Abstract
Glucose-dependent regulation of carbon metabolism is a subject of intensive studies. We have previously shown that the switch from gluconeogenesis to glycolysis is associated with ubiquitin-proteasome linked elimination of the key enzyme fructose-1,6-bisphosphatase. Seven glucose induced degradation deficient (Gid)-proteins found previously in a genomic screen were shown to form a complex that binds FBPase. One of the subunits, Gid2/Rmd5, contains a degenerated RING finger domain. In an in vitro assay, heterologous expression of GST-Gid2 leads to polyubiquitination of proteins. In addition, we show that a mutation in the degenerated RING domain of Gid2/Rmd5 abolishes fructose-1,6-bisphosphatase polyubiquitination and elimination in vivo. Six Gid proteins are present in gluconeogenic cells. A seventh protein, Gid4/Vid24, occurs upon glucose addition to gluconeogenic cells and is afterwards eliminated. Forcing abnormal expression of Gid4/Vid24 in gluconeogenic cells leads to fructose-1,6-bisphosphatase degradation. This suggests that Gid4/Vid24 initiates fructose-1,6-bisphosphatase polyubiquitination by the Gid complex and its subsequent elimination by the proteasome. We also show that an additional gluconeogenic enzyme, phosphoenolpyruvate carboxykinase, is subject to Gid complex-dependent degradation. Our study uncovers a new type of ubiquitin ligase complex composed of novel subunits involved in carbohydrate metabolism and identifies Gid4/Vid24 as a major regulator of this E3.
INTRODUCTION
Glucose is the main carbon source of many cells, providing energy and building blocks for a variety of essential cellular components. Glucose also has a pivotal role in cellular regulation. Its consumption via glycolysis and its regeneration via gluconeogenesis are central pathways of carbohydrate metabolism in many organisms. Regulation of both pathways occurs at three steps catalyzed by different reciprocally acting enzymes. In glycolysis, these steps include the phosphorylation of glucose by hexokinase, the phosphorylation of fructose-6-phosphate by phosphofructokinase, and the synthesis of pyruvate and ATP from phosphenolpyruvate by pyruvate kinase. In gluconeogenesis, these steps are circumvented by pyruvate carboxylase and phosphoenolpyruvate carboxykinase (PEPCK), fructose-1,6-bisphosphatase (FBPase), and glucose-6-phosphatase. Dysregulation of these antagonistic pathways in humans leads to type 2 diabetes (Wahren and Ekberg, 2007).
In the yeast Saccharomyces cerevisiae, major regulation events occur when cells previously grown in a glucose-deprived medium are supplied with this sugar. These events include gene expression and repression, alterations in the stability of certain mRNAs, and posttranslational modification of many gene products (Gancedo, 1998; Vaulont et al., 2000). FBPase is expressed when yeast cells are growing in media without a fermentable carbon source (growth on ethanol [EtOH] or acetate). After glucose addition, catabolite inactivation and degradation of FBPase occur (Gancedo, 1971; Holzer, 1976; Marcus et al., 1988; Hämmerle et al., 1998; Wolf, 2004). Catabolite inactivation encompasses repression of the FBP1 gene, decrease of the enzyme activity upon phosphorylation, and allosteric inhibition by fructose-2,6-bisphosphate and AMP (Mazon et al., 1982; von Herrath and Holzer, 1988). Subsequently, FBPase undergoes rapid degradation (Holzer, 1989). Similar mechanisms are described for PEPCK, cytosolic malate dehydrogenase, and isocitrate lyase (Holzer, 1976; Hämmerle et al., 1998). Degradation of FBPase blocks gluconeogenesis and prevents an otherwise ongoing futile cycle of ATP hydrolysis (Schork et al., 1994a,b, 1995; Wolf, 2004).
In addition to the previously identified GID1/VID30, GID2/RMD5, and GID3 genes, a genome-wide screen for FBPase stabilization upon glucose shift allowed the discovery of six further so-called glucose induced degradation deficient (GID) genes termed GID4 to GID9 (Regelmann et al., 2003). Among those genes, GID3 and GID6 were shown to encode the ubiquitin-conjugating enzyme Ubc8 and the deubiquitinating enzyme Ubp14 (Schüle et al., 2000; Regelmann et al., 2003). Gid1, Gid4, and Gid5 had previously been identified as Vid30, Vid24, and Vid28, respectively. They were implicated in a glucose-induced vesicular targeting of FBPase for vacuolar degradation observed when yeast cells were starved >48 h in media containing acetate as a nonfermentable carbon source (Hoffman and Chiang, 1996; Chiang and Chiang, 1998; Hung et al., 2004). Proteasome-dependent degradation of FBPase was obtained using different conditions: yeast cells were grown overnight in ethanol-containing media before glucose addition (Schüle et al., 2000; Regelmann et al., 2003; Hung et al., 2004). Detailed characterization of Gid2/Rmd5 demonstrated that it is not present as a monomeric protein within the cell. In a glycerol step gradient, it sediments at ∼600 kDa, suggesting that it is part of a soluble protein complex called Gid complex (Regelmann et al., 2003). Proteomic interaction studies demonstrated that seven of the Gid proteins belong to one and the same complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006). A similar complex, constituted from protein homologues to the Gid proteins, has been found in mammals, in which one of its subunits has been shown to be involved in proteasome-dependent degradation of α-catenin (Kobayashi et al., 2007; Suzuki et al., 2008). Except for Gid3/Ubc8, which is not integral to the complex, and Gid2/Rmd5, no function in proteasome-dependent catabolite degradation of FBPase has been described for the newly discovered Gid proteins (Regelmann et al., 2003).
Here, we further characterize the contribution of Gid2/Rmd5 to proteasomal degradation of FBPase, and we show that it confers E3 activity to the Gid complex. In addition, we show that degradation of PEPCK is dependent on Gid2/Rmd5. Moreover, we uncover Gid4/Vid24 as a key regulator of FBPase degradation by the proteasome.
MATERIALS AND METHODS
Yeast Strains and Media
Previously described standard methods were used for media preparation and genetic and molecular biological techniques (Guthrie and Fink, 1991; Ausubel et al., 1992). The S. cerevisiae strains used in these studies are summarized in Supplemental Table 1. Unless otherwise stated, all yeast strains were grown at 30°C. Precultures were grown 16 h in YPD medium containing 2% glucose, diluted 1:12.5 into YPD, and grown for additional 6–7 h. Thereafter, cells were resuspended in YPethanol (2%) and grown for 16 h to allow FBPase synthesis. For induction of FBPase degradation, cells were shifted onto YPD medium containing 2% glucose.
Construction of Epitope-tagged Gid Proteins and Plasmids
Gid1/Vid30, Gid5/Vid28, Gid6/Ubp14, Gid7, Gid8, and Gid9/Fyv10 were C-terminally labeled with a 3xHA-tag. Strain YJR12 (GID1-HA3) was constructed by chromosomal integration of a 1.6-kb polymerase chain reaction (PCR) fragment consisting of a 3xHA-tag and a Schizosaccharomyces pombe HIS5 marker (Cottarel, 1995) into W303-1B cells. The PCR fragment was generated using oligonucleotides YJR12fwd, YJR12rev and plasmid p3xHA-HIS5 (S. Munro, Cambridge, United Kingdom) as template. Strains YTP10 (GID7-HA3), YTP11 (GID5-HA3), YTP12 (GID9-HA3), YSA1 (GID8-HA3), and YBB1 (GID6-HA3) were generated by chromosomal integration of a 1.75-kb PCR fragment. Plasmid pFA6a-3HA-His3MX6 was described previously (Longtine et al., 1998). Gid4/Vid24 was N-terminally Myc9 tagged according to Gauss et al. (2005). For URA3 marker rescue with pSH63, instead of 1% raffinose and 1% galactose, the medium contained 2% galactose. The integration cassette was amplified from pOM22 (Gauss et al., 2005) by using primer YOS1fwd and YOS1rev and transformed into strain W303-1B. Correct integration was confirmed by Southern blot analysis or PCR. GID2-HA3 construction is described in Regelmann et al. (2003). The pOS2 plasmid was constructed by insertion of a Myc9-GID4 fragment in a StuI/SbfI-digested pCM184 plasmid (Euroscarf, Frankfurt, Germany). All oligonucleotides used are listed in Supplemental Table 2. The construction of FBPase C-terminal TAP fusion was conducted as described previously based on the homologous recombination of a PCR product at a specific gene locus on the chromosome (Puig et al., 2001). The plasmid expressing the FBPase-TAP fusion protein was obtained using a gap-repair strategy. Plasmid pRG6 (de la Guerra et al., 1988), which contains the FBP1 gene and its genomic flanking regions was digested with NcoI. The plasmid, lacking the 800 base pair NcoI fragment was then transformed in a yeast strain expressing a chromosomally C-terminally tagged FBPase. Cells able to survive on complete minimal (CM) media lacking uracil were selected, plasmid rescue was performed, and the obtained plasmids were analyzed for the presence of an FBPase-TAP coding sequence, under the native FBPase promoter.
