotic stimulation and under prolonged apoptotic treatment.
As shown in Figure 4 and Figure S2, ATP production is increased after 24–48 h of treatment with either tunicamycin or actinomycin D. Longer treatments induce a relative decrease in ATP levels; however, even after many hours from the apoptotic stimulus application, the level of ATP produced remains high if compared with wild-type ETNA cells. This demonstrates that apoptosome-deficient cells under apoptotic induction are metabolically active for a long time, even in absence of cytochrome c (Figure 4 and Figure S2). Ultimately, the cells start dying (after 120–144 h of treatment, depending on the cell line) as shown by very low ATP production (Figure S2) and loss of membrane integrity, as detected by trypan blue staining (Figure S3). By contrast, as expected, in wild-type ETNA cells there is a strong decrease in ATP production, starting already after a 48-h treatment (see Figure S2).
Figure 4.
Both ETNA−/− and C9DNETNA cells are metabolically active after prolonged apoptotic treatment as seen by ATP content and MTS assay. (A) ATP production in ETNA−/− and C9DNETNA cells treated with 3 μg/ml tunicamycin (Tun) or 20 μM actinomycin D (ActD) for the times indicated. A.U., arbitrary units. The ATP content has not been normalized for the amount of protein, but it has been evaluated after seeding and treating the same number of cells for each condition. (B) Reducing equivalent production measured by MTS-formazan assay in ETNA−/− and C9DNETNA cells treated with tunicamycin (Tun) or actinomycin D (ActD) for the times indicated. Values represent the mean ± SD of four experiments.
We also evaluated the production of NADH and NADPH by dehydrogenase enzymes (MTS assay, FACS analysis, and 340-nm absorbance measurement), this being an indication of metabolically active cells. Again, even though cells lose cytochrome c, a high metabolic activity is maintained for a long time in both ETNA−/− and C9DNETNA cells treated with tunicamycin or actinomycin D (Figure 4B and Figure S4). To confirm these findings, the production of reducing equivalents has been normalized both to the amount of protein and to the number of cells (Figure 4B and Figure S4A).
To discard the possible selection of clones highly resistant to caspase-independent cell death, we performed the experiments using another ETNA−/− clone (ETNA−/−C2) and another C9DNETNA clone (C9DNETNA-C6; Figures S2, S3, and S4A). We also performed the same experiments using another cell type (Apaf1−/− MEFs), obtaining similar results (Figure S4A).
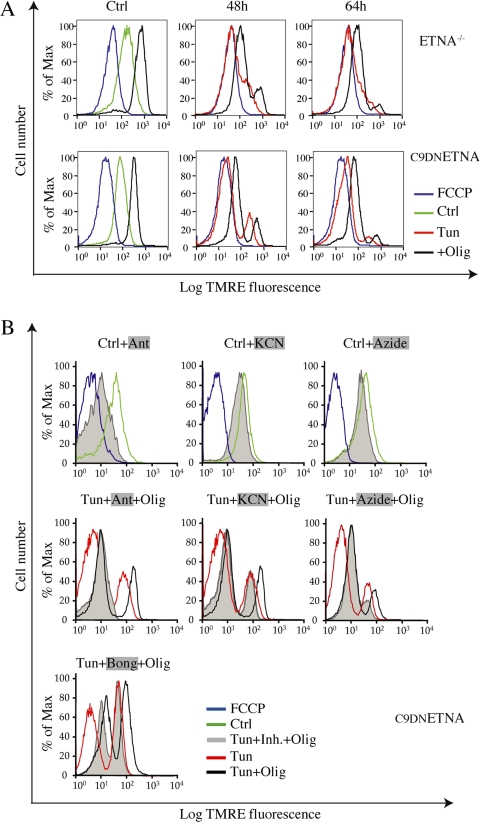
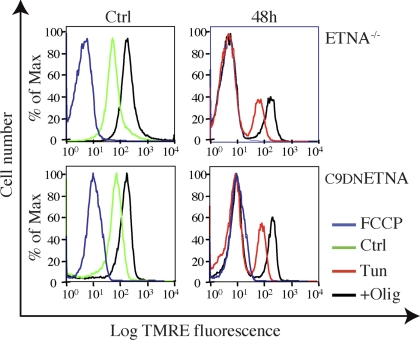
We wondered how ETNA−/− and C9DNETNA cells were able to produce a high amount of ATP in the absence of cytochrome c. To investigate the possibility that some cytochrome c might still be located within mitochondria and support the respiratory chain, we measured ΔΨm by incubation with TMRE and FACS analysis. We used FCCP, an uncoupler that dissipates the transmembrane potential by preventing the generation of a proton gradient, as a control for depolarized mitochondria. As reported in Figure 5A, during apoptotic induction there is a marked depolarization of mitochondria (Tun, red line). This is in contrast with other parameters, such as levels of ATP and reducing equivalents, that are well maintained. This fall in ΔΨm is progressive during both tunicamycin and actinomycin D treatment (Figure 5A and not shown).
Figure 5.
