Abstract
A subgroup of the AAA+ proteins that reside in the endoplasmic reticulum and the nuclear envelope including human torsinA, a protein mutated in hereditary dystonia, is called the torsin family of AAA+ proteins. A multiple-sequence alignment of this family with Hsp100 proteins of known structure reveals a conserved cysteine in the C-terminus of torsin proteins within the Sensor-II motif. A structural model predicts this cysteine to be a part of an intramolecular disulfide bond, suggesting that it may function as a redox sensor to regulate ATPase activity. In vitro experiments with OOC-5, a torsinA homolog from Caenorhabditis elegans, demonstrate that redox changes that reduce this disulfide bond affect the binding of ATP and ADP and cause an attendant local conformational change detected by limited proteolysis. Transgenic worms expressing an ooc-5 gene with cysteine-to-serine mutations that disrupt the disulfide bond have a very low embryo hatch rate compared with wild-type controls, indicating these two cysteines are essential for OOC-5 function. We propose that the Sensor-II in torsin family proteins is a redox-regulated sensor. This regulatory mechanism may be central to the function of OOC-5 and human torsinA.
INTRODUCTION
Dystonia is a neurological movement disorder characterized by involuntary muscle contractions, frequently forcing the body into twisting movements and abnormal postures. Early-onset torsin dystonia (or DYT1 dystonia, also known as Oppenheim's dystonia) is the most common and severe form among 14 different types of inherited dystonia syndromes (DYT1-14) (Kabakci et al., 2004). A GAG deletion in the DYT1 gene resulting in the loss of a single glutamate residue in the carboxy terminus of its gene product, torsinA, accounts for many cases of early-onset dystonia (Ozelius et al., 1997). TorsinA is the founding member of a poorly understood subclass of AAA+ proteins that includes torsinB (TOR1B) and at least two other proteins encoded by related genes, TOR2A and TOR3A, in humans, as well as OOC-5 in the nematode Caenorhabditis elegans (Ozelius et al., 1997, 1999; Basham and Rose, 2001).
Human torsinA is a 332-amino acid protein containing putative sites for glycosylation and phosphorylation, a signal sequence and a hydrophobic region at the amino-terminus that is presumably associated with the membrane of the endoplasmic reticulum (ER) (Breakefield et al., 2001; Callan et al., 2007). The better characterized AAA+ proteins fold into multidomain structures that self-associate, forming oligomers that catalyze the unfolding, disassembly, and/or translocation of a variety of substrates (Neuwald et al., 1999; Iyer et al., 2004). The signal sequence and the following hydrophobic region lead to the localization of torsinA within the lumen of the ER, placing it in position to perhaps act on important and unknown secreted (Liu et al., 2003; Hewett et al., 2007) or stress-related protein substrates (Hewett et al., 2003). Recent evidence suggested torsinA may also act on substrate(s) in the nuclear envelope (NE) as well as the ER (Naismith et al., 2004; Goodchild and Dauer, 2004, 2005). The DYT1 mutation of torsinA (ΔE302/303) presumably leads to dystonia due to a loss of its AAA+ function (Goodchild and Dauer, 2004). Little is known about this function. Loss of the activity of torsinA localized in the neuronal nuclear envelope disrupts neuronal maturation, resulting in neurodevelopment defects in animal models (Goodchild et al., 2005). This loss of function could be caused by direct alteration of catalytic or regulatory residues or by secondary effects such as inhibition of torsinA folding or oligomerization as required for assembly of an active AAA+ protein. TorsinA has also been shown to regulate trafficking of a G-protein–coupled receptor, dopamine transporters, and ion channels. Notably, the deletion mutant does not exhibit this function and acts as a dominant-negative via the association with wild-type protein (Torres et al., 2004).
Understanding torsinA's function is a key to establishing the molecular mechanism of dystonias, as torsinA's substrates likely play a role in the pathology of non-DYT1 dystonias as well. Here, a low-resolution structural model of human torsinA was deduced, based on a multiple alignment of torsin subfamily proteins with the closest homologue of known structure, ClpA from Escherichia coli. A very similar model based on ClpB was recently presented (Kock et al., 2006). Both proposed models suggest torsinA is composed of two subdomains, an N-terminal α/β subdomain and a C-terminal α-helical subdomain containing a dystonia-causing mutation (Neuwald et al., 1999; Iyer et al., 2004). Interestingly, six prominently conserved cysteine residues in torsin proteins distinguish this family from other AAA+ proteins. The ClpA-based model presented here predicts that these cysteines form at least two disulfide bonds in torsinA and perhaps other family members: one disulfide is located in the large N-terminal domain and the other in the C-terminal small domain. The most profound implication of the prediction is that it places the cysteine at position 319 of human torsinA in the Sensor-II motif of other AAA+ proteins (Erzberger and Berger, 2006; Kock et al., 2006). Generally, the Sensor-II motif of AAA+ proteins contains a conserved arginine that interacts with the γ-phosphate of ATP, as the amino acid sequence is GAR in E. coli ClpA, ClpB (Mogk et al., 2003; Kock et al., 2006) and others (Lee et al., 2003). However, the multiple sequence alignment presented in this work predicts that the corresponding sequence at this position is GCK in torsin proteins (Kock et al., 2006), suggesting that the interaction between this motif and nucleotide may be regulated by the redox status of Cys 319 of human torsinA. In this article, we directly test this hypothesis using the nematode protein, OOC-5.
OOC-5 is a torsinA homologue that plays an essential role in nematode oogenesis and embryo development (Basham and Rose, 1999). OOC-5 and torsinA both colocalize with ER markers, suggesting OOC-5 to be a good model protein for investigating the basic biophysical properties and biochemical function of torsin family proteins (Basham and Rose, 2001). Thus, to investigate the relationship between the redox and nucleotide status of the proteins and their function, as suggested by the structural model, in vitro and in vivo studies of OOC-5 were performed.
Based on these results, a hypothesis is proposed that in the torsin family OOC-5 and torsinA function as integrators of two input signals, cellular nucleotide and redox status, as required, to regulate specific actions of these proteins on unknown substrates for oocyte development and neuronal function, respectively.
MATERIALS AND METHODS
Bioinformatic Methods: Sequence Database Searches and Inferred Alignment of Human TorsinA to the E. coli ClpA Structure
Homologues of human torsinA were initially collected from a BLAST (Altschul et al., 1997) search of the NCBI nonredundant database (4/2/03, 1,396,568 sequences) using the full-length sequence of human torsinA (gi 4557541, range 1-332) as query. Nineteen significant hits (E ≤ 1e − 5) were obtained after removal of fragments and highly similar sequences. These 19 homologues were aligned using T-COFFEE (Notredame et al., 2000; v. 1.37, default parameters) without manual adjustment. Among these hits were the six sequences displayed under the human torsinA in the alignment of Figure 1A. Not included in these close homologous proteins, but having an E-value (4e − 4) just under the significance cutoff, was the ClpB sequence from Plasmodium berghi (gi 1197585, range 611-821). No significant structure hits were found at this stage that aligned with the extreme C-terminal portion of human torsinA, where the dystonia-associated Glu 302/303 deletion was located.
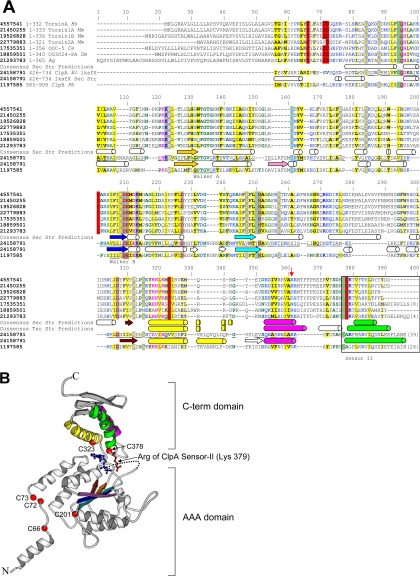
Figure 1.
General structural model of C. elegans OOC-5 and other members of the torsin family. (A) Alignment of torsin proteins with E. coli ClpA, a protein of known structure. C. elegans OOC-5, human torsinA, and five close homologues, aligned using T-COFFEE, are shown on the first seven lines. The consensus secondary structure prediction for these proteins using three methods, as described in the text, are displayed on the eighth line; predicted helices are shown as cylinders and predicted strands are shown as arrows. NCBI gi identifiers are shown in bold type, ranges of sequence numbering included in the alignment are given immediately thereafter, and species abbreviations are shown in italics. Sequences from E. coli ClpA and P. berghi ClpB, found using database searches as described in the text, are shown on lines 9 and 11, respectively. These two sequences have been aligned to human torsinA, and thus implicitly to OOC-5 and the other torsin proteins, through a shared conserved domain database (CDD) profile, as described in the text. The portions of the ClpA sequence that align to human torsinA are underlined. The experimentally determined secondary structure of ClpA (PDB 1ksfX) is shown below its corresponding sequence. Consensus tertiary structure predictions of the smaller C-terminal domain only, using ROSETTA, are displayed below the secondary structure predictions for this domain. Colors of secondary structure elements correspond to colors in B. The alignment was colored automatically using CHROMA; sites of predominately hydrophobic character are yellow, whereas sites of predominantly charged or polar character are blue. Six conserved cysteines predicted to form at least two disulfide bonds in the torsin proteins are highlighted in red. Positions 1-55 of the alignment (corresponding to position 1-34 of torsinA) are not reliably aligned. (B) Locations of six cysteine residues in the torsin family and the inferred Sensor-II motif mapped onto the E. coli ClpA structure (PDB 1ksfX). Conserved cysteines are shown as red spheres and are numbered as in A. The Arg 702 side chain of E. coli ClpA, aligned to the Lys of the C. elegans OOC-5 sequence at position 379 in Figure 1A, is explicitly shown in red. The ADP observed in the ClpA active site is shown in blue.