To construct the plasmid pFBPase, a genomic fragment encompassing the FBP1 gene together with 1000 base pairs of its upstream and 200 base pairs of its downstream sequences was amplified by PCR with primers pFBPase-fwd and pFBPase-rev (Supplemental Table 2) and inserted into a SpeI/ClaI-digested pRS316 plasmid (Sikorski and Hieter, 1989). The resulting plasmid was verified by enzymatic restriction and sequencing. The plasmid-expressed FBPase is functional and undergoes degradation as the chromosomally expressed enzyme.
Mutation of the Degenerated RING Domain of GID2/RMD5
A point mutation in the conserved Cys residue 379 of Gid2/Rmd5 was performed using the Transformer site-directed mutagenesis kit (Clontech, Mountain View, CA). The template plasmid was generated by digesting a YCP50 plasmid (Rose et al., 1987) using BamHI and SalI. The resulting fragment, bearing the GID2/RMD5 ORF with its endogenous promoter and terminator regions was inserted in pRS316. Oligonucleotides are listed in Supplemental Table 2. The mutated GID2/RMD5 was integrated in pRS306 digested with BamHI and SalI. Genomic integration was carried out by transforming the resulting plasmid in YTS3 yeast cells. Chromosomal DNA of colonies that lost the ability to grow on 5-fluorouracil containing medium was extracted, and the GID2/RMD5 gene was sequenced.
Western Blotting
Western blotting was performed as described in Schork et al. (1995). Extracts were prepared via alkaline lysis (Yaffe and Schatz, 1984) and finally resuspended in urea buffer (200 mM Tris-HCl pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, 1% 2-mercaptoethanol, and 0.05% bromphenol blue). We used 3 OD600 of cells for each sample. Antibodies used were obtained from BAbCO (Richmond, CA) (hemagglutinin [HA], clone 16B12) and Calbiochem (San Diego, CA) (Myc, clone 9E10); FBPase polyclonal antibody was obtained from K. D. Entian (Goethe Universität, Frankfurt, Germany) or was produced by rabbit immunization using a purified FBPase-glutathione transferase (GST).
Immunoprecipitation
For immunoprecipitations (IPs) cells were cultivated as described above for FBPase turnover assays and samples were withdrawn at the indicated time points. Cells (30 OD600) were harvested, washed with water, and resuspended in 600 μl of phosphate-buffered saline (PBS) buffer pH 7.4 (137 mM NaCl, 1.25g/l Na2HPO4, and 0.35g/l NaH2PO4) containing protease inhibitors (Complete; Roche Diagnostics, Mannheim, Germany; 1.1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 μg/ml each of antipain, pepstatin A, chymostatin, and leupeptin) and lysed at 4°C with glass beads for 20 min. After centrifugation, 500 μl of the supernatant was transferred to a new test tube. FBPase antibody was added, and the samples were gently agitated end over end for 2 h at 4°C. Immunoprecipitates were collected by adding 50 μl of 5% (wt/vol) protein A-Sepharose CL-4B (GE Healthcare, Little Chalfont, United Kingdom) and further incubated for 1.5 h. For IP, the Sepharose beads were centrifuged and washed five times with ice-cold PBS buffer. Proteins were released from Sepharose by boiling in 50 μl of urea buffer.
Glycerol Density Gradient Fractionation
Yeast cells were grown for 16 h in YPethanol. For analysis in gluconeogenic cells, 50 OD600 of cells were harvested; for analysis in glycolytic cells, cells were shifted on YPD for 20 min before 50 OD600 of cells were harvested. Cells were resuspended in 600 μl of 0.1 M KH2PO4, pH 7.0, in the presence of Complete, 1.1 mM PMSF, and protease inhibitors; lysed with glass beads (20 min; 4°C); and centrifuged at 10,000 × g for 15 min. Then, 200 μl aliquots of the resulting cell extracts were layered on top of a glycerol step gradient [450 μl each of 50%, 40%, 30%, 20 and 10% of glycerol in 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) buffer pH 6.8] and centrifuged at 55,000 rpm and 15°C for 4 h in a TLS-55 rotor. Thereafter, 200 μl fractions were collected, precipitated with trichloroacetic acid (10%), solubilized in urea buffer, and analyzed by Western blotting.
Polyubiquitination of FBPase
Polyubiquitination was assessed either by growing the cells containing the FBPase-tandem affinity purification (TAP)-tag encoding plasmid on CM medium without uracil, 2% glucose or growing the GID2-HA3 and GID2C379S-HA3 strains on YPD to OD600 3–4. After harvesting (500 × g; 5 min), cells were resuspended in the same medium containing 2% ethanol and left to grow for 6 h to allow derepression of FBPase. Then, 50 OD600 of yeast were harvested before and 25 min after addition of 2% glucose. After washing in 1 ml of water containing 20 mM N-ethylmaleimide, 20 mM NaN3, and 1 mM PMSF, cells were pelleted at 500 × g for 4 min at 4°C and resuspended in 600 μl of ice-cold PBS buffer containing protease inhibitors and lysed at 4°C with glass beads (300 μl; 0.4–0.6 mm in diameter) during 20 min. IP of FBPase was performed as described above, and the pellet was washed five times with PBS buffer containing 0.2% Triton X-100. Alternatively, FBPase-TAP was pulled down using 80 μl of 50% (vol/vol) immunoglobulin G (IgG)-Sepharose beads. After 3 h of incubation at room temperature, beads were washed four times with PBS added with 150 mM NaCl and 1% (vol/vol) Triton X-100. In both cases, beads were resuspended in 50 μl of urea buffer, boiled for 5 min at 95°C and used for immunoblotting with monoclonal anti-ubiquitin antibody (clone P4G7; Covance Research Products, Princeton, NJ).
In Vitro Polyubiquitination Assay
A GST-fusion protein of Gid2/Rmd5, and, as a positive control, the mammalian RING protein gp78 (309–643-amino acid fragment) were expressed in the Escherichia coli BL21 strain. Bacteria were grown to OD600 0.8–1 in 2× yeast tryptone containing 100 μg/ml ampicillin, and expression of the extrinsic protein was induced at 16°C with 0.5 mM isopropyl β-d-thiogalactoside during 6–12 h. Cells were harvested, resuspended in buffer A (50 mM Tris–HCl, pH 7.5, 250 mM NaCl, 5 mM dithiothreitol (DTT), 2 mM PMSF, and 1% Triton-X 100) and lysed with a French press. A typical 20 μl ubiquitination reaction was carried out by adding 0.25 μg of E1 (yeast), 0.6 μg of UbcH5b, 10 μg of HA-ubiquitin, 1 μl of energy regeneration solution (all BostonBiochem, Cambridge, MA), 2 μl of 10× ubiquitin reaction buffer (500 mM Tris-HCl, pH 7.5, 500 mM NaCl, 100 mM MgCl2, 10 mM DTT, and 250 μM ZnCl2), and 8 μl of Gid2/Rmd5 or gp78 cell lysate. The reactions were incubated at 30°C for 2 h, and ubiquitination of proteins was monitored by Western blotting using monoclonal anti-HA antibody.