Under apoptotic treatment, apoptosome-null cells progressively dissipate ΔΨm in the absence of cytochrome c. (A) ETNA−/− and C9DNETNA cells untreated (Ctrl, green line), treated with 10 μM FCCP (FCCP, blue line) as a control for mitochondrial depolarization, or treated with tunicamycin for 48 or 64 h (Tun, red line) were stained with TMRE (50 nM) for 10 min, washed, and then analyzed by FACS analysis for ΔΨm. ETNA−/− and C9DNETNA cells untreated or treated with tunicamycin for 48 or 64 h were incubated with oligomycin (2 μM) for 10 min (+Olig, black line) before TMRE staining. (B) Before TMRE staining (50 nM) and FACS analysis, C9DNETNA cells were left untreated (Ctrl, green line), treated with 10 μM FCCP (FCCP, blue line), or treated for 5 min with 20 μM antimycin A (Ctrl+Ant), 5 mM KCN (Ctrl+KCN), or 10 mM NaN3 (Ctrl+NaN3; Tun+Inh.+Olig, gray shadow). C9DNETNA cells treated with tunicamycin for 48 h (Tun, red line) were then treated for 2 min with 20 μM antimycin A (Tun+Ant+Olig), 5 mM KCN (Tun+KCN+Olig), and 10 mM NaN3 (Tun+NaN3+Olig). C9DNETNA cells treated with tunicamycin for 44 h (Tun, red line) were then treated for 2 min with 50 μM bongkrekic acid (Tun+Bong+Olig). In all cases cells were finally incubated with oligomycin (2 μM) for 10 min before TMRE staining (gray shadow).
In conclusion, if cells induced to die do not undergo apoptosis because of the absence of a functional apoptosome, they are still able to maintain a high metabolism (for many days before dying), as seen by measurement of ATP and reducing equivalent production. This occurs even though cytochrome c has been released and degraded and ΔΨm has been lost.
On Apoptotic Treatment, Cell Metabolism in the Absence of Cytochrome c Is Maintained by Glycolysis
Next, we wanted to determine how cells maintain high ATP levels and high metabolic activity despite a loss of cytochrome c and a null ΔΨm, both of which suggest a mitochondrial impairment. Thus, we decided to study the contribution of glycolytic and mitochondrial metabolism to ATP production. To address this issue, we blocked the F0F1-ATPase with oligomycin to inhibit the production of ATP based on the respiratory chain, and therefore we measured ATP produced by glycolysis only.
When we treated ETNA−/− and C9DNETNA cells under apoptotic stimulation with oligomycin, we did not observe any decrease in the ATP production (Figure 6 and Figure S5, ▩). This strongly suggests that in cells under apoptotic stimulation ATP is supplied mainly by the glycolytic metabolism. In fact, under the same conditions we also inhibited glycolysis by removing glucose and serum from the medium (to allow complete removal of intermediates of glycolysis) and by adding methylpyruvate (Mpyr, a cell permeable form of pyruvate) to sustain the electron-transport chain, and we detected a strong decrease in the amount of ATP produced starting after 48 h of tunicamycin treatment (Figure 6 and Figure S5, ■). After a 24-h treatment, ATP is still produced even under conditions of inhibited glycolysis by oxidative phosphorylation (Figure 6 and Figure S5, ■) because cytochrome c is still within mitochondria (data not shown). Tunicamycin treatment requires, in fact, at least 40 h to kill wild-type ETNA cells (Figure 1). Because the removal of serum could be stressful for the cells and could induce additional events that may interfere with the experiment, we repeated all the assays by blocking glycolysis using dialyzed serum, and we obtained exactly the same results (data not shown). To extend our findings to another cell type, we performed the same experiments in Apaf1−/− MEFs, obtaining similar results (Figure S6).
Figure 6.
On apoptotic treatment, ATP production is supported by glycolysis. (A) Gray bars, ATP production in wild-type ETNA cells grown in 10% FBS medium and left untreated (Ctrl) or treated with 3 μg/ml tunicamycin for the given time periods, and ATP production in ETNA cells under the same treatments and incubated for the last 20 min with 2 μM oligomycin (Olig) are shown. Black bars, ATP production in ETNA cells left untreated (Ctrl) or treated with 3 μg/ml tunicamycin for the indicated time periods in complete medium is shown. In this case, 2 h before ATP measurement, glycolysis was inhibited by incubating cells in a medium without glucose, without serum, and with 10 mM Mpyr (−gluc-serum+Mpyr) with (Olig) or without 2 μM oligomycin addition for the last 20 min. A.U., arbitrary units. The experiment was done by seeding and treating the same number of cells for each condition. (B) Same as A but in ETNA−/− cells. (C) Same as A but in C9DNETNA cells. Data shown represent the mean ± SD of three experiments.
This experiment demonstrates that, in both ETNA cell systems and in another apoptosome-deficient cell system, during the apoptotic treatment there is a production of ATP almost exclusively due to the glycolytic metabolism.
Similarly, the reducing equivalent production in ETNA−/− and C9DNETNA treated with tunicamycin and measured by MTS assay was not affected by oligomycin (Figure 7). This indicates that such production depends on glycolysis. In Figure S5 the ATP content was normalized both to the number of cells and to the amount of protein to strengthen the data.
Figure 7.
On apoptotic treatment, glycolysis sustains the production of NADH and NADPH by dehydrogenase enzymes (MTS assay). MTS assay in ETNA−/− and C9DNETNA cells grown in 10% FBS medium and left untreated (Ctrl) or treated with 3 μg/ml tunicamycin (Tun) for the indicated time periods with or without 2 μM oligomycin addition for the last 20 min (Olig). Normalization was done to the amount of protein. Data shown represent the mean ± SD of three experiments.
Mitochondria Are Not Irreversibly Impaired under Apoptotic Treatment, and the Recovery of ΔΨm Is Dependent on Active Glycolytic Metabolism
Next, we wanted to assess if depolarized mitochondria of cells induced to apoptosis and surviving mainly by glycolysis were irreversibly impaired.