To obtain a structural link to the C-terminal region, a more sensitive RPS-BLAST search of the NCBI conserved domain database (Marchler-Bauer et al., 2003; v.1.61, 10927 position-specific scoring matrices) was performed using as query the portion of the P. berghi ClpB sequence found during the initial BLAST search. The top hit revealed a highly significant match to the ClpA profile (E = 3e − 85), which contained the E. coli ClpA sequence (gi 24158791) with known structure (Guo et al., 2002; PDB identifier 1ksfX; Berman et al., 2000). The ClpA profile therefore allowed a confident alignment between human torsinA with the ClpA structure through the intermediary ClpB sequence (denoted by the underlined regions of the ClpA sequence in Figure 1A). This result indicated that the human torsinA protein adopted an overall fold similar to that of the E. coli ClpA protein, a member of the AAA+ superfamily (Neuwald et al., 1999; Iyer et al., 2004). In addition, the alignment suggested a structural location of six conserved cysteine residues (Figure 1, A and B) and the location of the arginine of Sensor-II in E. coli ClpA (Figure 1, A and B).
Additional support for this alignment was provided by evidence obtained from secondary and tertiary structure predictions. Secondary structure predictions of the full-length torsins were performed using three independent methods: J-Pred (Cuff et al., 1998), PHD (Rost and Sander 1993), and SSpro (Pollastri et al., 2002). A consensus secondary structure prediction for the torsins alignment was constructed as follows. For each of the three methods, the predicted secondary structure type (helix, strand, or other) that was found in the majority (i.e., 10 or more) of the 19 sequences in the torsin alignment was taken to be the predicted secondary structure for that method at that alignment position. Then, the secondary structure type found in the majority (i.e., at least two of the three) prediction methods was taken to be the overall prediction at that position. Tertiary structure predictions of the C-terminal domain using ROSETTA (Bonneau et al., 2001) were performed using the C-terminal 62 residues of human torsinA (start from position 314 in Figure 1A, i.e., position 271 in human torsinA). A consensus secondary structure prediction was derived from these data as follows. 2000 candidate structures were generated, and the secondary structure type found in the majority of structures (i.e., 1001 or more) was taken to be the predicted secondary structure type for that residue.
Materials
Restriction enzymes were purchased from New England BioLabs (Beverly, MA) except BalI from Promega (Madison, WI). Ultrapure GdmCl was obtained from ICN Biochemicals (Irvine, CA). IPTG was from RPI Corp (Mount Prospect, IL). Iodoacetate (IAA) was from Pierce (Rockford, IL). 5,5′-dithiobis-(2-nitrobenzoic acid (DTNB) was from Molecular Probes (Eugene, OR). Oxidized (GSSG) and reduced glutathione (GSH), Bis-Tris Propane, dithiothreitol (DTT), EDTA, Tris(2-carboxyethyl)phosphine (TCEP), urea, NADPH, glutathione reductase, AMP, ADP, ATP, AMPPNP, AMPPCP, ATPγS, iodoacetamide (IAM), trypsin (TPCK treated from bovine pancreas), and α-chymotrypsin (TLCK treated from bovine pancreas) were from Sigma-Aldrich (St. Louis, MO). His·bind resin and prepacked columns were from Amersham Biosciences (Piscataway, NJ). Twice-deionized water was used throughout.
Bacterial Expression and Purification of Soluble Lumenal Domain of OOC-5
The lumenal domain of OOC-5 (44OOC-5) without signal sequence and hydrophobic region was amplified and cloned into the pSmt3 expression vector (Mossessova and Lima, 2000), a generous gift from C. Lima (Cornell University, New York). The E179Q point mutation was generated by QuikChange Site-Directed mutagenesis (Stratagene, La Jolla, CA).
The His/Smt3-44OOC5 or His/Smt3-44OOC5E179Q fusion proteins were expressed in Rosetta-gami B (DE3) pLacI cell (Novagen, Madison, WI). Cultures were grown at 37°C to an OD600 of 1.0 and then cooled down to 15°C, induced by the addition of 0.25 mM IPTG, and allowed to express for 20 h. The cells were harvested and lysed by sonication in buffer A (25 mM Bis-Tris Propane, 150 mM NaCl, 10% glycerol, pH 9.0). The purification of the fusion proteins was followed using standard nickel-affinity chromatography. The fusion protein was cleaved with Ulp1 protease (vol/vol: 500/1) and then dialyzed against 20 mM Bis-Tris Propane, 50 mM NaCl, pH 9.0. The His-tagged Smt3, the fusion protein and His-tagged Ulp1 were removed by running through a HiTrap Q HP (5 ml) and a second nickel column. Finally, the purified 44OOC5 or 44OOC5E179Q were obtained by collecting the monodisperse peak eluted from a size-exclusion column, HiPrep Sephacryl S-200 HR, with buffer B (20 mM Tris, 200 mM NaCl, pH 8.5). Protein concentrations were determined from the absorbance at 280 nm, using a molar extinction coefficient of 23,850 M−1 cm−1 in 6 M GdmCl for both 44OOC-5 wild type and the E179Q mutant proteins.
Circular Dichroism Measurements
Far-UV circular dichroism (CD) spectra were measured at 20°C in a 0.1-cm path-length quartz cuvette using Jasco J-810 Circular Dichroism spectropolarimeter equipped with a PFD-425S Peltier temperature controller. The protein concentration was 0.18 mg/ml (5 μM) in 20 mM phosphate buffer, pH 8.5. Reductants, such as TCEP and DTT, and nucleotides were added and incubated at 20°C for 30 min before measurement. Spectra were collected from 260 to 195 nm with a data pitch of 0.2 nm. A bandwidth of 1 nm was used with a detector response of 1 s and scanning speed of 50 nm/min. Percentage of protein secondary structure was estimated by online software “K2d: Prediction of percentages of protein secondary structure from CD spectra” (http://www.embl-heidelberg.de/∼andrade/k2d/). The thermal dependence of the structure was monitored at 222 nm between 20 and 70°C with a data pitch of 0.5°C and a temperature slope of 2°C/min.
Determination of Disulfide Bonds in 44OOC-5 by Double Trapping with IAA and IAM
The integral number of free cysteine residues and disulfide bonds present in the protein was determined by sequentially adding both acidic IAA and neutral IAM (Creighton, 1984; Zander et al., 1998). Free sulfhydryls in 44OOC-5 (if there is any) were first trapped by incubation in 30 mM IAA in 50 mM Tris-HCl, pH 8.2, and 1 mM EDTA in the presence of 8 M urea for 15 min at 37°C. The reaction was stopped by precipitation with 10 vol of a cold acetone-1 N HCl mix (98:2, vol/vol). The precipitate was recovered by centrifugation, and washed three times by resuspension in a cold acetone-1 N HCl–H2O mix (98:2:10, vol/vol:vol) and recentrifugation. The pellet was dissolved in 8 M urea, 10 mM DTT, 50 mM Tris-HCl, pH 8.2, 1 mM EDTA, with incubation at 37°C for 30 min to reduce the remaining disulfides. The newly generated free sulfhydryls were then alkylated by the addition of 50 mM IAM and incubated at 37°C for 10 min.
Unfolding and reducing markers were prepared by incubation in 8 M urea and 10 mM DTT in 50 mM Tris-HCl, pH 8.2, and 1 mM EDTA at 37°C for 30 min and then were alkylated by 50 mM IAM, 50 mM IAA, or mixtures of the two for 15 min at room temperature. Double trapped species and the unfolded and reduced markers were subjected to electrophoresis in 6% polyacrylamide gels containing 8 M urea. IAA leaves behind the charged carboxymethyl group when it reacts with a cysteine residue, whereas IAM leaves behind an uncharged group. The total number of IAA molecules incorporated can be determined because migration in urea gels is dependent on charge. The number of distinct bands present in the marker lanes should correspond to one more than the number of half-cystine present in the protein molecules (Creighton, 1984).
Disulfide Mapping by Protease Digestion followed by LC-MS/MS
The complete trypsin digestion followed by Nano-LC-MS/MS experiment was conducted by Protein Chemistry Technology Center at University of Texas Southwestern Medical Center at Dallas. Wild-type OOC-5 (10 μg) in 20 mM Tris-HCl and 200 mM NaCl, pH 8.5, was digested with trypsin overnight at 37°C. The tryptic digest of the sample was fractionated using a Dionex LC-Packings HPLC. Peptides were first desalted on a 300-μm × 1-mm PepMap C18 trap column with 0.1% formic acid in HPLC grade water at a flow rate of 20 μl/min. After desalting for 5 min, peptides were flushed onto a LC Packings 75 μm × 15 cm C18 nanocolumn (3 μm, 100 Å) at a flow rate of 200 nl/min. A 45-min gradient was used for the HPLC separation, with the acetonitrile concentration increased from 2 to 45%. Eluates were analyzed with a QSTAR XL mass spectrometer (Applied Biosystems, Foster City, CA). Data were acquired in Information Dependent Acquisition (IDA) mode where the top three precursor ions were selected per cycle for MS/MS experiment. Mass ranges for the MS survey scan and MS/MS were m/z 300-1800 and m/z 50-1800, respectively. Raw data were processed by Analyst QS to generate the PKL file. Data were searched by using Mascot search engine (Matrix Science, London, United Kingdom) against the SwissProt database and the home-built database containing OOC-5 protein sequence.