Gid4/Vid24 Expression in Gluconeogenic Cells
gid4Δ/vid24Δ yeast cells were transformed with the pOS2 plasmid encoding Gid4/Vid24 under control of the TetR promoter or the corresponding control vector. After 16 h of growth on CM-Trp media containing 2% glucose and 40 μg/ml doxycycline, cells were transferred into the same medium lacking cysteine and methionine for 16-h growth. Five OD600 of yeast were pelleted washed once with distilled water and resuspended in 2% EtOH-CM-Trp medium without cysteine and methionine, but containing 40 μl/ml doxycycline. After 3 h of incubation; radiolabeling was achieved by addition of 9.25 MBq of [35S]methionine and a further 2 h of incubation time (pulse). Consecutively, cells were centrifuged, washed with water and resuspended in 6.5 ml of CM-Trp-EtOH (2%) containing 10 mM nonradioactive methionine (chase). Samples were harvested every hour during 3 h. After FBPase immunoprecipitation and 10% SDS-polyacrylamide gel electrophoresis, gels were overlaid with Phosphor screen and scanned with Storm 860 (GE Healthcare) for protein quantification.
Degradation of Gid4/Vid24
To monitor Gid4/Vid24 degradation, cells were grown as described above for FBPase turnover assays. After 16 h of growth on YPethanol (2%); cells were shifted onto YPD for 30 min to induce GID4/VID24 expression. Cells were then shifted onto fresh YPD with 100 μg/ml cycloheximide. For the proteasome inhibition experiment, along with cycloheximide treatment, 60 μM MG-132 were added 30 min ahead of harvesting. This treatment was repeated every 30 min to ensure a proper proteasome inhibition. To monitor Gid4/Vid24 stability; 1.5 OD600 of cells were taken at each time point and analyzed by immunoblotting.
Sequence Analysis
Sequence database searches were carried out with a nonredundant data set assembled from current releases of Uniprot and GenPept (Benton, 1990; Bairoch and Apweiler, 1997). Multiple alignments were calculated by MUSCLE (Edgar, 2004), using excised domains instead of the entire protein sequences. Generalized profile construction (Bucher et al., 1996), and searches were run locally using the pftools package, version 2.1. Generalized profiles were constructed using the BLOSUM45 substitution matrix (Henikoff and Henikoff, 1992) and default penalties of 2.1 for gap opening and 0.2 for gap extension. Profile matches were analyzed for statistical significance by means of the score distribution of a randomized database (Hofmann, 2000). Only sequence matches that were detected with a probability p lower than 0.01 were included in subsequent rounds of iterative profile refinement.
RESULTS
Expression Profiles and Sedimentation Pattern of Gid Proteins
In a genomic screen, the previously discovered GID1/VID30, GID2/RMD5, and GID3/UBC8 genes were identified along with six additional GID genes to be necessary for glucose-induced FBPase degradation (Schork et al., 1995; Hämmerle et al., 1998; Schüle et al., 2000; Regelmann et al., 2003). Further characterization of Gid2/Rmd5 suggested that it is part of a complex of at least 600 kDa (Regelmann et al., 2003). Mass spectrometry analyses of proteins interacting with either Gid7 or Gid1/Vid30 uncovered the Gid proteins as subunits of the same complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006).
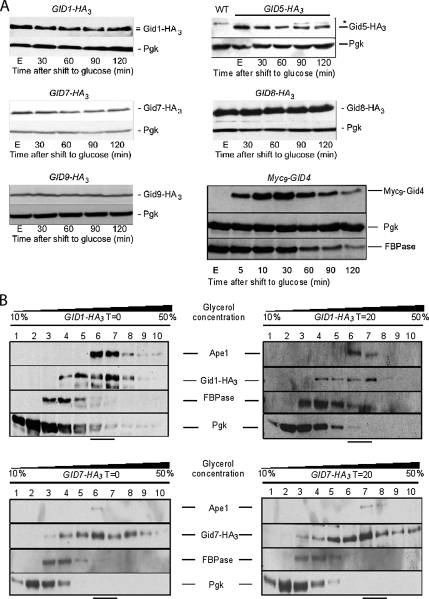
To be able to detect the Gid proteins immunologically, their genes were chromosomally tagged with either 3xHA or 9xMYC coding sequences. The resulting strains were tested for their ability to degrade FBPase. Except for cells expressing the Gid9-HA3 protein, all strains displayed wild-type FBPase degradation kinetics (data not shown). The expression of the Gid proteins was monitored in ethanol-growing cells and up to 120 min after shift to glucose-containing media. Besides Gid2/Rmd5 (Regelmann et al., 2003), five additional Gid proteins (Gid1/Vid30, Gid5/Vid28, Gid7, Gid8, and Gid9/Fyv10) were already present in ethanol-grown (gluconeogenic conditions) cells, and their levels remained constant over 120 min after shift to glucose (Figure 1A). An additional Gid protein, Gid4/Vid24 was undetectable when cells were grown in ethanol-containing medium. However, as soon as cells were transferred to glucose-containing medium, the Gid4/Vid24 protein rapidly occurred, its levels culminating ∼30 min after shift. Thereafter, Gid4/Vid24 levels decreased in parallel with those of FBPase (Figure 1A). Gid4/Vid24 synthesis upon glucose addition to cells growing on a nonfermentable carbon source had already been described, whereas no subsequent degradation of the protein had been observed under the inactivation conditions used (Chiang and Chiang, 1998).
Figure 1.
Gid complex subunits are present in gluconeogenic cells. (A) Strains expressing chromosomally tagged Gid proteins with the HA or Myc epitope were grown 16 h in YPethanol (E) and thereafter treated with 2% glucose. Samples were taken at the indicated time points and processed as described in Material and Methods. Proteins were visualized by immunoblotting. *, cross reaction; WT, wild type, control for cross-reaction. (B) Strains bearing a chromosomally expressed HA-tagged Gid protein (Gid1/Vid30 and Gid7) were grown 16 h in YPethanol. Samples were harvested before (T = 0) or 20 min after shift to glucose (T = 20), and proteins were separated on a glycerol step gradient. Ten fractions were collected, and proteins were visualized by immunoblotting (Pgk, 3-phosphoglycerate kinase; Ape1, aminopeptidase I).
Using glycerol step gradient analysis, Regelmann et al. (2003) found that Gid2/Rmd5 exists in a complex of ∼600 kDa. As shown in Regelmann et al. (2003) for the Gid2/Rmd5 protein, the other six Gid proteins (Gid1/Vid30, Gid4/Vid24, Gid5/Vid28, Gid7, Gid8, and Gid9/Fyv10) also sediment at high molecular mass (data not shown). Indeed, mass spectrometry analyses identified a complex in which the seven Gid proteins reside (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006). All these analyses were performed in glucose-containing media. Because Gid4/Vid24 is absent under gluconeogenic conditions, we tested its requirement for the formation of a high molecular mass Gid complex. Glycerol step gradients were performed to observe the sedimentation patterns of Gid1-HA3 and Gid7-HA3 in gluconeogenic conditions (Gid4/Vid24 absent; Figure 1A), and 20 min after shift to glucose, where Gid4/Vid24 is present (Figure 1A). As shown in Figure 1B, no significant differences in the sedimentation patterns of Gid1-HA3 (116 kDa) and Gid7-HA3 (92 kDa) before (T = 0) and 20 min (T = 20) after glucose shift could be seen: both proteins sediment with molecular masses in the range of 600 kDa, as shown by their cofractionation with the 600 kDa aminopeptidase I homodecamer complex (Ape1). Although it cannot be completely excluded that the binding partners within the Gid-complex vary between ethanol- and glucose-grown cells, this experiment suggests that the tested Gid proteins are already present in a complex when cells grow on a nonfermentable carbon source and do not dissociate to their respective monomers when Gid4/Vid24 is absent from the cell.