Treatment with oligomycin is able to hyperpolarize mitochondria in control cells because the proton gradient is not used to produce ATP (Figure 5A). Impaired mitochondria cannot undergo oligomycin-induced hyperpolarization. Under tunicamycin treatment, cells display a bimodal distribution with respect to TMRE fluorescence. One population is formed by cells with a normal ΔΨm and another population (growing in number during the treatment) exhibits depolarized mitochondria. When such tunicamycin-treated cells are incubated with oligomycin, both populations are able to hyperpolarize. Not only cells displaying normal ΔΨm, but even cells that have lost ΔΨm are able to regain it (Figure 5A). These data suggest that depolarized mitochondria have not reached a point-of-no-return, and are capable of repolarization.
We observed that, in tunicamycin-related cells, oligomycin-induced regeneration of ΔΨm is insensitive to anti-mycin A, which impairs the respiratory chain by blocking complex III, and to KCN or NaN3, which are both inhibitors of complex IV (Figure 5B and Figures S7 and S8). This suggests that ΔΨm might be regenerated in the absence of respiratory chain activity. A similar mechanism, allowing ΔΨm in the absence of a functional electron transport chain, has been shown to function in rho zero cells (Buchet and Godinot, 1998). Interestingly, the adenine nucleotide translocator (ANT) inhibitor, bongkrekic acid, partially impairs oligomycin-induced regeneration of ΔΨm in tunicamycin-treated cells (Figure 5B).
When we blocked ATPase with oligomycin under conditions of inhibited glycolysis, we observed hyperpolarization of control cells (Figure 8, black line, Ctrl) and of the cell population displaying intact ΔΨm under tunicamycin treatment (Figure 8, black line, 48 h). In contrast with the previous experiment, done in presence of serum and glucose (Figure 5A, black line), depolarized cells were not able, in this case, to recover their ΔΨm (Figure 8, black line).
Figure 8.
Recovery of ΔΨm, under apoptotic treatment of apoptosome-null cells, is dependent on active glycolytic metabolism. ETNA−/− cells and C9DNETNA cells untreated (Ctrl, green line), treated with 10 μM FCCP (FCCP, blue line) or treated with tunicamycin for 48 h (Tun, red line) were deprived of glucose and serum and were supplied with Mpyr 2 h before TMRE staining and FACS analysis. ETNA−/− cells and C9DNETNA cells untreated or treated with tunicamycin (Tun) for 48 h, were incubated with oligomycin (2 μM) for 10 min (+Olig, black line) before TMRE staining.
This experiment indicates that without glycolysis, apoptotic treatment and opening of mitochondrial outer membrane irreversibly affect mitochondria. However, when glycolysis is active, mitochondria are depolarized but are still capable of repolarization.
Glycolysis Inhibition Induces an Increased Production of ROS and Blocks Cell Recovery
Glycolysis might be important to allow mitochondrial recovery because it supplies the ATP necessary to allow ΔΨm regeneration by a mechanism similar to the one hypothesized for rho zero cells (Buchet and Godinot, 1998). However, this might not be the only role of glycolysis. To assess why glycolysis protects mitochondrial function, we have measured ROS production. For this purpose, we used the ROS-sensitive probe DCF. We observed that when glycolysis is inhibited, there is a high production of ROS both in untreated (Ctrl) and in tunicamycin-treated cells (48 h) independently on the cell line we analyzed (Figure 9). Because ROS impair mitochondrial function (Ott et al., 2007), the increased ROS production might contribute to explaining (besides the lack of ATP) why ΔΨm, in the absence of glycolysis, is unable to recover.
Figure 9.
ROS production increases in absence of glycolysis. ETNA−/−, C9DNETNA ETNA−/−C2, C9DNETNA-C6, and Apaf1−/− MEFs untreated (Ctrl), or treated for 48 h with tunicamycin (Tun), were cultured in complete medium (black line) or were deprived of glucose and serum and were supplied with Mpyr (gray shadow) 2 h before ROS measurement by FACS analysis. One hour before FACS analysis, cells were incubated with the ROS-sensitive probe DCF (30 μM). As negative control, cells cultured in complete medium were not incubated with DCF (red line).
We also found out that, if glycolysis is functional, apoptosome-deficient cells after cytochrome c release might recover and proliferate but only if the apoptotic stimulus is removed within a certain time range after induction (Figure 10). After 48 h of treatment, when the large majority of cells had released cytochrome c, tunicamycin was washed out. Medium was replaced with complete medium without tunicamycin, and cells were kept in these conditions for additional 24 h (72 h-washed), 48 h (96 h-washed), 96 h (120 h- washed), and 120 h (144 h-washed). Mitochondrial recovery (indicated by mitochondrial cytochrome c localization), cell survival, and cell proliferation were analyzed. After 72–96 h of tunicamycin treatment, cells appeared shrunk and were mostly detached. Their cytochrome c was released and degraded (Figure 10A). At 120–144 h, cells were all detached, and they incorporated trypan blue, which stains only dead cells (Figure S3). By contrast, cultures washed at 48 h and observed at 72, 96, and 120 h showed 1) an increasing number of cells containing cytochrome c in their mitochondria, 2) absence of cell shrinkage (72 h, 96 h washed compared with not washed cells), and 3) an increasing number of cells attached to the dish. However, waiting 72 h to wash the cells, or blocking glycolysis, inhibited cell recovery (Figure 10B and data not shown).
Figure 10.