At a measured redox potential of −217 mV, produced by a defined ratio of reduced-to-oxidized glutathione (10 mM GSH/0.6 mM GSSG; Ferguson et al., 2006), wild-type OOC-5 was incubated at room temperature for 3 h and then alkylated by 50 mM IAA for 15 min at room temperature. After buffer exchange on a Zeba desalting column (Pierce) to remove glutathione and IAA, the protein was analyzed at the Protein Chemistry Technology Center at University of Texas Southwestern Medical Center at Dallas by complete protease digestion and Nano-LC-MS/MS.
Redox Potential of the Purified 44OOC-5
The equilibrium redox potential of 44OOC-5 in vitro was determined by trypsin digestion under titrated redox conditions. Eight micrograms of 44OOC-5 in 20 mM Tris-HCl, 150 mM NaCl, pH 8.5, 1 mM EDTA, and various concentrations of reduced and oxidized glutathione (Ferguson et al., 2006), was incubated at room temperature for 3 h and then digested by trypsin (50:1, m:m) at room temperature for 30 min. The effect of nucleotide on the redox potential was performed as above except that ADP or ATP was incubated with protein in the prepared redox buffer. Cleaved samples were resolved by 10% Tris-Tricine SDS-PAGE under reducing conditions and stained by Coomassie blue.
The concentration of GSH in the redox buffer was determined by the total thiol content spectrophotometrically (ε412 = 14150 M−1 cm−1) using DTNB (Riddles et al., 1983) before and after the incubation. The equilibrium concentration of GSSG was quantified enzymatically using yeast glutathione reductase coupled to NADPH oxidation at 340 nm (Zander et al., 1998). The redox potential was calculated using the Nernst equation as described (Aslund et al., 1997).
Generation and Analysis of Transgenic C. elegans
Wild-type (N2) and ooc-5 mutant strains (KK696) were maintained under standard conditions at 20°C (Brenner, 1974). A minigene construct of ooc-5 under control of the ooc-5 promoter was created for transformation rescue assays by using ooc-5 cDNA sequences to replace exons 2–4 of a genomic DNA construct capable of rescuing ooc-5 mutants (Basham and Rose, 2001). This plasmid, pAH83-unc119, also contains the unc-119 gene. Cysteines 287 and 329 were mutated to serine in pHis/Smt3-44OOC-5 using QuikChange Site-Directed mutagenesis kit (Stratagene). The BalI-digested ooc-5 cDNA fragment from the pHis/Smt3-44OOC-5 cysteine-to-serine mutant (237-1022 base pairs of ooc-5 cDNA (total 1071 base pairs) containing 859-861 for Cys 287 and 985-987 for Cys 329) was cloned into the BalI site of pAH83-unc119 to create pAH83-unc119CS. Plasmids were introduced into the strain KK696 (ooc-5(it145) unc-4 (e120)/mnC1) using the complex array procedure, with pRF4 (carrying the dominant rol-6 mutation) as a cotransformation marker (Kelly et al., 1997). F1 Roller worms were picked to separate plates to identify heritable lines and two independent lines carrying pAH83-unc119 and pAH83-unc119CS as extrachromosomal arrays were obtained (referred to as ooc-5(+) and ooc-5(C287S,C329S)), respectively). To test for embryonic viability, L4 stage Roller worms homozygous for ooc-5 unc-4 were picked to separate plates. After 48 h, the worm was removed, and the number of larvae and unhatched embryos was counted 24 h later. Non-Rol ooc-5 unc-4 siblings (which have lost the transgene) were also analyzed as a control. Because ooc-5 is a maternal effect lethal mutation, homozygous ooc-5 worms are viable, but produce virtually all dead embryos unless rescued by the transgene (see Table 3).
Table 3.
Cysteine to serine mutations in the C-terminal domain of OOC-5 disrupt the rescuing ability of an ooc-5 transgene
| Transgenea | Worms holding embryos (n)b | Worms laying embryos (n)c | Embryonic viabilityd (%) |
|---|---|---|---|
| ooc-5 (it145); ooc-5 (+) | |||
| Line 1 | 4 | 15 | 72.04 |
| Line 2 | 0 | 17 | 20.90 |
| ooc-5 (it145); ooc-5 (C287S,C329S) | |||
| Line 1 | 17 | 20 | 5.11 |
| Line 2 | 10 | 10 | 3.26 |
| ooc-5 (it145) (no transgene) | |||
| Line 1 | 10 | 8 | 0.45 |
| Line 2 | 18 | 7 | 0.75 |
a Worms containing the wild-type ooc-5 transgene, the mutant ooc-5(C287S,C329S) transgene, or no transgene as indicated, were tested for their ability to rescue worms homozygous for the ooc-5(it145) mutation (see Materials and Methods). Because ooc-5 is a maternal effect lethal mutation, homozygous ooc-5 worms are viable, but produce virtually all dead embryos unless rescued by the transgene. Some homozygous worms also hold the majority of their eggs instead of laying them on the plate.
b Worms that laid fewer than 20 embryos.
c Worms that laid at least 20 embryos.
d L4 worms were picked to individual plates and removed after 48 h. Viability was calculated 24 h later from the number of larvae hatched divided by the total number of larvae and unhatched embryos. Viability was only scored from those worms laying at least 20 embryos.
RESULTS
Relationship of OOC-5 and TorsinA to Other AAA+ Proteins
Based on substantial sequence similarity, as well as secondary and tertiary structure predictions, the human torsinA sequence is predicted to adopt an overall fold similar to the second AAA+ module (D2 large and small domains) of the E. coli ClpA AAA+ protein (PDB identifier 1ksfX). As described in Materials and Methods, the boundary between the large and small domains was predicted to occur approximately at position 313/314 of the alignment in Figure 1A (position 270/271 of the human torsinA sequence), which is consistent with the alignment of torsinA with the C-terminal AAA+ domain of ClpB from Thermus thermophilus (the C-terminal subdomain is from residues 272–332; Kock et al., 2006). The secondary structure of the small C-terminal domain (Figure 1B) was predicted to be predominantly α-helical, which placed the structural location of the dystonia-associated Glu 302/303 deletion at a surface-exposed position at the C-terminus of the second helix in the small domain (Figure 1A, red arrow). This is distinct from the model based on the alignment of torsinA with ClpB, because the location of the Glu 302/303 deletion is at the N-terminus of the second helix, whereas the third helix in these two alignments is located in the same position (Kock et al., 2006). In addition, six conserved cysteine residues found in the torsin family proteins (red highlighted columns in Figure 1A) were observed to be spatially proximate when mapped onto their aligned positions in the ClpA structure (Figure 1B). These predicted structural features of human torsinA were useful in guiding experimental investigation, as described below.
These invariant cysteine residues are a unique sequence signature of the torsin proteins that separates them from other AAA+ proteins. Given the proposed overall structural similarity between the torsin proteins and ClpA, the proposed domain boundary described above, and the location of these proteins in the oxidizing environment of the ER lumen, the conservation of these Cys residues could be explained by the existence of at least two disulfide bonds. First, the Cys (position 201 in Figure 1A) located N-terminal to the third β-strand (dark blue) would be topologically near a Cys (position 66) that is located near the N-terminus (Figure 1B). Second, the Cys located just after the proposed domain boundary at position 323 is predicted by the model to form a disulfide in the small C-terminal domain with the Cys at position 378 (Figure 1B). Although automatic profile alignments were unable to confidently align torsinA with ClpA in this extreme C-terminal region (after position 362), two additional pieces of evidence supported the alignment of these latter Cys residues. First, the patterns of hydrophobicity, as well as the conserved Gly at position 377, matched well between the predicted and actual α-helices (green cylinders in Figure 1A). Second, consensus secondary structure predictions placed the third α-helix of the domain (green cylinders) at this location.
This predicted structure model placed the Cys at the position 378 from our alignment equivalent to Ala 701 in ClpA (i.e., the Cys at position 319 of human torsinA or the Cys at position 329 of OOC-5) in the Sensor-II motif (Figure 1A). This aspect is also consistent with the alignment of torsinA with the second AAA+ domain from T. thermophilus ClpB (Kock et al., 2006). We hypothesize that the Sensor-II motif in torsin proteins acts as a redox-regulated sensor (RRSII) that is coupled to the nucleotide binding of the N-terminal AAA domain and modulates the function of this group of AAA+ proteins. In the following experiments, we utilize the nematode homologue of torsinA, OOC-5, to test this hypothesis.