Gid1/Vid30 Interacts with FBPase
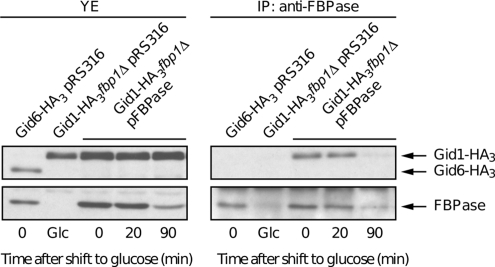
A direct involvement of the Gid complex in FBPase degradation requires binding of FBPase to Gid proteins. We therefore examined whether Gid1-HA3, as an example, coimmunoprecipitates with FBPase. We used a GID6-HA3 strain as a control for the interaction of FBPase with the Gid complex. GID6 was shown previously to encode Ubp14 deubiquitinating enzyme, its deletion prevents the cleavage of polyubiquitin chains released from proteasome substrates into single ubiquitin moieties. This deletion leads to competition between uncleaved polyubiquitin chains and polyubiquitinated proteins for the proteasome (Amerik et al., 1997), which has been shown to impair proteasomal degradation of FBPase (Eisele et al., 2006). Thus, the effect of GID6 deletion on FBPase degradation is unspecific. Moreover, Gid6/Ubp14 was not detected as being part of the Gid complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006). YJJ9 (GID1-HA3, fbp1Δ) and YBB1 (GID6-HA3) strains were transformed with a plasmid expressing a wild-type FBPase (pFBPase) or pRS316 as a vector control. YJJ9 strain transformed with pFBPase and YBB1 strain transformed with pRS316 were grown in YPethanol and shifted to YPD. The YJJ9 strain transformed with pRS316, for which the endogenous FBPase is not replaced by a plasmid-encoded enzyme, is unable to grow on nonfermentable carbon sources and was therefore grown on YPD only. Samples were withdrawn at the indicated time points, and FBPase was precipitated using specific anti-FBPase antibodies (Figure 2). The precipitates were monitored by immunoblotting with FBPase and HA-antibodies. Figure 2 shows that FBPase strongly coimmunoprecipitates with Gid1-HA3 in YPethanol at the time point “0” and after 20 min of glucose addition to cells. A similar result was obtained when a strain expressing FBPase chromosomally was used for the immunoprecipitation (data not shown). No interaction between FBPase and Gid6-HA3/Ubp14-HA3 could be observed, demonstrating that no unspecific interaction occurs between FBPase and the HA-tag. Moreover, Gid1-HA3 was not immunoprecipitated when no FBPase was present, establishing that the anti-FBPase antibody does not bind unspecifically to Gid1-HA3.
Figure 2.
FBPase binds to Gid1/Vid30. Strains expressing HA-tagged Gid proteins (Gid1/Vid30 and Gid6/Ubp14) and transformed with a plasmid expressing a wild-type FBPase (pFBPase) or its corresponding vector control (pRS316) were grown 16 h on YPethanol. Samples were harvested 0, 20, and 90 min after glucose treatment, and proteins were extracted. Coimmunoprecipitation was performed with FBPase antibody. Protein immunoblot was carried out with FBPase and HA antibodies. The noninteracting Gid6/Ubp14 protein serves as a control (YE, extracts; IP, immunoprecipitations). The FBP1 deletion mutant transformed with the vector control was grown on glucose and provides a control for the specificity of the anti-FBPase antibody.
Gid2/Rmd5 Confers Ubiquitin Ligase (E3) Activity
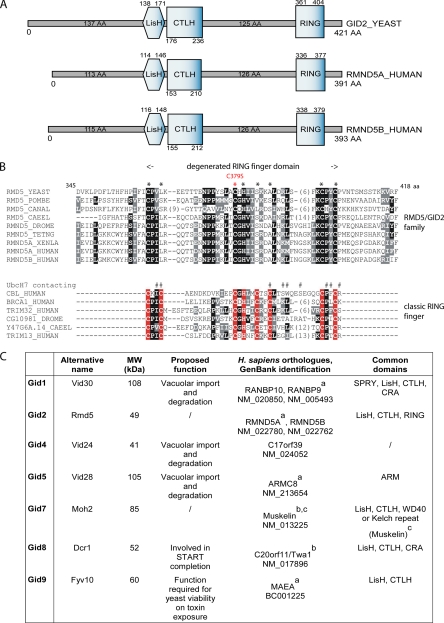
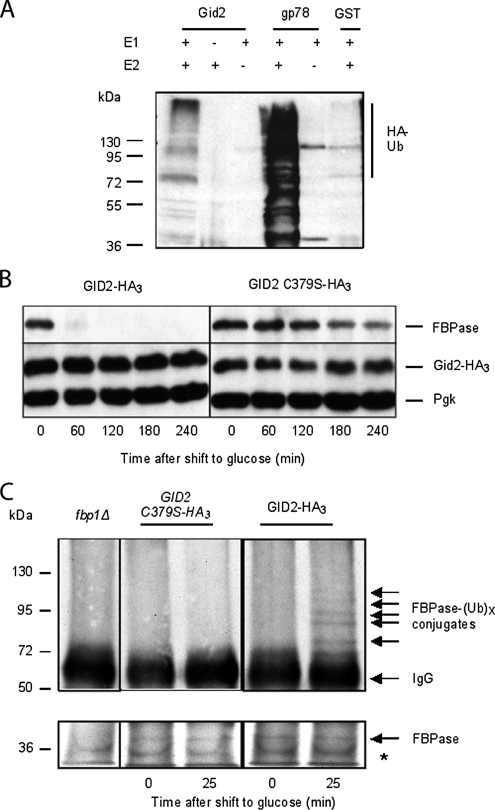
Gid2/Rmd5 contains a Lissencephaly type-1-like homology motif (LisH), a C-terminal to LisH motif (CTLH), and a degenerated RING finger domain, an organization shared with its human orthologues (Figure 3, A and C). Many ubiquitin ligases contain a RING-finger domain bearing eight Cys/His coordination sites to complex Zn2+ ions necessary for activity (Lorick et al., 1999; Fang and Weissman, 2004). A complete cysteine and histidine pattern in a RING domain is not necessarily critical for the E3 function, as the U-box domain family shows. Although the U-box domain still adopts the same structure as the RING domain, it is an extreme example of degeneration of zinc-coordinating residues, where none of them are retained (Ohi et al., 2003). In Gid2/Rmd5 the degenerated RING domain is characterized by an incomplete series of Zn2+ ion-coordinating residues compared with the classic RING finger (Figure 3B, compare top and bottom alignments). The canonical RING domain encompasses eight residues coordinating two Zn cations. The first Zn2+ ion is coordinated by the first, second, fifth, and sixth residue; the second by the remaining residues three, four, seven, and eight (Lorick et al., 1999; Fang and Weissman, 2004). In Gid2/Rmd5 besides the first cysteine, the four residues coordinating the second Zn2+ ion are conserved (Figure 3B), which strongly suggests that one ion is retained in this degenerated RING domain. This prompted us to suspect that Gid2/Rmd5 may bear an ubiquitin ligase activity. The ubiquitin system is present in all eukaryotes but is not found among bacteria. A heterologous expression of a GST-Gid2 fusion protein in E. coli led to polyubiquitination of proteins in the bacterial lysate when E1 and E2 enzymes, ATP and HA-ubiquitin were added (Figure 4A). When only the GST protein was expressed, no polyubiquitination could be detected (Figure 4A). We further determined the contribution of the Gid2/Rmd5 degenerated RING finger to FBPase polyubiquitination in vivo by mutating the conserved cysteine residue at position 379 of the putative Gid2/Rmd5 RING domain to serine. The HA-tagged version of the mutated gene was introduced into the yeast genome. As can be seen in Figure 4B, degradation of FBPase is dramatically impaired in cells carrying the GID2C379S mutation. The strain expressing the mutant protein is unable to polyubiquitinate FBPase in vivo (Figure 4C). We conclude that Gid2/Rmd5 bears an E3 activity necessary for degradation of FBPase.