Relocalization of cytochrome c, recovery and proliferation of tunicamycin-treated cells upon removal of the apoptotic induction. (A) ETNA−/− cells were treated with 3 μg/ml tunicamycin (Tun, 72 and 96 h). In parallel, ETNA−/− cells treated with 3 μg/ml tunicamycin were washed after 48 h of treatment and medium was replaced with complete medium without tunicamycin (Tun+wash). Cells were kept in these conditions for additional 24 h (72 h-washed), 48 h (96 h-washed), 96 h (120 h-washed), and 120 h (144 h-washed). (B) ETNA−/− cells were treated with 3 μg/ml tunicamycin; after 48 h of treatment, cells were washed, and medium was replaced with medium deprived of glucose and serum and were supplied with Mpyr, without tunicamycin (Tun+wash, no glycolysis). Cells were kept in these conditions for additional 24 h (72 h-washed-no glyc) and 48 h (96 h-washed-no glyc). Under these conditions, cells appeared shrunk and most of them were detached. After additional 96 h, cells were completely detached (not shown). Detection by immunofluorescence of cytochrome c localization (green) and nuclear DAPI staining (blue) is shown.
The Glycolytic Metabolism of ETNA−/− and C9DNETNA Cells under Apoptotic Induction Is Based on Beclin 1–dependent Autophagy
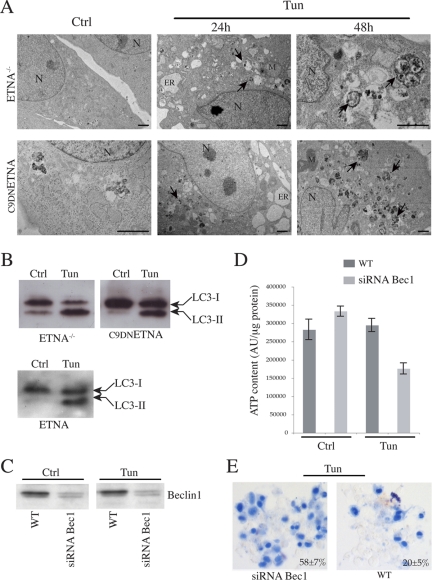
Autophagy is a self-degradative process involved both in basal turnover of cellular components and in response to nutrient starvation or organelle damage in a wide range of eukaryotes (Levine and Yuan, 2005). Because ETNA−/− and C9DNETNA cells produce ATP through glycolysis when cytochrome c is released, and because autophagy is a way to resist death, we decided to test if autophagy played a role in the maintenance of ATP production under these conditions. The occurrence of autophagy was analyzed by assessing 1) the increased number of autophagosomes (autophagic vesicles) by electron microscopy and 2) the conversion of microtubule-associated protein 1 light-chain 3 (LC3)-I (18 kDa) in the lipidated form LC3-II (16 kDa), essential for its binding to the membranes of autophagosomes (Ferraro and Cecconi, 2007). We observed that wild-type ETNA, ETNA−/−, and C9DNETNA cells under apoptotic treatment clearly undergo autophagy. In fact, in contrast with untreated cells, we detected many autophagic vesicles in both cell lines treated with tunicamycin (Figure 11A) or actinomycin D (not shown). In addition, we observed an increase of the LC3-II band, whose relative amount reflects the abundance of the autophagosomes (Figure 11B). Both these autophagic markers are detectable soon after the apoptotic treatment. Very interestingly, when autophagy is suppressed by siRNA, silencing the proautophagic gene Beclin 1 in tunicamycin-treated ETNA−/− and C9DNETNA cells, there is a marked decrease in the production of ATP (Figure 11, C and D, and data not shown) and an earlier death of apoptosome-deficient cells, as suggested by the increased trypan blue staining (Figure 11E and data not shown).
Figure 11.
The glycolytic metabolism of ETNA−/− and C9DNETNA cells under apoptotic treatment is based on Beclin 1–dependent autophagy. (A) Ultrastructural features by means of electron microscopy were observed in ultrathin sections from ETNA−/− and C9DNETNA cells untreated (Ctrl) or treated with 3 μg/ml tunicamycin (Tun; 24 h). Numerous autophagic vacuoles (black arrows) and dilation of endoplasmic reticulum (ER) are visible in the cytoplasm of tunicamycin-treated cells. The prolonged treatment (Tun; 48 h) enhances autophagic vacuole formation (black arrows). N, nucleus; M, mitochondrion; ER, endoplasmic reticulum. Scale bar, 1 μm. (B) Western blot analysis of LC3-I to LC3-II conversion in total lysates of ETNA−/−, C9DNETNA, and wild-type ETNA cells untreated (Ctrl) or treated with 3 μg/ml tunicamycin for 33 h (Tun). (C) Western blot analysis of Beclin 1 amount in ETNA−/− cells (WT) and in ETNA−/− cells transiently transfected for 48 h with siRNA duplex against Beclin 1 mRNA (siRNABec1) treated with 3 μg/ml tunicamycin for further 48 h (Tun) or left untreated for the same time (Ctrl). (D) ATP content in ETNA−/− cells (WT) and in ETNA−/− cells transiently transfected with siRNA duplex against Beclin 1 (siRNABec1) and treated with 3 μg/ml tunicamycin for 48 h (Tun) or left untreated (Ctrl) for the same time was measured. Values in D represent the mean ± SD of three experiments. (E) Representative images of trypan blue staining of C9DNETNA cells (WT) and C9DNETNA cells transiently transfected with siRNA duplex against Beclin 1 (siRNABec1) and treated with 3 μg/ml tunicamycin for 96 h (Tun) or left untreated (Ctrl) for the same time. Percentage of trypan blue–incorporating cells were determined by counting ∼1000 cells (per each condition and per each experiment) and calculating the average of three independent experiments ± SD.