Cysteine 329 in Sensor-II Motif Forms a Disulfide with Cysteine 287 in the C-Terminus of OOC-5
Based on the multiple sequence alignment of Figure 1A, the lumenal domain of OOC-5 (44OOC-5) was constructed and expressed in E. coli with a His/Smt3 tag, and purified as a single band shown by SDS-PAGE (Supplemental Figure S1A).
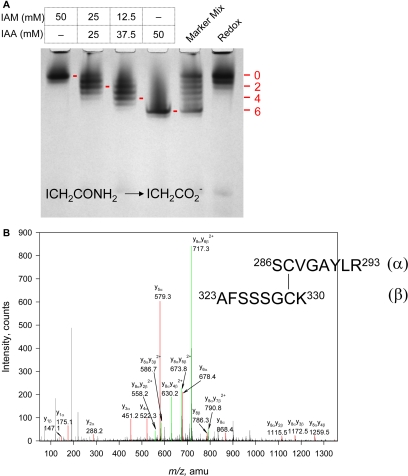
The redox status of cysteine residues in OOC-5 was determined by protein electrophoretic mobility shift assay. Briefly, free sulfhydryls were first trapped by adding a negative charge with IAA in the presence of urea, and then all disulfides were reduced to free thiol groups with DTT and blocked using the uncharged IAM. The calibrating marker was generated by reducing the purified protein and then modifying with various mixtures of IAM and IAA. The IAM-treated proteins (Figure 2A, left lane) have the lowest electrophoretic mobility due to the lack of charge introduced by IAM. Molecules of OOC-5 with 1–6 acidic carboxymethyl groups are generated from left to right (Figure 2A, lanes 2–4) by reacting with 1:1, 1:3 and 0:1 mixture of IAM and IAA. The sample in lane 5 is a mixture of the four marker samples. The in vitro redox state of OOC-5 was determined by the above double trapping with IAA and IAM. As shown in the right lane of Figure 2A, purified OOC-5 exists primarily as a fully oxidized form (the six conserved cysteines form three disulfides), except that a minor fraction of the protein has two free, reduced cysteines. This result is consistent with the model which predicted that at least two intramolecular disulfide bonds exist in the torsin proteins.
Figure 2.
Disulfide bond formation in purified OOC-5. (A) IAA/IAM double trapping to determine the redox state of OOC-5. Protein was first treated with IAA, reduced by DTT, and then treated by IAM. Mobility standards, corresponding to fully oxidized OOC-5 (lane 1), fully reduced OOC-5 (lane 4), or all seven oxidation states of OOC-5 (lane 5, bands with 0–6 acidic groups on the 6 Cys thiols) are prepared by treating protein with DTT and either IAM, IAA, or a mixture of IAA and IAM (lanes 2 and 3), respectively. Numbers 0, 2, 4, and 6 represent the number of free Cys thiol groups alkylated by acidic IAA. Urea is present during all steps of the procedure to ensure that all thiols are accessible to alkylating and reducing agents. (B) MS/MS analysis of the signal at m/z 1650.75 obtained from trypsin digestion of 44OOC-5 at oxidized condition (see Table 1). The singly charged fragments are shown in red. The peaks of doubly charged ion are shown in green. Ser(286)-Arg(293) and Ala(323)-Lys(330) are linked between Cys-287 and Cys-329.
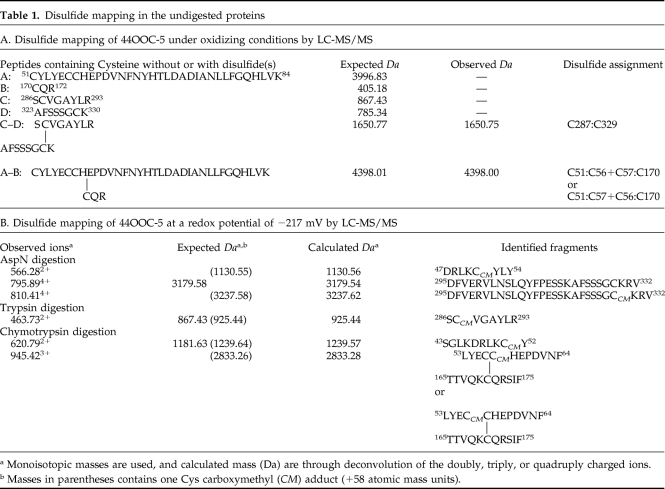
To identify which cysteines form disulfide pairs under oxidizing conditions, trypsin digestion followed by Nano-LC-MS/MS experiment was performed. Complete digestion of 44OOC-5 should generate four peptides containing cysteine residues (A, B, C, and D) if there were no disulfides present in the undigested protein (Table 1A). However, only a peptide with Mr of 1650.75 was found from the MS data (data not shown), corresponding to the linked peptides C and D with a single disulfide. ESI-MS/MS spectrum of the Mr of 1650.75 peptide (Figure 2B) demonstrates that a disulfide is formed between Cys 287 and Cys 329 of OOC-5, also consistent with the model (Figure 1A). Another peptide with Mr of 4398.00 was observed by MS and expected for linked peptides A and B with two pairs of disulfide. However, MS/MS data of the Mr of 4398.00 peptide did not give clear structural information. Therefore, the four conserved cysteines in the N-terminal AAA domain, i.e., Cys 51, Cys 56, Cys 57, and Cys 170, probably form two pairs of disulfides (Table 1A), consistent with the results of double-trap alkylation experiment (Figure 2A). This is partially consistent with the prediction of one disulfide formed in the N-terminal domain, but the possibility of two disulfides in the AAA domain cannot be ruled out by structural prediction.
Table 1.
Disulfide mapping in the undigested proteins
a Monoisotopic masses are used, and calculated mass (Da) are through deconvolution of the doubly, triply, or quadruply charged ions.
b Masses in parentheses contains one Cys carboxymethyl (CM) adduct (+58 atomic mass units).
Effect of Redox on the Structure, Stability, and Nucleotide Binding of OOC-5
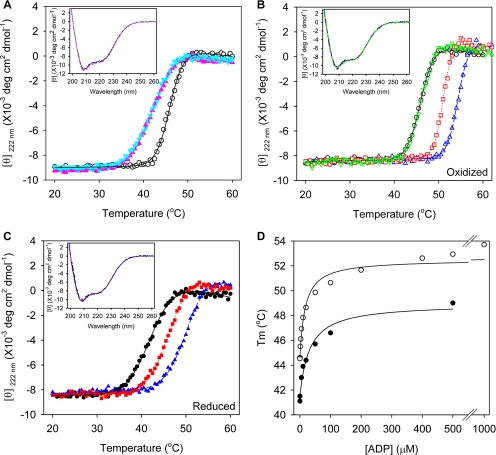
The results above revealed that purified OOC-5 exists predominately in a fully oxidized form. When a far-UV CD spectrum was collected to determine the secondary structure of OOC-5, two negative peaks at 222 and 208 nm of the CD spectrum were observed in the absence of any reductant, indicating that OOC-5 assumed an α-helical structure as predicted. The mean residue ellipticity at 222 nm is about −8.5 × 103 deg cm2 dmol−1 (Figure 3A inset, solid line), indicating that the lumenal domain is 30% α-helix secondary structure, consistent with the predicted structure (Figure 1B). Notably, the CD spectrum of OOC-5 does not measurably change if the protein is pretreated with reducing agents, such as DTT or TCEP (Figure 3A, inset, dotted and dashed lines), demonstrating that disulfide formation is not required for maintaining the secondary structure of OOC-5. However, temperature melt experiments show that the midpoint of the thermal stability transition (Tm), as detected by the ellipticity at 222 nm, shifts from 45 to 42°C in the presence of TCEP or DTT (Figure 3A and Table 2), showing that OOC-5 is marginally stabilized by oxidation.
Figure 3.
Purified OOC-5 binds ADP and ATP, but not AMP. (A) Temperature melts under oxidized and reduced conditions. 44OOC-5 in 20 mM phosphate buffer, 200 mM NaCl, pH 8.5, with a protein concentration of 5 μM monomer, in the absence (○) or presence of 0.5 mM TCEP (▲), or 1 mM DTT (▼). Inset, far-UV CD spectra at all these conditions at 20°C before melting. Black solid line is for protein in the absence of reductant, pink dashed for the presence of TCEP, and cyan dotted for the presence of DTT. (B and C) Temperature melts in the presence of different nucleotides and under oxidized and reduced conditions. Inset, far-UV CD spectra before melting. Black solid line is for protein in the absence of nucleotide, green dash-dot for the presence of AMP, blue dashed for the presence of ADP, and red dotted for the presence of ATP. Five micromolar monomer of 44OOC-5 in 20 mM phosphate buffer, 200 mM NaCl, pH 8.5, was incubated in the absence or presence of 500 μM nucleotide or was incubated in 0.5 mM TCEP without or with nucleotide. ○ and ●, protein alone; ▽, protein with AMP; △ and ▲, protein with ADP; and □ and ■, protein with ATP. (D) ADP titration by temperature melting under oxidized and reduced conditions. Five micromolar monomer of 44OOC-5 in 20 mM phosphate buffer, 200 mM NaCl, pH 8.5, was incubated in different concentrations of ADP in the absence (○) or in the presence of 0.5 mM TCEP (●). Kd (ox) = 17 ± 3 μM; Kd (red) = 31 ± 7 μM.