Figure 3.
Domain structures of Gid proteins. (A) Structure similarities between the Gid2/Rmd5 (S. cerevisiae) and RMND5A/B (Homo sapiens) proteins. (B) Alignment of the RING domains found in the Gid2/Rmd5 protein family and classic RING-finger proteins. Positions with invariant or conservatively replaced residues in at least 50% of the sequences are printed on black or gray background, respectively (not all sequences analyzed are shown). The leftmost column describes the sequence name and contains the species abbreviation: YEAST, S. cerevisiae; POMBE, S. pombe; CANAL, Candida albicans; HUMAN, H. sapiens; XENLA, Xenopus laevis; DROME, Drospophila melanogaster; CAEEL, Caenorhabditis elegans; TETNG, Tetraodon nigroviridis. Larger insertions are not shown; the number of omitted residues is indicated in parentheses. Asterisks above the Gid2/Rmd5 family alignment correspond to positions homologous to zinc coordinating residues in classic RING finger domains (red). The mutation introduced in the Gid2/Rmd5 degenerated RING domain is highlighted (C379S, red). Above the classic RING-finger proteins alignment, residues of Cbl in contact with UbcH7 are marked by # (Zheng et al., 2000). (C) Overview of the Gid proteins and their counterparts in H. sapiens. The table summarizes the common features in Gid proteins and their human counterparts. aOrthologues identified in a CTLH complex by Kobayashi et al. (2007); bOrthologues found to interact together by Umeda et al. (2003).cMuskelin bears CTLH and Kelch domains, which makes its overall structure similar to Gid7. Other functions and names, SGD, www.yeastgenome.org.
Figure 4.
Gid2/Rmd5 shows E3 activity. (A) Gid2/Rmd5 ubiquitinates protein in vitro. Lysates of E. coli expressing either Gid2/Rmd5 or the mammalian RING-finger protein gp78 as a positive control and GST as a negative control were incubated with E1, E2 (UbcH5b), ATP and HA-ubiquitin for 2 h at 30°C. To assess the specificity of the reaction, same lysates were incubated without E1 or E2. Polyubiquitination was detected using monoclonal HA antibody. (B) GID2-HA3 and its mutated C379S counterpart-expressing cells were grown 16 h in YPethanol at 25°C and shifted to YPD. Samples were taken at indicated time points and FBPase degradation was monitored by immunoblotting. Pgk, 3-phosphoglycerate kinase, loading control. (C) Point mutation in the Gid2/Rmd5 degenerated RING domain abolishes FBPase polyubiquitination. Chromosomally tagged GID2-HA3, which shows a wild-type phenotype, and a strain bearing the C379S point mutation in a GID2-HA3 strain were grown 16 h in YPethanol at 25°C and shifted to YPD. Samples were taken at indicated time points, and FBPase was immunoprecipitated. Polyubiquitination was detected using monoclonal ubiquitin antibody. fbp1Δ: FBPase deletion. Presence of FBPase in the immunoprecipitates was controlled by immunoblotting with FBPase antibody. *, cross-reaction.
Role of Gid4/Vid24 in the FBPase Catabolite Degradation
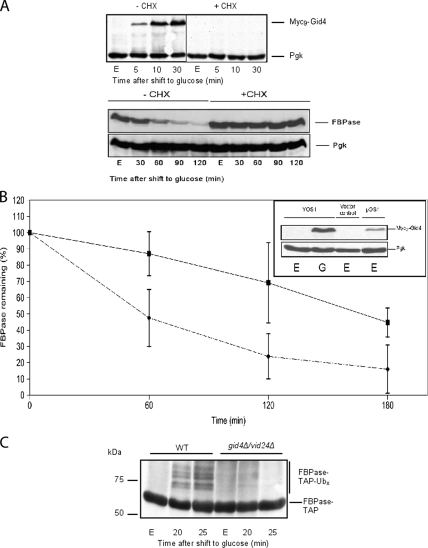
Gid4/Vid24 is necessary for FBPase degradation (Regelmann et al., 2003). The protein is absent in ethanol-growing cells and occurs early after glucose addition to the medium (Figure 1A). Thus, blocking de novo protein synthesis is likely to prevent Gid4/Vid24 appearance and subsequent FBPase breakdown by the proteasome. To test this hypothesis, cells were treated with the translation inhibitor cycloheximide when they were shifted from gluconeogenic to glycolytic conditions. Indeed, cells that underwent cycloheximide treatment were unable to express Gid4/Vid24 and therefore to degrade FBPase (Figure 5A). Because Gid3/Ubc8 and other Gid proteins, except Gid4/Vid24, are present in ethanol-growing cells (Figure 1, Regelmann et al. (2003)), this result suggests that Gid4/Vid24 might trigger FBPase degradation. If Gid4/Vid24 were a switch, initiation of FBPase elimination should take place when Gid4/Vid24 is expressed in ethanol-growing cells. FBP1 gene expression is repressed when cells grow on a glucose-containing medium; on the contrary, when grown on a nonfermentable carbon source, yeast cells readily express FBPase (Zaragoza and Gancedo, 2001). Thus, a pulse-chase experiment, where FBPase was radioactively labeled, has been carried out to monitor FBPase degradation when Gid4/Vid24 expression is forced in ethanol-growing cells. Indeed, expression of Gid4/Vid24 in YPethanol growing cells lowers the half-life of FBPase by approximately threefold (Figure 5B). Although one cannot exclude that additional modifications are necessary to signal FBPase degradation, this result strongly suggests that Gid4/Vid24 is the only protein whose de novo synthesis is required to trigger FBPase elimination.
Figure 5.
Gid4/Vid24 promotes FBPase degradation. (A) De novo protein synthesis is necessary for FBPase degradation and Myc9-Gid4 expression. Wild-type cells were grown 16 h in YPethanol (E) and shifted to YPD with (+CHX) or without (−CHX) cycloheximide (100 μg/ml). FBPase and Myc9-Gid4 levels were monitored at indicated times via immunoblotting with anti-FBPase or anti-Myc antibodies, respectively. (B) Gid4/Vid24 expression triggers FBPase degradation. Myc9-GID4 was cloned under a TetR promoter and expressed in cells growing on YPethanol. Pulse-chase analysis of FBPase in cells bearing either the Myc9-GID4 expressing plasmid (♦) or the respective vector control (■) was carried out (means of three independent experiments, ±confidence interval, α = 0.05). Inset, immunoblot showing the steady-state levels of Myc9-Gid4 in glucose-inactivated cells (30 min, YOS1, G) and in ethanol grown cells (pOS1, E). (C) Gid4/Vid24 is necessary for FBPase-TAP polyubiquitination. A plasmid expressing a FBPase-TAP fusion protein was transformed into W303 (WT) and gid4Δ/vid24Δ strains. Samples were taken at indicated time points, and FBPase was pull-downed using IgG-Sepharose. Polyubiquitination was detected using monoclonal ubiquitin antibody.