DISCUSSION
The apoptotic process is very well regulated at different levels to prevent uncontrolled cell death; one of the strategies used by the cell to control programmed cell death is to hide proapoptotic factors inside mitochondria. Protein stability is another key regulatory mechanism in the control of several cellular processes including apoptosis. Several apoptotic proteins are, indeed, degraded by the ubiquitin-proteasome pathway (Argentini et al., 2000; Suzuki et al., 2001; Wilson et al., 2002; Zhang et al., 2004). During mitochondria turnover there could be risk of release of small amount of proapoptotic factors residing in the intermembrane space, including cytochrome c. This would prime an unwanted apoptotic pathway. To avoid this, it has been proposed that there must be a threshold of cytochrome c to be reached in order to activate the apoptosome (Waterhouse et al., 2002). Indeed, in our model we have observed a proteasome-dependent degradation of cytochrome c released in the cytosol. In this scenario, the degradation of cytochrome c once in the cytosol may represent a mechanism needed to prevent accidental apoptosis, as already reported for other mitochondrial proapoptotic factors such as Smac/DIABLO (MacFarlane et al., 2002).
Moreover, it has been reported that, in the absence of caspase activation, mitochondria remain functional after mitochondrial permeabilization by using the low levels of cytochrome c released into the cytosol to regenerate ΔΨm (Waterhouse et al., 2001). In glucose-deprived and Mpyr-provided cells, the maintenance of ΔΨm ensured mitochondrial ATP generation, making it possible for these cells to survive after MOMP. However, this effect was only temporary because ATP levels dropped after more than 12 h of actinomycin D treatment. In this context, the inhibition of cytochrome c degradation could help cells to maintain mitochondrial ATP production and to recover from the apoptotic insult. In fact, it has been shown that readdition of cytochrome c in digitonin-permeabilized cells was able to restore ΔΨm in cells induced to undergo apoptosis in the presence of caspase inhibitors (Waterhouse et al., 2001).
Considering the importance of cytochrome c in the metabolic activity of cells and hence in their survival, we set out to establish whether in mammalian cells holo-cytochrome c was degraded after ubiquitination and, if so, to identify the ubiquitin attachment sites on the protein. The inhibition of cytochrome c ubiquitination and degradation upon its release into the cytosol during apoptotic stimulation could be a tool to support the survival of cells. By immunoprecipitating cytochrome c from cell lysates and then performing Western blot analysis with an anti-ubiquitin antibody, we detected a band whose size suggests that cytochrome c is biubiquitinated even though determination of the number of ubiquitins by electrophoretic positions is known to be inaccurate (Jennissen, 1995; Pearce and Sherman, 1997). We performed a mutational analysis, so excluding that Lys6, Lys8, and Lys9 are the attachment sites for ubiquitin. Further studies will be necessary to identify the ubiquitination sites in mammalian cytochrome c.
It has been already reported that cells might have the potential to recover after cytochrome c release (Martinou et al., 1999; Deshmukh et al., 2000; Goldstein et al., 2000; Waterhouse et al., 2001; Cozzolino et al., 2004). This matches the observation that Apaf1 and caspase-9 (which act downstream of MOMP) deficiency can contribute to oncogenic transformation. The overproliferation of Apaf1−/− and caspase-9−/− proneurons in knockout mice indicates that those cells, triggered to apoptosis, not only do not die but are even able to proliferate (Cecconi et al., 1998; Kuida et al., 1998; Soengas et al., 1999). In these studies, the point-of-no return in cell death is considered the loss of ΔΨm. The maintenance of ΔΨm after MOMP is necessary to provide the cell with a continued source of energy and to maintain mitochondrial functions. During the period over which ΔΨm is maintained, cells could be able to recover. However, it has also been reported that this is possible only for a short period after apoptotic induction; ΔΨm loss eventually happens and this precedes a drop in ATP and cell death (Waterhouse et al., 2001).
We wanted to extend over time the analysis of apoptosome-impaired cells induced to die. We observed that after both tunicamycin and actinomycin D treatment there is an increase in ATP production. The increase in ATP production after an apoptotic insult might be very likely due to need of ATP for the apoptotic machinery (Zamaraeva et al., 2005). In fact, even though in both ETNA−/− and C9DNETNA cells the apoptotic machinery is inhibited downstream of apoptosome formation, the apoptotic pathway proceeds regularly until the step of cytochrome c release. On more extended observation, we noted that such cells are able to maintain a normal metabolic activity (ATP generation and production of reducing equivalents) even after a long treatment, although ΔΨm is apparently dissipated (Figures 4 and 5). Therefore, they can actively survive for about 6 d before dying (Figures S2, S3, and S4). Under these conditions, ATP generation and production of reducing equivalent are maintained by glycolysis (Figure 6, B and C). In fact, even though oligomycin is added, cells keep in producing ATP, whereas oligomycin addition, followed by glucose removal, abolishes ATP production (Figure 6, B and C). The measurement of mitochondrial respiration indicates that oligomycin addition does not inhibit, but only reduces respiration at a higher extent in control cells (∼35%) than in tunicamycin-treated cells (∼22%; see Figure S8).