Table 2.
Thermal stability for 44OOC-5 wild type and the E179Q mutant in the presence of nucleotide under different redox conditions
| Nucleotidea | 44OOC-5 |
44OOC-5E179Q |
||
|---|---|---|---|---|
| Oxidizedb | Reducedc | Oxidizedb | Reducedc | |
| None | 45.2 ± 0.3 | 42.0 ± 0.2 | 44.7 ± 0.2 | 41.7 ± 0.2 |
| AMP | 44.7 ± 0.7 | — | 44.9 | — |
| ADP | 53.5 ± 0.4 | 50.7 ± 0.6 | 52.4 ± 0.7 | 50.4 ± 0.1 |
| ATP | 50.2 ± 0.4 | 46.1 ± 0.2 | 52.1 ± 0.5 | 48.1 ± 0.2 |
| ATPγS | 50.4 ± 0.7 | 46.8 | 52.1 | — |
| AMPPNP | 45.7 | — | — | — |
| AMPPCP | 44.9 | — | — | — |
Thermal stability, Tm (°C), was measured as a midpoint of the transition temperature by monitoring far UV-CD ellipticity at 222 nm. The data shown here are the mean value ± SE calculated from at least three different experiments setting from two different purification preps, except the measured value only from single experiment.
a Nucleotide without Mg2+ was added to protein solution under oxidized or reduced conditions to 500 μM concentration before measurement.
b Wild type or E179Q 44OOC-5 was 5 μM monomer concentration in 20 mM phosphate buffer, 200 mM NaCl, pH 8.5, in the absence of reductant (oxidized).
c Reduced protein was incubated in 0.5 mM TCEP for 30 min at 20°C before measurement.
Torsin proteins were expected to bind nucleotides as do other AAA+ proteins. As shown in Figure 3B, inset, the far UV-CD spectrum of OOC-5 does not change in the presence of any of the nucleotides tested, including ATP, ADP, and AMP, demonstrating no significant change in the secondary structure. When OOC-5 is oxidized, ATP and ADP shift the Tm from 45°C to a temperature higher than 50°C as detected by the ellipticity at 222 nm (Figure 3B and Table 2), consistent with ligand induced stabilization. Interestingly, ATPγS, but not other ATP analogues such as AMPPNP or AMPPCP, increased Tm to 50°C with a saturated concentration of 0.5 mM (Table 2). When OOC-5 is reduced, ATP and ADP shift Tm from 42°C to higher temperatures (Figure 3C and Table 2). These results indicate that both oxidized and reduced forms of OOC-5 bind ATP and ADP.
To determine if redox status affected the affinity for nucleotide, OOC-5 was titrated with ADP under both oxidized and reduced conditions, and binding was detected by temperature melt shifts. As shown in Figure 3D, the binding of ADP to OOC-5 is saturated at 0.5 mM under both oxidized and reduced conditions. The Kd values for ADP of oxidized and reduced OOC-5 are 17 ± 3 and 31 ± 7 μM, respectively.
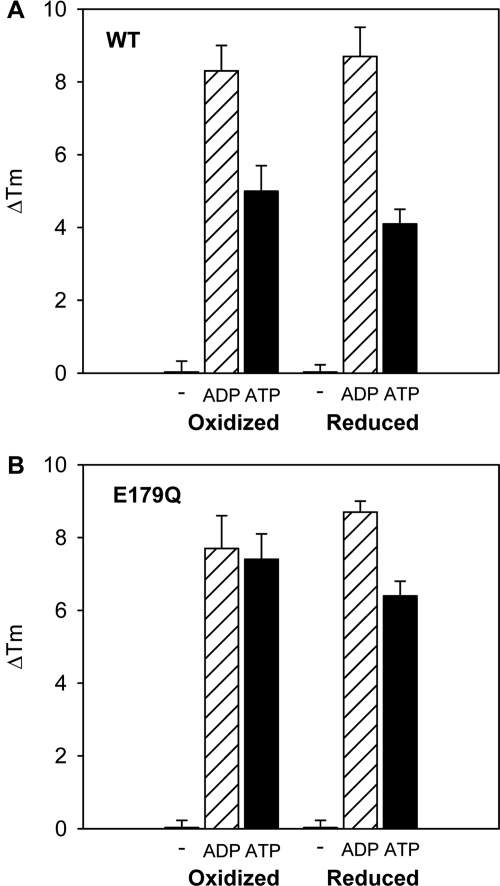
It is known that the glutamate residue in the Walker B motif is responsible for ATP hydrolysis and the substitution of glutamine for glutamate blocks hydrolysis and produces a “substrate trap” mutant (Weibezahn et al., 2003; Naismith et al., 2004; Goodchild and Dauer, 2004, 2005). OOC-5 containing this mutation (E179Q) has features similar to that of wild type, including the same far-UV CD spectrum (data not shown) and the same Tm under both oxidized and reduced conditions (Table 2). However, when a shift of temperature melt in Tm (ΔTm) is probed to compare the differences in nucleotide binding between wild-type OOC-5 and the E179Q mutant, both oxidized and reduced conformations of wild-type OOC-5 interact with ADP more tightly than ATP (Figure 4A). By contrast, this preference is not seen for the E179Q mutant under oxidized conditions (Figure 4B). Moreover, the preference of reduced conformation for ADP over ATP is diminished for the E179Q mutant relative to the wild-type OOC-5 (Figure 4B). These results suggest that the substrate trap mutant binds ATP more tightly than does wild-type OOC-5.
Figure 4.
Nucleotide binding differences under oxidizing and reducing conditions for both wild type and the E179Q mutant. Tm increase in the presence of nucleotides ADP and ATP relative to protein alone, under oxidizing and reducing conditions for wild type (A) and the E179Q mutant (B). ΔTm data are calculated from the experiments shown in Table 2.
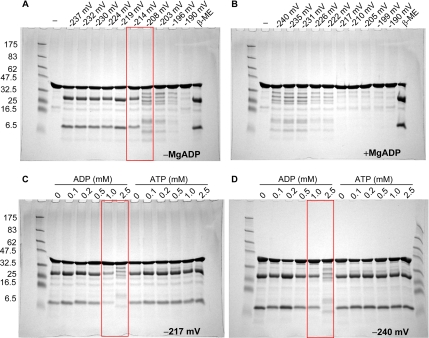
AAA+ proteins, including Clp/Hsp100 family proteins, typically form a hexameric ring (ClpA, Guo et al., 2002; ClpB, Lee et al., 2003; ClpX, Kim and Kim, 2003; HslU, also known as ClpY, Bochtler et al., 2000; Sousa et al., 2000) in the presence of ATP to exert their functions (Maurizi and Xia, 2004). Analytical gel filtration analysis indicates 44OOC-5 produces a symmetric peak with MW of 36 kDa under oxidized conditions, which is very close to the calculated MW of a monomer (Supplemental Figure S2A). Under reducing conditions in the presence of TCEP, the retention volume of OOC-5 did not change (Supplemental Figure S2A), indicating that disulfide reduction has no impact on the global conformation of its monomeric structure and does not induce oligomerization. The E179Q mutant was also analyzed by gel filtration. Neither the wild-type OOC-5 nor the E179Q mutant exhibited a detectable change in retention volume on gel filtration column in the presence of ATP or ADP (Supplemental Figure S2, A and B). This suggests that OOC-5 exists predominantly as monomers in solution, independent of nucleotide and redox state, when its hydrophobic membrane association sequence has been removed.
A Redox-sensitive Conformational Change Is Coupled to ADP Binding
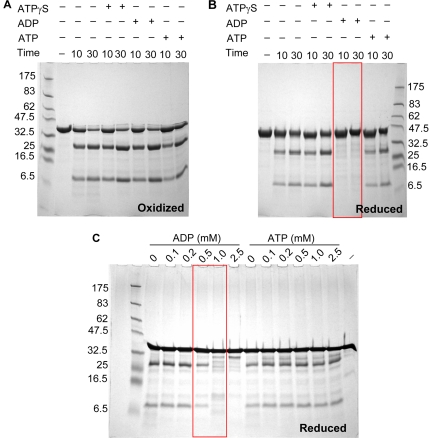
Limited proteolysis was used to probe the conformation of purified OOC-5 in the absence and in the presence of nucleotide and under oxidizing or reducing conditions. Under oxidizing conditions, but without nucleotide, two major fragments are produced by trypsin digestion, one corresponding to the large N-terminal AAA domain and the other the smaller C-terminal domain that contains the disease-causing GAG deletion in torsinA (Supplemental Figure S1B). Similar results are obtained with chymotrypsin (Supplemental Figure S1C). ESI-MS and Edman degradation analyses demonstrate that the proteolytic sites for trypsin and chymotrypsin are located at the unstructured loop between amino acids 251-265 of OOC-5 (Supplemental Figure S1, B–D), which is consistent with predicted structure (Figure 1A). In addition, the presence of nucleotide, such as ATP, ADP, or ATPγS, under the same conditions does not alter the digestion pattern of OOC-5 (Figure 5A). Interestingly, under reducing conditions, accessibility to the cleavage site of OOC-5 is ADP dependent (Figure 5B). Thus, the nucleotide-binding site and the redox status of the conserved cysteine residues are coupled by a local conformational change that is reflected in proteolytic cleavage. By contrast, neither ATP nor ATPγS has significant effect on the conformation of OOC-5 (Figure 5B). OOC-5 was titrated with ADP under reducing condition and probed by limited proteolysis, with ATP as a negative control. As shown in Figure 5C, the coupling of ADP binding and redox sensing only occurs at very high concentration of ADP, i.e., 1 mM, higher than the Kd calculated by temperature melt (Figure 3D).