The fact that Gid4/Vid24 is a molecular switch triggering FBPase degradation even in gluconeogenic conditions led us ask whether the Gid2/Rmd5-dependent polyubiquitination of FBPase would depend on Gid4/Vid24. The TAP tag allows to purify proteins (Puig et al., 2001). Because it bears a protein A sequence, it is able to strongly bind the IgG constant fragment. This property allowed us to use the tag to perform IgG-Sepharose pull-downs. Moreover, when fused to an otherwise poorly detectable protein, it easily facilitates protein detection. Immunoprecipitation of FBPase was often complicated by the fact that the enzyme migrates only slightly faster on SDS-polyacrylamide gels than IgGs. Therefore, a plasmid expressing a catalytically active C-terminally TAP-tagged FBPase was transformed in W303 (WT) and gid4Δ/vid24Δ strains to ensure a good detection of the enzyme and its polyubiquitinated forms. Pull-down with IgG-Sepharose and subsequent detection with an anti-ubiquitin antibody revealed that Gid4/Vid24 is indeed necessary for FBPase polyubiquitination (Figure 5C). Together, these results suggest that Gid4/Vid24 is a switch allowing the polyubiquitination of FBPase before its degradation by the proteasome.
Elimination of Gid4/Vid24 Depends on the Ubiquitin–Proteasome System and the Gid Complex
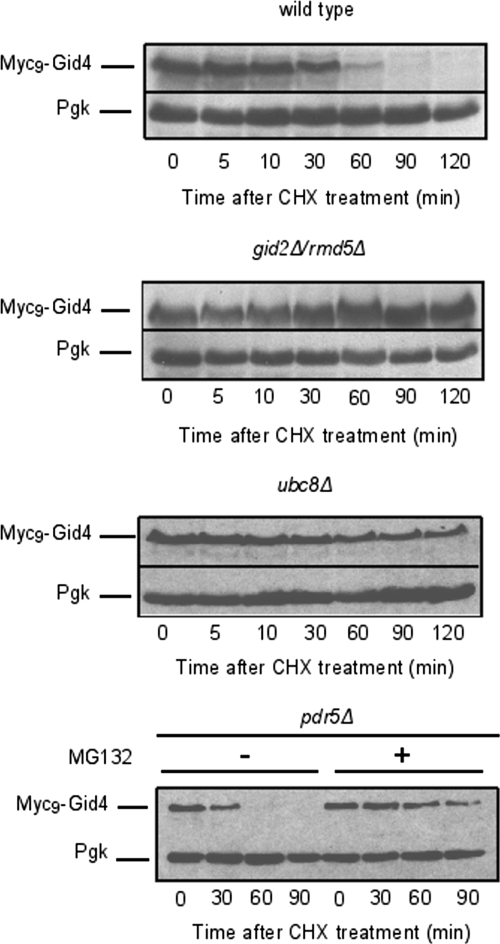
As shown in Figure 1A, Myc9-Gid4 levels increase with time when cells are shifted from YPethanol to YPD. After reaching a maximum at ∼30 min after shift to glucose, the Myc9-Gid4 signal diminishes in parallel with FBPase. To test whether this diminution is dependent on the ubiquitin-conjugating enzyme Ubc8, components of the Gid complex, and the proteasome, we deleted GID2/RMD5, UBC8, and, for proteasome inhibition by the proteasome inhibitor MG-132, PDR5 in a strain expressing Myc9-Gid4. PDR5 encodes an ATP-binding cassette transporter, which prevents accumulation of hydrophobic compounds in the yeast cytosol (Balzi et al., 1994; Bissinger and Kuchler, 1994). Its deletion is thus necessary to prevent inhibitor efflux from the cell. After overnight growth in ethanol-containing medium, the tested strains were shifted to glucose-containing medium for 30 min to allow Myc9-Gid4 expression before cycloheximide treatment. Samples were collected at the indicated time points after cycloheximide treatment. Figure 6 shows that, similar to elimination of FBPase, degradation of Gid4/Vid24 depends on Ubc8 (E2) and the proteasome. Interestingly, degradation of Gid4/Vid24 also depends on components of the Gid complex because deletion of GID2/RMD5 stalls its degradation (Figure 6). Elimination of Myc9-Gid4 is not due to the Myc-tag as native, endogenous Gid4/Vid24 shows identical behavior (Josupeit and Wolf, unpublished).
Figure 6.
Gid4/Vid24 degradation depends on Gid2/Rmd5, Ubc8 and the proteasome. W303-1B expressing Myc9-Gid4 was deleted for UBC8, GID2/RMD5, or PDR5 (ubc8Δ, gid2Δ, pdr5Δ, respectively). Strains were grown 16 h on YPethanol, shifted to YPD, and expression of Gid4/Vid24 was allowed to proceed during 30 min. Thereafter, cells were treated with cycloheximide and samples were harvested at the indicated times. Proteins were visualized via immunoblotting. Proteasome involvement in Gid4/Vid24 degradation was analyzed in a pdr5Δ strain using the proteasome inhibitor MG-132.
Catabolite Degradation of PEPCK Requires Subunits of the Gid Complex
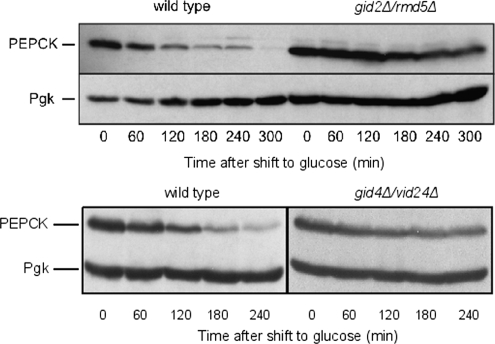
The highly exergonic last reaction step in glycolysis, the formation of pyruvate and ATP by pyruvate kinase is circumvented in gluconeogenesis by the action of pyruvate carboxylase and PEPCK. As for FBPase, PEPCK synthesis is repressed by glucose and subjected to catabolite degradation when cells are shifted from gluconeogenic to glycolytic conditions (Holzer, 1976; Muller et al., 1981; Mercado and Gancedo, 1992; Yin et al., 2000). This allowed us to monitor the degradation of PEPCK after shift of cells from YPethanol to YPD. Interestingly, deletions of GID2/RMD5 or GID4/VID24 stall degradation of PEPCK, indicating the requirement of the Gid complex for its catabolite degradation (Figure 7). This suggests that the Gid complex plays a general role in catabolite degradation.
Figure 7.
GID deletions impair PEPCK degradation. Wild type, gid2Δ/rmd5Δ, or gid4Δ/vid24Δ cells were grown 16 h on YPethanol, and thereafter shifted onto YPD. Cells were harvested every hour, and extracts were prepared. Proteins were visualized via immunoblotting. Pgk, 3-phosphoglycerate kinase, loading control.