In control ETNA−/− and C9DNETNA cells, treatment with oligomycin acutely hyperpolarizes mitochondria because the protonic gradient is not used to produce ATP, because of the inhibition of the F0F1-ATPase by oligomycin (Figure 5A). Surprisingly, we observed that oligomycin is able to induce a recovery of ΔΨm, not only in control cells (Figure 5A, Ctrl) but also in apoptosome-impaired cells under extended apoptotic treatment (Figure 5A, 48 h and 64 h). Because ΔΨm maintenance requires an intact inner membrane (Mootha et al., 2001), our studies suggest that the mitochondrial inner membrane is not irreversibly affected. There could still be the possibility of restoring the proton gradient across the mitochondrial inner membrane even though cytochrome c has been released and mitochondria are depolarized.
How could we explain the oligomycin-induced ΔΨm regeneration in tunicamycin-treated cells? This effect of oligomycin is not due to inhibition of ATP synthesis, because these cells do not produce ATP by oxidative phosphorylation (Figure 6). Moreover, oligomycin allows a partial restoration of ΔΨm in cells treated with tunicamycin even though we block the respiratory chain with complex III (antimycin A) or complex IV (KCN or NaN3) inhibitors (Figure 5B and Figures S7 and S8). In principle, ΔΨm might be regenerated in the absence of respiratory chain activity by a mechanism similar to the one functioning in rho zero cells. These cells are depleted of mitochondrial DNA and, therefore, they do not have a functional electron transport chain. It is generally accepted that the ΔΨm of mtDNA-depleted cells arises from the electrogenic exchange of glycolytic ATP4− for ADP3− via the adenine nucleotide carrier ANT. Within the mitochondrial matrix, ATP4− deriving from glycolysis is hydrolyzed by an intramitochondrial ATPase, probably an incomplete F1-ATPase (F0 is not functional in rho zero cells) providing ADP3−, which is then exchanged with glycolytic ATP4− by ANT (Buchet and Godinot, 1998; Appleby et al., 1999; Chandel and Schumacker, 1999; Arnould et al., 2003). Oligomycin (which binds and inhibits only the F0 portion of F0F1-ATPase) does not eliminate ATPase activity and ΔΨm in rho zero cells, because they are devoid of F0 (Buchet and Godinot, 1998 Appleby et al., 1999). Based on this hypothesis, the main source of ΔΨm regeneration by oligomycin in tunicamycin-treated cells would be the glycolytic ATP (Figure 8); we can speculate that the F1 subcomplex (insensitive to oligomycin) might dissociate from the integral F0 portion (sensitive to oligomycin) of the F0F1-ATPase, thus rendering F1 capable of ATP hydrolysis and F0 capable of oligomycin-sensitive proton permeability. It is noteworthy that, in absence of oligomycin, mitochondria energization by a rho zero-like mechanism in tunicamycin-treated cells is masked, probably by proton leak through mitochondrial inner membrane (Figure 5). Oligomycin can reveal this potential by inhibiting the leak. However, further experiments are needed to fully identify the cause of ΔΨm regeneration in tunicamycin-treated cells in the presence of oligomycin.
We have observed that (in contrast with antimycin and KCN/NaN3) an ANT inhibitor, bongkrekic acid, reduces membrane potential regeneration upon tunicamycin/oligomycin treatment (Figure 5B). However, it does not abolish completely mitochondria re-energization (Figure 5B, left peak), so implying that other electrogenic pumps may also be involved in this rho zero-like mechanism of ΔΨm generation, as already proposed by other authors (Pereira-da-Silva et al., 1993; Buchet and Godinot, 1998; Appleby et al., 1999; Arnould et al., 2003).
Our study reveals that, in apoptosome-impaired cells even after a prolonged toxic insult, depolarized mitochondria do not reach a point-of-no-return. The inner mitochondrial membrane remains potentially functional after cytochrome c release and even after ΔΨm loss; it can still compensate for ion leakage and allow a possible re-energization of mitochondria. Vice versa, oligomycin addition upon apoptotic treatment cannot repolarize mitochondria when also glycolysis is inhibited, even though Mpyr is supplied (Figure 8, 48 h). Glycolysis is thus important for maintaining the mitochondria potentially able to recover their ΔΨm, presumably because (under apoptotic stress and after cytochrome c degradation) it is the only source of ATP, which is necessary for ΔΨm generation through a rho zero-like mechanism. Our conclusions concern both kinds of cell type, ETNA−/− and C9DNETNA cells. This means that there is no difference regarding the analyzed metabolic parameters when either Apaf1 or caspase-9 are impaired within the apoptosome.
We have found out that the absence of glycolysis induces an increased ROS production (Figure 9). ROS impair mitochondrial function by inactivating mitochondrial respiratory chain complexes and by stimulating lipid peroxidation. Lipid peroxides affect respiration, oxidative phosphorylation, maintenance of ΔΨm, and inner membrane barrier properties. In tunicamycin-treated cells (in which mitochondria are already affected and depolarized) this can lead to changes in the structure and function of the mitochondria consistent with increased proton leakage and uncoupling of mitochondria (Ott et al., 2007). Therefore, another reason for the need of glycolysis to protect mitochondrial function, besides the need of ATP to sustain ΔΨm regeneration, is the negative control of increased ROS production.
We have shown that, after cytochrome c release, the recovery of the cell is also possible, but only when glycolysis is active and the apoptotic stimulus is removed within a certain time range after induction (Figure 10), similarly to what recently reported by Colell et al. (2007).