Figure 5.
The conformational change of 44OOC-5 is dependent on ADP only under reducing condition. Limited proteolysis of 44OOC-5 wild type by trypsin in the presence of nucleotides and under different redox conditions. Protein was first incubated in 20 mM Tris-HCl, 200 mM NaCl, pH 8.5, 4% glycerol with nucleotide (2.5 mM with 5 mM MgCl2) in oxidized (A) or reduced buffer with 2 mM TCEP (B) for 30 min at 20°C and then digested by trypsin at 20°C for 10 and 30 min. The digested samples were resolved by a reducing 10% Tris-Tricine SDS-PAGE and Coomassie blue stain. (C) Nucleotides ADP and ATP were titrated in reduced buffer with 2 mM TCEP for wild-type 44OOC-5 before trypsin digestion.
Torsin proteins localize in the lumen of the ER where the redox potential is around −170 to −185 mV, significantly more oxidizing than the cytoplasm with a redox potential about −240 mV (Hwang et al., 1992; Moriarty-Craige and Jones, 2004). Thus, it is reasonable to propose that torsin proteins may be regulated by the redox state of ER or involved in the redox regulation of other ER proteins. The sensitivity of the RRSII would be expected to occur in this range. The biological redox buffer, GSH and GSSG, was used to determine the equilibrium redox potential of OOC-5. Using protease susceptibility to monitor conformational changes, the equilibrium redox potential of OOC-5 was calculated to be −210 mV in the absence of nucleotide (Figure 6A). This value is more reduced than the redox potential of protein disulfide isomerase (−175 mV; Lundstrom and Holmgren, 1993; Gilbert, 1995), which is another ER-resident protein. However, when ADP was present, the trypsin digestion pattern showed that the redox potential changes from −210 mV to less than −240 mV (Figure 6B), which is the redox potential of GSH itself (Gilbert, 1995). This result indicates the conformational change of OOC-5 is altered by both redox potential and ADP binding in a physiologically reasonable range. In this regard the ADP dependent conformational change of OOC-5 at the redox potential of −217 mV (Figure 6A) occurs at an ADP concentration of 1 mM and higher (Figure 6C), which is very close to the affinity measured by limited proteolysis under complete reducing condition in the presence of TCEP (Figure 5C). At the redox potential of the cytoplasm, i.e., −240 mV, the coupling of ADP binding and redox sensing also occur at the same concentration of ADP of 1 mM (Figure 6D), suggesting the coupling is dependent on the reduced condition of the critical OOC-5 cysteines.
Figure 6.
The effect of nucleotide on the equilibrium redox potential of 44OOC-5. The redox potential of 44OOC-5 in the absence (A) or in the presence of ADP (B) was determined by trypsin proteolysis. Aliquots of 44OOC-5 were equilibrated in buffers containing defined ratios of reduced-to-oxidized glutathione or treated with β-mercaptoethanol in 20 mM Tris-HCl, 200 mM NaCl, pH 8.5, with or without 2.5 mM ADP and 5 mM MgCl2 at 20°C before the addition of trypsin. The digested samples were resolved by reducing 10% Tris-Tricine SDS-PAGE. Using the Nernst equation, the equilibrium redox potential of 44OOC-5 in the absence of nucleotide was calculated to be −210 mV. The concentrations of GSH and GSSG calculated by weight, shown in B, are typically 2–3 mV less negative than the values measured spectrophotometrically as shown in A. (C and D) Nucleotides ADP and ATP were titrated in redox buffer at −217 mV (10 mM GSH, 0.6 mM GSSG) and at −240 mV (10 mM GSH, 0.1 mM GSSG) for wild-type 44OOC-5 before trypsin digestion.
To determine which cysteines are redox sensitive, disulfide mapping experiments were performed by carboxymethylating under specific redox potentials followed by Nano-LC-MS/MS. At a redox potential of −217 mV, when the conformational change of OOC-5 is coupled to ADP binding (Figure 6C), cysteines at 287 and 329 are reduced and carboxymethylated by iodoacetic acid detected by MS/MS (Table 1B). This indicates that the disulfide formed between Cys 287 and Cys 329 is reduced at redox conditions where the functional activity of nucleotide binding changes. Protection of Cys 51 is also lost. By contrast, Cys 170 is still oxidized at −217 mV and forms disulfide with either Cys 56 or Cys 57 (Table 1B).
Disruption of the Critical Disulfide Adjacent to Sensor-II Interferes with OOC-5 Function in C. elegans
Cysteine 329 in the Sensor-II motif was demonstrated to be a part of a disulfide by in vitro experiments, and the redox state of this cysteine was shown to be coupled to nucleotide binding. To test whether this RRSII was required for the biological function of OOC-5 in vivo, an ooc-5 transgene bearing mutations of cysteines 287 and 329 (ooc-5(C287S,C329S)) was introduced into C. elegans to examine its ability to rescue the maternal effect embryonic lethal phenotype of ooc-5(it145) mutants (Basham and Rose, 1999). Two independent transgenic lines were generated for the mutant ooc-5(C287S,C329S) and the wild-type ooc-5(+) control. Homozygous ooc-5(it145) worms carrying no transgene produce <2% hatching embryos (Table 3), whereas wild-type ooc-5(+) control lines have 20–72% embryo hatching (Table 3), consistent with previous rescue experiments (Basham and Rose, 2001). By contrast, the mutant ooc-5(C287S,C329S) lines only show 3–5% hatching embryos (Table 3), indicating that disruption of the RRSII interferes with the function of OOC-5. Therefore, the redox sensing cysteine 329 in the RRSII of OOC-5 is essential for OOC-5 function during oocyte or early embryo development in C. elegans (Basham and Rose, 1999, 2001). This is consistent with the above in vitro experimental results and suggests that OOC-5 acts through coupling redox sensing to nucleotide binding.
DISCUSSION
The torsin protein family represents a novel group of AAA+ proteins, which reside in the ER and the NE and are predicted to contain a conserved cysteine residue in the nucleotide Sensor-II motif in their C-terminal region. The representative protein in this family, torsinA, is mutated in human patients suffering from severe hereditary dystonia. Understanding the structure and function of torsin family proteins is necessary to elucidate the molecular pathology of the disease. OOC-5 from C. elegans is the only member with an established biological function. It shares 40% sequence identity with human torsinA and plays an essential role in nematode oogenesis and embryonic development. Utilizing OOC-5 as a model protein, our results demonstrate that cysteine 329 in Sensor-II is part of a critical, conserved disulfide bond. Disruption of this disulfide has no impact on the gross tertiary or secondary structure or oligomeric state. The redox potential of OOC-5 in the absence of nucleotide was determined as −210 mV, which is between that of disulfide reductant thioredoxin (−270 mV) and protein disulfide isomerase (−175 mV; Lundstrom and Holmgren, 1993; Gilbert, 1995). Although the enzyme is likely never nucleotide free in cells and the ADP concentration in the lumen of the ER is uncertain, it can be expected that high ADP levels are only achieved under conditions of significant metabolic stress. Notably, the redox potential became more reduced if ADP was added to the system, indicating that the nucleotide binding state is coupled to the redox status of OOC-5. Thus, we propose that the Sensor-II in the torsin protein family is a RRSII, which provides a potential regulatory function of the OOC-5 and torsin proteins.
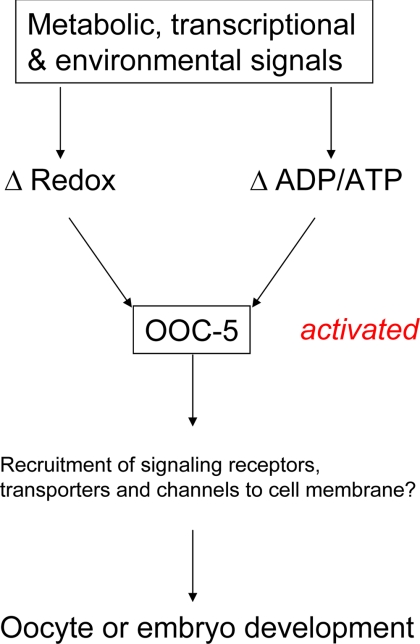
From the results in transgenic worms, the mutation of two cysteine residues in the C-terminus of OOC-5 leads to very low embryo hatch rate. Thus, the oxidization of the cysteine in RRSII is critical for the function of OOC-5. On the basis of these results, we hypothesize that OOC-5 functions during oocyte or early embryo development as an integrator of redox and nucleotide concentrations, two key parameters reflecting the metabolic status of the cell (Figure 7).
Figure 7.
A hypothetical model for OOC-5 as an integrator of redox status and nucleotide state during oocyte or embryo development. Metabolic, transcriptional, and environmental signals alter the redox status and the nucleotide concentrations inside the cells, which are predicted to change the redox state and nucleotide binding affinity of OOC-5. This coupled change of redox status and nucleotide binding would then activate OOC-5, leading to the recruitment of the substrates necessary for oocyte or embryo development. These substrates could be receptors, transporters, or channels as proposed previously (Torres et al., 2004).