DISCUSSION
FBPase is essential for yeast growth on nonfermentable carbon sources such as acetate or ethanol. When a fermentable carbon source becomes available, the FBP1 gene expression is repressed and the enzyme is inactivated by allosteric inhibition before being degraded (Holzer, 1976; von Herrath and Holzer, 1988). The site of degradation (vacuole or proteasome) has been subject to controversy. Now, it seems rather clear that the type of gluconeogenic conditions influence the degradation pathway (Schork et al., 1994b; Chiang and Chiang, 1998; Hämmerle et al., 1998; Hung et al., 2004). Vacuolar import and degradation of FBPase occurs when cells are grown 2–3 d on an acetate-containing medium. Such conditions trigger autophagy, which is not likely to be relieved by the sole addition of glucose to the medium. Thus, one may speculate that yeast cells use the pre-existing degradation machinery to specifically degrade FBPase when growth conditions favor this autophagic process. On the contrary, overnight growth on ethanol, a nonfermentable but natural carbon source for S. cerevisiae triggers FBPase degradation by the proteasome. A genome-wide screen revealed nine genes whose deletion impaired proteasomal-dependent FBPase degradation upon shift to glucose-containing medium (Regelmann et al., 2003). Among these genes, Gid1/Vid30, Gid4/Vid24, Gid5/Vid28 were first implicated in a vacuolar-dependent FBPase degradation (Chiang and Chiang, 1998). Moreover, GID3 and GID6 genes were shown to encode Ubc8 and Ubp14, respectively (Schüle et al., 2000; Regelmann et al., 2003). It was demonstrated that Gid2/Rmd5 belongs to a soluble complex of ∼600 kDa (Regelmann et al., 2003). Also, Gid1/Vid30, Gid4/Vid24, Gid5/Vid28, Gid7, Gid8, and Gid9/Fyv10 belong to a soluble complex (data not shown). Systematic interaction studies identified those Gid proteins as units of the same complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006). Epitope tagging allowed us to monitor the expression of the Gid proteins. Interestingly, Gid4/Vid24 is the only Gid protein that cannot be detected in ethanol-growing cells. This led us to suspect that Gid4/Vid24 might be an activator of the Gid complex. In glycerol step gradient analysis, no differences between the sedimentation patterns of Gid1-HA3 and Gid7-HA3 before (Gid4/Vid24 absent) and 20 min after shift to glucose (Gid4/Vid24 present) could be revealed (Figure 1B). Although it cannot be excluded that the binding partners of Gid1/Vid30 or Gid7 differ between gluconeogenic and glycolytic conditions, both proteins remain in a complex even when Gid4/Vid24 is absent, suggesting that this Gid protein has no influence on the assembly of monomeric Gid proteins to a higher molecular mass complex. The Gid proteins are involved in the proteasomal degradation of FBPase. Whether the Gid complex is part of an upstream signaling pathway or whether it is directly involved in FBPase poyubiquitination and degradation remained to be clarified. We show that Gid1/Vid30 interacts with FBPase. As Gid1/Vid30 is part of the Gid complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006), this result indicates that FBPase and the Gid complex interact.
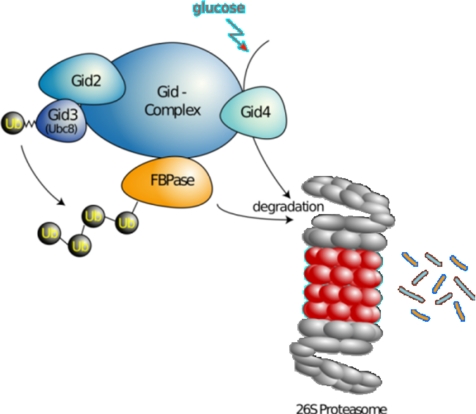
A GID2/RMD5 deletion prevents polyubiquitination of FBPase (Regelmann et al., 2003). Alignments of the Gid2/Rmd5 protein sequence with known RING-finger E3s revealed that it bears a so-called degenerated RING-finger where Zn2+ coordination residues are missing (Figure 3B). Although the U-box domain family lacks all Zn2+ coordinating residues, it still polyubiquitinates proteins (Hatakeyama et al., 2001 Ohi et al., 2003). This prompted us to test whether this would also be the case for Gid2/Rmd5. Indeed, Gid2/Rmd5 polyubiquitinates proteins in vitro. Moreover, destroying a remaining Zn2+ coordination site in Gid2/Rmd5 leads to a failure to polyubiquitinate FBPase in vivo (Figure 4). These results imply that the Gid complex is an ubiquitin ligase (E3), whose activity is provided by the Gid2/Rmd5 subunit (Figure 8).
Figure 8.
The Gid complex, a novel ubiquitin ligase (E3) required for the degradation of the key gluconeogenic enzyme fructose-1,6-bisphosphatase. The Gid complex binds to FBPase, when S. cerevisiae cells are growing on an ethanol-containing medium. On shift of cells to glucose, Gid4/Vid24 occurs and activates the complex, which then polyubiquitinates FBPase before its degradation by the proteasome. Gid4/Vid24 is itself degraded by the proteasome.
We further characterized the function of the Gid4/Vid24 subunit within the new E3 complex. Gid4/Vid24 had previously been implicated in targeting FBPase from vesicles to the vacuole and was shown to occur only upon glucose addition to acetate-starved cells where it remained stable (Chiang and Chiang, 1998). We here show that also in cells growing on ethanol, a natural carbon source of yeast, Gid4/Vid24 occurs upon glucose shift (Figure 1A). Because Gid4/Vid24 becomes detectable as fast as 5 min after shift of cells from ethanol to glucose, it is unlikely that GID4/VID24 gene expression depends on transcription followed by translation. Indeed, GID4/VID24 mRNA levels do not vary significantly when cells grow on these two different media (DeRisi et al., 1997). Thus, the regulation of GID4/VID24 expression might be carried out at the posttranscriptional level. Hung et al. (2004) confirmed our data that Gid4/Vid24 partakes in proteasome-dependent FBPase degradation. The different expression patterns of Gid4/Vid24 observed during the vacuolar degradation process of FBPase in which it remains stable (Chiang and Chiang, 1998) and the proteasomal degradation of the enzyme in which Gid4/Vid24 is degraded might reflect the different requirements of Gid4/Vid24 in these processes. Because Gid4/Vid24 has no influence on sedimentation patterns of two Gid proteins known to be part of the Gid complex (Figure 1B), it seems more likely that Gid4/Vid24 activates the Gid complex rather than stabilizing it. As shown for acetate-starved cells (Chiang and Chiang, 1998), cycloheximide treatment of cells grown overnight on an ethanol-containing medium prevents Gid4/Vid24 expression on glucose (Figure 5A).
Our result suggests that Gid4/Vid24 might be a switch activating the Gid complex to degrade FBPase. Indeed, forced expression of Gid4/Vid24 under conditions where FBPase is actively expressed and stable, triggered its degradation (Figure 5B). Moreover, lack of Gid4/Vid24 inhibits polyubiquitination of an FBPase-TAP fusion protein (Figure 5C). Together, these results suggest that Gid4/Vid24 activates the Gid complex by promoting Gid2/Rmd5-dependent FBPase polyubiquitination. The mechanism of Gid complex activation by Gid4/Vid24 remains to be elucidated. Because FBPase interacts with the Gid complex already in ethanol-grown cells (Figure 2), Gid4/Vid24 is not likely to recruit FBPase to the complex. Gid4/Vid24 binds to the Gid complex (Ho et al., 2002; Krogan et al., 2006; Pitre et al., 2006); thus, one hypothesis is that it alters the complex conformation to activate it (Figure 8).
Gid4/Vid24 is degraded during elimination of FBPase by the proteasome (Figure 1A). Its degradation depends on the ubiquitin conjugating enzyme Ubc8 and the E3 component Gid2/Rmd5 (Figure 6). Moreover, degradation of Gid4/Vid24 requires the proteasome (Figures 6, 8). Obviously, Gid4/Vid24 undergoes a regulatory loop exerted by components of the Gid complex: it might limit the activity of the Gid complex to a certain type of substrates like FBPase or prepare cells for new growth on nonfermentable carbon sources.
PEPCK, another gluconeogenic enzyme, is also subject to catabolite degradation (Holzer, 1976; Muller et al., 1981). As Figure 7 shows, it is stabilized in GID2/RMD5 and GID4/VID24 deleted cells. Thus, Gid complex-dependent degradation is not restricted to FBPase but plays a more general role within the regulation of carbohydrate metabolism.
Gid protein homologues were also found to form a complex in mammals (Kobayashi et al., 2007). Although no function for this CTLH complex has been described, one subunit has been implicated in proteasomal degradation of α-catenin (Suzuki et al., 2008). This suggests that the CTLH complex, like the Gid complex, might also bear E3 activity.