Finally, we observed that apoptosome-deficient cells undergo autophagy soon after the apoptotic treatment. Most important, we found that the role of glycolysis in contributing to cell survival for prolonged periods after cytochrome c release depends on Beclin 1–regulated autophagy, this being highly relevant for survival of apoptosome-deficient proneurons under apoptotic stress. Autophagy plays a key role in the bioenergetic management of starvation by generating metabolic substrates that can be recycled and further processed to maintain cellular ATP production (Levine and Kroemer, 2008; Kotoulas et al., 2006). It has very recently been shown that, in the absence of caspase activation, increased glycolysis by GAPDH overexpression enhances recovery from MOMP and, consequently, cell survival (Colell et al., 2007). This is probably due to the fact that increased ATP production by glycolysis contributes in maintaining ΔΨm. In addition, the authors observe a GAPDH-enhanced autophagy that contributes to cell survival probably by promoting the removal of terminally damaged mitochondria (Colell et al., 2007).
In particular, our experiments demonstrate that autophagy, under cytotoxic stress, is necessary for the production of ATP and for cell survival (Figure 11). This suggests that recycling of cellular constituents through autophagy (followed by their enzymatic breakdown in simple subunits) is necessary to support glycolysis, for example, through degradation of glycoproteins and glycogen autophagy (Elbein, 1991; Kotoulas et al., 2006). Moreover, lactic acid, most amino acids, and glycerol can be converted into glucose via the gluconeogenic pathway (occurring also in the brain); also, degradation of nucleic acids releases ribose-1-phosphate, which can be transformed into intermediates of glycolysis (glyceraldehyde-3-phosphate and fructose-6-phosphate) by the pentoso-phosphate pathway (Gaitonde et al., 1977; Rawn, 1998). Our findings help in clarifying the issue of survival and recovery of apoptosome-inactivated cells after mitochondrial permeabilization. Unraveling this issue could have important implications for oncogenesis, because recovery of transformed cells could be enhanced by apoptosome inactivation if autophagy is allowed. Moreover, such discovery could have important consequences regarding the survival and overproliferation of apoptosome-deficient neurons, both of which, we have shown, depend not only on the absence of the apoptosome but also on autophagic mechanisms to sustain cell metabolism. This is important because therapies based on transplantation of neural precursor cells need to take into account not only the role of apoptosis but also that of autophagy in their survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank Giuseppe Filomeni, Alberto Ferri, and Adamo Diamantini for the helpful discussion and excellent technical assistance and Martin W. Bennet for the valuable editorial work. This work was partially supported by grants from Fondazione Telethon, Compagnia di San Paolo, Associazione Italiana Ricerca sul Cancro, the Italian Ministry of University and Research (MUR), and the Italian Ministry of Health. E.F. is a fellow of Fondazione Santa Lucia, and F.C. is an Associate Telethon Scientist.
Abbreviations used:
- ΔΨm
mitochondrial transmembrane potential
- ANT
adenine nucleotide translocator
- C9DN
caspase-9 dominant negative
- DCF
2′,7′-dichlorodihydrofluorescein
- ETNA−/−
embryonic telencephalic naïve Apaf1−/−
- FCCP
carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone
- MEFs
mouse embryonic fibroblasts
- MOMP
mitochondrial outer membrane permeabilization
- Mpyr
methylpyruvate
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- ROS
reactive oxygen species
- TMPD
N,N,N′,N′-tetramethyl-p-phenylenediamine
- TMRE
tetramethylrhodamine ethyl ester.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0858) on June 11, 2008.
REFERENCES
- Appleby R. D., Porteous W. K., Hughes G., James A. M., Shannon D., Wei Y. H., Murphy M. P. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- Argentini M., Barboule N., Wasylyk B. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene. 2000;19:3849–3857. doi: 10.1038/sj.onc.1203737. [DOI] [PubMed] [Google Scholar]
- Arnould T., Mercy L., Houbion A., Vankoningsloo S., Renard P., Pascal T., Ninane N., Demazy C., Raes M. mtCLIC is up-regulated and maintains a mitochondrial membrane potential in mtDNA-depleted L929 cells. FASEB J. 2003;17:2145–2147. doi: 10.1096/fj.03-0075fje. [DOI] [PubMed] [Google Scholar]
- Buchet K., Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J. Biol. Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- Cecconi F., Alvarez-Bolado G., Meyer B. I., Roth K. A., Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Chandel N. S., Schumacker P. T. Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett. 1999;454:173–176. doi: 10.1016/s0014-5793(99)00783-8. [DOI] [PubMed] [Google Scholar]
- Colell A., et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- Cozzolino M., et al. Apoptosome inactivation rescues proneural and neural cells from neurodegeneration. Cell Death Differ. 2004;11:1179–1191. doi: 10.1038/sj.cdd.4401476. [DOI] [PubMed] [Google Scholar]
- Deshmukh M., Kuida K., Johnson E. M., Jr. Caspase inhibition extends the commitment to neuronal death beyond cytochrome c release to the point of mitochondrial depolarization. J. Cell Biol. 2000;150:131–143. doi: 10.1083/jcb.150.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S., Hajek P., Lin C., Shin S. K., Attardi G., Chomyn A. Mitochondrial outer membrane permeability change and hypersensitivity to digitonin early in staurosporine-induced apoptosis. J. Biol. Chem. 2003;278:1346–1353. doi: 10.1074/jbc.M209269200. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Martins L. M., Kaufmann S. H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Ekert P. G., Read S. H., Silke J., Marsden V. S., Kaufmann H., Hawkins C. J., Gerl R., Kumar S., Vaux D. L. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J. Cell Biol. 2004;165:835–842. doi: 10.1083/jcb.200312031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein A. D. Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 1991;5:3055–3063. doi: 10.1096/fasebj.5.15.1743438. [DOI] [PubMed] [Google Scholar]