Domain Boundaries in Torsin Family Proteins
The structural hallmark of the AAA+ class of proteins is a module composed of two domains: a large domain composed of five parallel β-strands decorated by helices of varying length and a small domain based on a three-helix bundle, with possible extensions to additional domains (Neuwald et al., 1999). The alignment of Figure 1A, constructed using profile alignment methods that are generally more sensitive and accurate than the more commonly used pairwise alignment methods (such as BLAST), suggested a likely position of the domain boundary that is in contrast to earlier predictions (Neuwald et al., 1999). Although a recently published alignment of torsinA with the second AAA+ domain of T. thermophilus ClpB showed the similar prediction of the domain boundary in human torsinA, they did not test this conclusion experimentally (Kock et al., 2006). Our alignment was supported by six pieces of evidence. First, sequence comparisons of the ClpA profile with the torsinA profile showed that a conserved PFXPL motif in the torsins spanning the domain boundary was also conserved to a lesser extent in the ClpA conserved domain database profile (data not shown). Second, the consensus secondary structure predictions placed a strand at this position (Figure 1A, brown arrow), which marks the end of the large domain in homologous AAA+ structures. Third, the predicted helix following this strand (Figure 1A, yellow cylinders), matched the helix of the ClpA structure well in both start position and hydrophobicity pattern. (Although the length of the predicted helix in the torsins was shorter than the helix found in ClpA, the homologous structure of the patch clamp loader, PDB 1iqpA [Oyama et al., 2001], also exhibits a shortened first helix at this location, data not shown.) Fourth, the secondary structure derived from the ROSETTA tertiary structure predictions (Figure 1A) were consistent with a helix starting at this position when the sequence of the small domain was considered in isolation. Fifth, sites susceptible to proteolysis were identified in the predicted linker (Figure 1A and Supplemental Figure S1). Finally, the model predicted specific disulfide bonds that were experimentally verified (Figure 2).
Oligomerization and ATPase Activity of Torsin Family Proteins
Purified OOC-5 eluted as single symmetric peak from gel filtration chromatography, corresponding to a monomeric species. This result is similar to those of Δ20aa torsinA, which existed primarily as monomers in the presence of ATP or ADP (Kustedjo et al., 2003). In most cases, oligomerization of AAA+ proteins is ATP dependent, especially for Clp/Hsp100 family members that contain two AAA+ domains within a polypeptide chain (Vale, 2000). However, another AAA ATPase containing a single AAA domain as torsin proteins, Vps4p from baker's yeast, can exist as a dimer in the absence of ATP or in the presence of ADP (Babst et al., 1998). Our results do not rule out the possibility that torsinA or OOC-5 oligomerize transiently or stably at a high effective concentration when associated with ER membranes.
Using either a coupled enzymatic assay or a malachite green colorimetric assay, ATPase activity of purified OOC-5 is undetectable, regardless of redox condition (data not shown). Published data of purified human torsinA showed that Δ20aa torsinA hydrolyzed ATP with a Vmax of 0.11 ± 0.01 nmol min−1 μg−1 and Km of 1.1 ± 0.2 mM (Kustedjo et al., 2003); Maltose-binding protein (MBP)-torsinA with a 52-amino acid N-terminus deletion has a reported specific activity of 20 pmol min−1 μg−1 (Pham et al., 2006), both of which were detected by [α-32P]ATP and polyethyleneimine-cellulose thin-layer chromatography. His-NusA-His-S-tagged torsinA purified from E. coli inclusion bodies catalyzed ATP with a Vmax of 70 nmol Pi min−1 mg−1 and Km of 15 μM using Pi assay (Konakova and Pulst, 2005). Noticeably, these three reports are not consistent with each other and no negative control, such as the E171Q mutant in which the catalytic glutamate is mutated to glutamine, was tested to confirm ATP hydrolysis.
There are at least three significant differences in conserved motifs between torsin proteins, including OOC-5, and other AAA+ proteins. First, the Walker A motif (GxxGS/TGKN) lacks the serine/threonine after lysine 116 (Lys 108 in human torsinA) which typically interacts with the Mg2+ ion in most AAA+ ATPases (Pye et al., 2006). Second, no arginine-finger is found in OOC-5 and human torsinA, which is also believed necessary to coordinate nucleotide hydrolysis (Pye et al., 2006). Finally, as mentioned earlier, the Sensor-II motif usually contains an arginine that interacts with the γ-phosphate of ATP (Erzberger and Berger, 2006). The sequence of this motif is GAR in E. coli ClpA and ClpB, but is GCK in OOC-5 and torsinA (Kock et al., 2006), in which the Cys participates in the redox sensitive disulfide in the C-terminus of torsinA/OOC-5. The above three differences in torsin family proteins may explain the lack of ATPase activity detected in our purified OOC-5 preparation. However, we cannot rule out that torsinA/OOC-5 is an ATPase. Most AAA+ proteins function as an ATPase when assembled into oligomers and are stimulated by the presence of substrate, e.g., ClpB by α-casein (Mogk et al., 2003) and/or by cofactors, e.g., α-SNAP stimulates ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF; Steel and Morgan, 1998). Purified OOC-5 is a monomer and its substrate(s) is unknown. Therefore, the basal ATPase activity of torsinA/OOC-5 may be stimulated by their unknown substrates or regulated by their unknown partners.
Redox Regulation on the Potential Substrates for Torsin Family Proteins
The redox potential of OOC-5 in the absence of nucleotide is very close to that of the lumen of the ER under normal conditions, which places it in a position to respond to the changes in ER redox state and supports a role for OOC-5 as a redox regulating protein inside the lumen of the ER. More interestingly, the nucleotide-binding state of OOC-5 is coupled to its redox status. There are two possibilities for the redox regulation of OOC-5. One possibility is the ATPase activity and/or substrate binding of OOC-5 and, thus, torsin proteins is regulated by the redox state of the ER, consistent with their proposed role as ER chaperone proteins involved in processing membrane proteins through the secretory pathway (Torres et al., 2004) and putative involvement in the ER stress response (Hewett et al., 2007). In this regard, the ER is known to mount profound responses to reducing conditions (Tu et al., 2000). In addition, local heterogeneities in redox potential in the ER remain an unexplored possibility. It is reasonable to assume that the redox state of the ER can change to more reducing conditions either locally or under some stress conditions where the nucleotide binding state of OOC-5 is coupled to its redox status. Another possibility is that redox changes between the ER and the NE drive the torsin family proteins to move within the contiguous compartment of the ER and NE and interact with different substrates in different locations within this compartment (Goodchild and Dauer, 2005). In addition, it is possible that the torsin family proteins, like protein disulfide isomerase (Park et al., 2006), may be directly involved in the redox regulation of other ER or secretory proteins for subsequent processes.
The low embryonic hatch rate in transgenic worms containing the mutation of two cysteine residues in the C-terminus of OOC-5 indicates the oxidization of the cysteine in RRSII in the C-terminus of OOC-5 is critical for oocyte or early embryo development. At present, the downstream targets of this signaling, the physiological substrates of OOC-5, remain to be identified. OOC-3 is a potential target of OOC-5, because it is required for the ER localization of OOC-5 in all cells of the early embryo (Basham and Rose, 2001). In addition, OOC-3 is a novel putative transmembrane protein localizing to the ER, the NE and cell–cell boundaries, and it is required for the same aspects of embryonic development as OOC-5 (Basham and Rose, 1999, 2001; Pichler et al., 2000). Lamina-associated polypeptide 1 (LAP1) and the homologous lumenal domain like LAP1 (LULL1) are the binding partners of torsinA in mammalian cells (Goodchild and Dauer, 2005). The interaction between torsinA and LAP1 or LULL1 is significantly diminished when the two cysteines in the C-terminus of torsinA but not the cysteines in the N-terminal domain are mutated to serines (L. Zhu and P. J. Thomas, unpublished data), consistent with the results above with OOC-5 in transgenic worms. Other suggested potential targets for OOC-5 and torsin family proteins could be secreted membrane receptors, transporters or ion channels (Torres et al., 2004).
Regarding the human torsinA and its dystonia causing mutation, the sequence alignment places the Glu affected by the dystonia-associated GAG deletion at the end of the second helix in the C-terminal domain (arrow, Figure 1, A and B). This position suggests that deletion of glutamate may alter the helical register and the ability to form the critical disulfide thereby disrupting the coupling of redox status and nucleotide binding and leading to the loss of the function of torsinA on its substrates.