In conclusion (Figure 8), our study shows that the Gid complex is a new ubiquitin ligase with novel types of subunits involved in catabolite degradation of gluconeogenic enzymes in yeast. We also identify Vid24/Gid4 as an important regulator of its ubiquitin ligase activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Sommer, M. Tyers, S. Munro and A. Weissman for plasmids and K. D. Entian for FBPase antibodies. We also thank Antje Schäfer for invaluable advice and critical reading of the manuscript. The help of E. Tosta during preparation of this manuscript is gratefully acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, SFB 495; and the Fonds der Chemischen Industrie, Frankfurt.
Abbreviations used:
- CTLH
C-terminal to LisH motif
- FBPase
fructose-1,6-bisphosphatase
- GST
Glutathione transferase
- LisH
Lissencephaly type-1-like homology motif
- ORF
Open reading frame
- PEPCK
phospho-enol-pyruvate carboxykinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0328) on May 28, 2008.
REFERENCES
- Amerik A., Swaminathan S., Krantz B. A., Wilkinson K. D., Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Kingston R. E., Seidman F. G., Struhl K., Moore D. D., Brent R., Smith F. A. Current Protocols in Molecular Biology. New York: Greene; 1992. [Google Scholar]
- Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25:31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- Benton D. Recent changes in the GenBank On-line Service. Nucleic Acids Res. 1990;18:1517–1520. doi: 10.1093/nar/18.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissinger P. H., Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- Bucher P., Karplus K., Moeri N., Hofmann K. A flexible motif search technique based on generalized profiles. Comput. Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- Chiang M. C., Chiang H. L. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J. Cell Biol. 1998;140:1347–1356. doi: 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel G. The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations, respectively: new selectable markers and new multi-purpose multicopy shuttle vectors, pSP3 and pSP4. Curr. Genet. 1995;28:380–383. doi: 10.1007/BF00326437. [DOI] [PubMed] [Google Scholar]
- de la Guerra R., Valdes-Hevia M. D., Gancedo J. M. Regulation of yeast fructose-1,6-bisphosphatase in strains containing multicopy plasmids coding for this enzyme. FEBS Lett. 1988;242:149–152. doi: 10.1016/0014-5793(88)81004-4. [DOI] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F., Braun B., Pfirrmann T., Wolf D. H. Mutants of the deubiquitinating enzyme Ubp14 decipher pathway diversity of ubiquitin-proteasome linked protein degradation. Biochem. Biophys. Res. Commun. 2006;350:329–333. doi: 10.1016/j.bbrc.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Fang S., Weissman A. M. A field guide to ubiquitylation. Cell Mol. Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo C. Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J. Bacteriol. 1971;107:401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Trautwein M., Sommer T., Spang A. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 2005;22:1–12. doi: 10.1002/yea.1187. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:21–37. [PubMed] [Google Scholar]
- Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hoffman M., Chiang H. L. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–1566. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. Sensitive protein comparisons with profiles and hidden Markov models. Brief. Bioinform. 2000;1:167–178. doi: 10.1093/bib/1.2.167. [DOI] [PubMed] [Google Scholar]
- Holzer H. Catabolite inactivation in yeast. Trends Biochem. Sci. 1976;1:178–181. [Google Scholar]
- Holzer H. Proteolytic catabolite inactivation in Saccharomyces cerevisiae. Revis. Biol. Celular. 1989;21:305–319. [PubMed] [Google Scholar]
- Hung G. C., Brown C. R., Wolfe A. B., Liu J., Chiang H. L. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J. Biol. Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- Hämmerle M., Bauer J., Rose M., Szallies A., Thumm M., Dusterhus S., Mecke D., Entian K. D., Wolf D. H. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:25000–25005. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Yang J., Ueda A., Suzuki T., Tomaru K., Takeno M., Okuda K., Ishigatsubo Y. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus F., Rittenhouse J., Moberly L., Edelstein I., Hiller E., Rogers D. T. Yeast (Saccharomyces cerevisiae) fructose-1,6-bisphosphatase. Properties of phospho and dephospho forms and of two mutants in which serine 11 has been changed by site-directed mutagenesis. J. Biol. Chem. 1988;263:6058–6062. [PubMed] [Google Scholar]
- Mazon M. J., Gancedo J. M., Gancedo C. Inactivation of yeast fructose-1,6-bisphosphatase. In vivo phosphorylation of the enzyme. J. Biol. Chem. 1982;257:1128–1130. [PubMed] [Google Scholar]
- Mercado J. J., Gancedo J. M. Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 1992;311:110–114. doi: 10.1016/0014-5793(92)81379-z. [DOI] [PubMed] [Google Scholar]
- Muller M., Muller H., Holzer H. Immunochemical studies on catabolite inactivation of phosphoenolpyruvate carboxykinase in Saccharomyces cerevisiae. J. Biol. Chem. 1981;256:723–727. [PubMed] [Google Scholar]
- Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Chazin W. J., Gould K. L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitre S., et al. PIPE: a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. BMC Bioinformatics. 2006;7:365. doi: 10.1186/1471-2105-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Regelmann J., Schüle T., Josupeit F. S., Horak J., Rose M., Entian K. D., Thumm M., Wolf D. H. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell. 2003;14:1652–1663. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Schork S., Bee G., Thumm M., Wolf D. H. Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 1994a;349:270–274. doi: 10.1016/0014-5793(94)00668-7. [DOI] [PubMed] [Google Scholar]
- Schork S., Bee G., Thumm M., Wolf D. H. Site of catabolite inactivation. Nature. 1994b;369:283–284. doi: 10.1038/369283a0. [DOI] [PubMed] [Google Scholar]
- Schork S. M., Thumm M., Wolf D. H. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Schüle T., Rose M., Entian K. D., Thumm M., Wolf D. H. Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase in yeast. EMBO J. 2000;19:2161–2167. doi: 10.1093/emboj/19.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Ueda A., Kobayashi N., Yang J., Tomaru K., Yamamoto M., Takeno M., Ishigatsubo Y. Proteasome-dependent degradation of alpha-catenin is regulated by interaction with ARMc8alpha. Biochem. J. 2008;411:581–591. doi: 10.1042/BJ20071312. [DOI] [PubMed] [Google Scholar]
- Umeda M., Nishitani H., Nishimoto T. A novel nuclear protein, Twa1, and Muskelin comprise a complex with RanBPM. Gene. 2003;303:47–54. doi: 10.1016/s0378-1119(02)01153-8. [DOI] [PubMed] [Google Scholar]
- Vaulont S., Vasseur-Cognet M., Kahn A. Glucose regulation of gene transcription. J. Biol. Chem. 2000;275:31555–31558. doi: 10.1074/jbc.R000016200. [DOI] [PubMed] [Google Scholar]
- von Herrath M., Holzer H. Sensitivity of fructose-1,6-biphosphatase from yeast, liver and skeletal muscle to fructose-2,6-biphosphate and 5′-adenosine monophosphate. Z. Lebensm. Unters. Forsch. 1988;186:427–430. doi: 10.1007/BF01127304. [DOI] [PubMed] [Google Scholar]
- Wahren J., Ekberg K. Splanchnic Regulation of Glucose Production. Annu. Rev. Nutr. 2007;27:329–345. doi: 10.1146/annurev.nutr.27.061406.093806. [DOI] [PubMed] [Google Scholar]
- Wolf D. H. From lysosome to proteasome: the power of yeast in the dissection of proteinase function in cellular regulation and waste disposal. Cell Mol. Life Sci. 2004;61:1601–1614. doi: 10.1007/s00018-004-4134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Hatton L., Brown A. J. Differential post-transcriptional regulation of yeast mRNAs in response to high and low glucose concentrations. Mol. Microbiol. 2000;35:553–565. doi: 10.1046/j.1365-2958.2000.01723.x. [DOI] [PubMed] [Google Scholar]
- Zaragoza O., Gancedo J. M. Elements from the cAMP signaling pathway are involved in the control of expression of the yeast gluconeogenic gene FBP1. FEBS Lett. 2001;506:262–266. doi: 10.1016/s0014-5793(01)02922-2. [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.