- Ferraro E., Cecconi F. Autophagic and apoptotic response to stress signals in mammalian cells. Arch. Biochem. Biophys. 2007;462:210–219. doi: 10.1016/j.abb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gaitonde M. K., Wharton J., Holt E. 14C-Labelled amino acids and glucose in rat brain and liver after injection of [2-14C]propionate. J. Neurochem. 1977;29:127–133. doi: 10.1111/j.1471-4159.1977.tb03934.x. [DOI] [PubMed] [Google Scholar]
- Goldstein J. C., Waterhouse N. J., Juin P., Evan G. I., Green D. R. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- Hakem R., et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- Ho A. T., Li Q. H., Hakem R., Mak T. W., Zacksenhaus E. Coupling of caspase-9 to Apaf1 in response to loss of pRb or cytotoxic drugs is cell-type-specific. EMBO J. 2004;23:460–472. doi: 10.1038/sj.emboj.7600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennissen H. P. Ubiquitin and the enigma of intracellular protein degradation. Eur. J. Biochem. 1995;231:1–30. [PubMed] [Google Scholar]
- Kluck R. M., Bossy-Wetzel E., Green D. R., Newmeyer D. D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- Kotoulas O. B., Kalamidas S. A., Kondomerkos D. J. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Kuida K., Haydar T. F., Kuan C. Y., Gu Y., Taya C., Karasuyama H., Su M. S., Rakic P., Flavell R. A. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- Levine B., Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;116:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu H., Xu C. J., Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- MacFarlane M., Merrison W., Bratton S. B., Cohen G. M. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- Martinou I., Desagher S., Eskes R., Antonsson B., Andre E., Fakan S., Martinou J. C. The release of cytochrome c from mitochondria during apoptosis of NGF-deprived sympathetic neurons is a reversible event. J. Cell Biol. 1999;144:883–889. doi: 10.1083/jcb.144.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V. K., Wei M. C., Buttle K. F., Scorrano L., Panoutsakopoulou V., Mannella C. A., Korsmeyer S. J. A reversible component of mitochondrial respiratory dysfunction in apoptosis can be rescued by exogenous cytochrome c. EMBO J. 2001;20:661–671. doi: 10.1093/emboj/20.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Gogvadze V., Orrenius S., Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- Pearce D. A., Sherman F. Differential ubiquitin-dependent degradation of the yeast apo-cytochrome c isozymes. J. Biol. Chem. 1997;272:31829–31836. doi: 10.1074/jbc.272.50.31829. [DOI] [PubMed] [Google Scholar]
- Pereira-da-Silva L., Sherman M., Lundin M., Baltscheffsky H. Inorganic pyrophosphate gives a membrane potential in yeast mitochondria, as measured with the permeant cation tetraphenylphosphonium. Arch. Biochem. Biophys. 1993;304:310–313. doi: 10.1006/abbi.1993.1355. [DOI] [PubMed] [Google Scholar]
- Rawn J. D. Biochemistry. Hillsborough, NC: Neil Patterson Publishers/Carolina Biological Supply Company; 1989. [Google Scholar]
- Saleh A., Srinivasula S. M., Acharya S., Fishel R., Alnemri E. S. Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J. Biol. Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- Sherman F. The importance of mutation, then and now: studies with yeast cytochrome c. Mutat. Res. 2005;589:1–16. doi: 10.1016/j.mrrev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Sokolik C. W., Cohen R. E. The structures of ubiquitin conjugates of yeast Iso-2-cytochrome c. J. Biol. Chem. 1991;266:9100–9107. [PubMed] [Google Scholar]
- Sonegas M. S., Alarcón R. M., Yoshida H., Giaccia A. J., Hakeem R., Mak T. W., Lowe S. W. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Susin S. A., Zamzami N., Castedo M., Hirsch T., Marchetti P., Macho A., Daugas E., Geuskens M., Kroemer G. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J. Exp. Med. 1996;184:1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Nakabayashi Y., Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse N. J., Goldstein J. C., von Ahsen O., Schuler M., Newmeyer D. D., Green D. R. Cytochrome c maintains mitochondrial transmembrane potential and ATP generation after outer mitochondrial membrane permeabilization during the apoptotic process. J. Cell Biol. 2001;153:319–328. doi: 10.1083/jcb.153.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse N. J., Ricci J. E., Green D. R. And all of a sudden it's over: mitochondrial outer-membrane permeabilization in apoptosis. Biochimie. 2002;84:113–121. doi: 10.1016/s0300-9084(02)01379-2. [DOI] [PubMed] [Google Scholar]
- Wilson R., Goyal L., Ditzel M., Zachariou A., Baker D. A., Agapite J., Steller H., Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., Wang X. Prevention of apoptosis by Bcl-2, release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- Zamaraeva M. V., Sabirov R. Z., Maeno E., Ando-Akatsuka Y., Bessonova S. V., Okada Y. Cells die with increased cytosolic ATP during apoptosis: a bioluminescence study with intracellular luciferase. Cell Death Differ. 2005;12:1390–1397. doi: 10.1038/sj.cdd.4401661. [DOI] [PubMed] [Google Scholar]
- Zhang H. G., Wang J., Yang X., Hsu H. C., Mountz J. D. Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene. 2004;23:2009–2015. doi: 10.1038/sj.onc.1207373. [DOI] [PubMed] [Google Scholar]
- Zou H., Li Y., Liu X., Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.