Anatomical analysis in DYT1 dystonia patients suggested there is not structural damage but rather cellular dysfunction in the neurons. Dopaminergic neurons might be particularly vulnerable to dysfunction of chaperones because they are vulnerable to oxidative stress (Walker and Shashidharan, 2003). Published data indicate torsinA functions in response to oxidative stresses but not other ER stresses (Hewett et al., 2003), and overexpression of torsinA protects against oxidative stress in COS-1 and PC12 cells (Kuner et al., 2003; Shashidharan et al., 2004). Recently, torsinA was found to modulate cellular levels of the dopamine transporter (DAT) (Torres et al., 2004) and may mediate protection from α-synuclein–induced neurodegeneration in dopaminergic neurons through down-regulation of DAT-1 (Cao et al., 2005). Therefore, human torsinA might function as a redox-dependent molecular chaperone in the neurons in a manner similar to that proposed for DJ-1 which inhibits α-synuclein aggregation (Shendelman et al., 2004). This hypothesis needs to be tested by investigation with suitable substrates at a variety of redox conditions.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank G. N. DeMartino, H. F. Gilbert, and members of the Thomas lab for helpful discussions and the Protein Chemistry Technology Center at University of Texas Southwestern Medical Center at Dallas for their Mass Spectrometry service. This work was supported by grants from the Dystonia Medical Research Foundation to L.Z. and P.J.T. and by grants from the National Institutes of Health to P.J.T. and L.S.R.
Abbreviations used:
- AAA
ATPases associated with cellular activities
- CD
circular dichroism
- DTNB
(5,5′-dithiobis-(2-nitrobenzoic acid)
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ESI
electrospray ionization
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- IAA
iodoacetate
- IAM
iodoacetamide
- LC
liquid chromatography
- MS
mass spectrometry
- NE
nuclear envelope
- RRS
redox-regulated sensor
- TCEP
tris(2-carboxyethyl) phosphine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0015) on June 11, 2008.
REFERENCES
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslund F., Berndt K. D., Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham S. E., Rose L. S. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev. Biol. 1999;215:253–263. doi: 10.1006/dbio.1999.9447. [DOI] [PubMed] [Google Scholar]
- Basham S. E., Rose L. S. The Caenorhabditis elegans polarity gene ooc-5 encodes a Torsin-related protein of the AAA ATPase superfamily. Development. 2001;128:4645–4656. doi: 10.1242/dev.128.22.4645. [DOI] [PubMed] [Google Scholar]
- Berman M. H., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M., Hartmann C., Song H. K., Bourenkov G. P., Bartunik H. D., Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bonneau R., Tsai J., Ruczinski I., Chivian D., Rohl C., Strauss C. E., Baker D. Rosetta in CASP4, progress in ab initio structure prediction. Proteins Suppl. 2001;5:119–126. doi: 10.1002/prot.1170. [DOI] [PubMed] [Google Scholar]
- Breakefield X. O., Kamm C., Hanson P. I. TorsinA: movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/s0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan A. C., Bunning S., Jones O. T., High S., Swanton E. Biosynthesis of the dystonia-associated AAA+ ATPase torsinA at the endoplasmic reticulum. Biochem. J. 2007;401:607–612. doi: 10.1042/BJ20061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Gelwix C. C., Caldwell K. A., Caldwell G. A. Torsin-medicated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J. Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bond formation in proteins. Methods Enzymol. 1984;107:305–329. doi: 10.1016/0076-6879(84)07021-x. [DOI] [PubMed] [Google Scholar]
- Cuff J. A., Clamp M. E., Siddiqui A. S., Finlay M., Barton G. J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Erzberger J. P., Berger J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Ferguson A. D., Labunskyy V. M., Fomenko D. E., Arac D., Chelliah Y., Amezcua C. A., Rizo J., Gladyshev V. N., Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J. Biol. Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- Gilbert H. F. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- Goodchild R. E., Dauer W. T. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild R. E., Dauer W. T. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild R. E., Kim C. E., Dauer W. T. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Guo F., Maurizi M. R., Esser L., Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hewett J., et al. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J. Neurosci. Res. 2003;72:158–168. doi: 10.1002/jnr.10567. [DOI] [PubMed] [Google Scholar]
- Hewett J. W., Tannous B., Niland B. P., Nery F. C., Zeng J., Li Y., Breakefield X. O. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc. Natl. Acad. Sci. USA. 2007;104:7271–7276. doi: 10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C., Sinskey A. J., Lodish H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPase. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Kabakci K., et al. Mutations in DYT1, extension of the phenotypic and mutational spectrum. Neurology. 2004;62:395–400. doi: 10.1212/01.wnl.0000113024.84178.f7. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Kim K. K. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kock N., Naismith T. V., Boston H. E., Ozelius L. J., Corey D. P., Breakefield X. O., Hanson P. I. Effects of genetic variations in the dystonia protein torsinA: identification of polymorphism at residue 216 as protein modifier. Hum. Mol. Genet. 2006;15:1355–1364. doi: 10.1093/hmg/ddl055. [DOI] [PubMed] [Google Scholar]
- Konakova M., Pulst S. M. Dystonia-Associated forms of TorsinA are deficient in ATPase activity. J. Mol. Neurosci. 2005;25:105–118. doi: 10.1385/JMN:25:1:105. [DOI] [PubMed] [Google Scholar]
- Kuner R., Teismann P., Trutzel A., Naim J., Richter A., Schmidt N., von Ahsen O., Bach A., Ferger B., Schneider A. TorsinA protects against oxidative stress in COS-1 and PC12 cells. Neurosci. Lett. 2003;350:153–156. doi: 10.1016/s0304-3940(03)00904-2. [DOI] [PubMed] [Google Scholar]
- Kustedjo K., Deechongkit S., Kelly J. W., Cravatt B. F. Recombinant expression, purification, and comparative characterization of torsinA and its torsion dystonia-associated variant Delta E-torsinA. Biochemistry. 2003;42:15333–15341. doi: 10.1021/bi0349569. [DOI] [PubMed] [Google Scholar]
- Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., Tsai F. T. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zolkiewska A., Zolkiewski M. Characterization of human torsinA and its dystonia-associated mutant form. Biochem. J. 2003;374:117–122. doi: 10.1042/BJ20030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom J., Holmgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi M. R., Xia D. Protein binding and disruption by Clp/Hsp100 chaperones. Structure. 2004;12:175–183. doi: 10.1016/j.str.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Mogk A., Schlieker C., Strub C., Rist W., Weibezahn J., Bukau B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- Moriarty-Craige S. E., Jones D. P. Extracellular thiols and thiol/disulfide redox in metabolism. Annu. Rev. Nutr. 2004;24:481–509. doi: 10.1146/annurev.nutr.24.012003.132208. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Lima C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Naismith T. V., Heuser J. E., Breakefield X. O., Hanson P. I. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Oyama T., Ishino Y., Cann I. K., Ishino S., Morikawa K. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell. 2001;8:455–463. doi: 10.1016/s1097-2765(01)00328-8. [DOI] [PubMed] [Google Scholar]
- Ozelius L. J., et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Ozelius L. J., et al. The TOR1A (DYT1) gene family and its role in early-onset torsion dystonia. Genomics. 1999;62:377–384. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- Park B., Lee S., Kim E., Cho K., Riddell S. R., Cho S., Ahn K. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell. 2006;127:369–382. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Pham P., Frei K. P., Woo W., Truong D. D. Molecular defects of the dystonia-causing torsinA mutation. Neuroreport. 2006;17:1725–1728. doi: 10.1097/WNR.0b013e3280101220. [DOI] [PubMed] [Google Scholar]
- Pichler S., Gonczy P., Schnabel H., Pozniakowski A., Ashford A., Schnabel R., Hyman A. A. OOC-3, a novel putative transmembrane protein required for establishment of cortical domains and spindle orientation in the P(1) blastomere of C. elegans embryos. Development. 2000;127:2063–2073. doi: 10.1242/dev.127.10.2063. [DOI] [PubMed] [Google Scholar]
- Pollastri G., Przybylski D., Rost B., Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Pye V. E., Dreveny I., Briggs L. C., Sands C., Beuron F., Zhang X., Freemont P. S. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Shashidharan P., Paris N., Sandu D., Karthikeyan L., McNaught K. S., Walker R. H., Olanow C. W. Overexpression of torsinA in PC12 cells protects against toxicity. J. Neurochem. 2004;88:1019–1025. doi: 10.1046/j.1471-4159.2003.02233.x. [DOI] [PubMed] [Google Scholar]
- Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclei aggragate formation. PLoS Biol. 2004;2:1764–1773. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M. C., Trame C. B., Tsuruta H., Wilbanks S. M., Reddy V. S., McKay D. B. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Steel G. J., Morgan A. Selective stimulation of the D1 ATPase domain of N-ethylmaleimide-sensitive fusion protein (NSF) by soluble NSF attachment proteins. FEBS Lett. 1998;423:113–116. doi: 10.1016/s0014-5793(98)00072-6. [DOI] [PubMed] [Google Scholar]
- Torres G. E., Sweeney A. L., Beaulieu J. M., Shashidharan P., Caron M. G. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc. Natl. Acad. Sci. USA. 2004;101:15650–15655. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B. P., Ho-Scheleyer S. C., Travers K. J., Weissman J. S. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- Walker R. H., Shashidharan P. Developments in the molecular biology of DYT1 dystonia. Mov. Disord. 2003;18:1102–1107. doi: 10.1002/mds.10549. [DOI] [PubMed] [Google Scholar]
- Weibezahn J., Schlieker C., Bukau B., Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- Vale R. D. AAA proteins. Lords of the ring. J. Cell Biol. 2000;150:13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander T., Phadke N. D., Bardwell J. C. Disulfide bond catalysts in Escherichia coli. Methods Enzymol. 1998;290:59–74. doi: 10.1016/s0076-6879(98)90007-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.