Abstract
Intramolecular interaction between myosin heads, blocking key sites involved in actin-binding and ATPase activity, appears to be a critical mechanism for switching off vertebrate smooth-muscle myosin molecules, leading to relaxation. We have tested the hypothesis that this interaction is a general mechanism for switching off myosin II–based motile activity in both muscle and nonmuscle cells. Electron microscopic images of negatively stained myosin II molecules were analyzed by single particle image processing. Molecules from invertebrate striated muscles with phosphorylation-dependent regulation showed head–head interactions in the off-state similar to those in vertebrate smooth muscle. A similar structure was observed in nonmuscle myosin II (also phosphorylation-regulated). Surprisingly, myosins from vertebrate skeletal and cardiac muscle, which are not intrinsically regulated, undergo similar head–head interactions in relaxing conditions. In all of these myosins, we also observe conserved interactions between the ‘blocked’ myosin head and the myosin tail, which may contribute to the switched-off state. These results suggest that intramolecular head–head and head-tail interactions are a general mechanism both for inducing muscle relaxation and for switching off myosin II–based motile activity in nonmuscle cells. These interactions are broken when myosin is activated.
INTRODUCTION
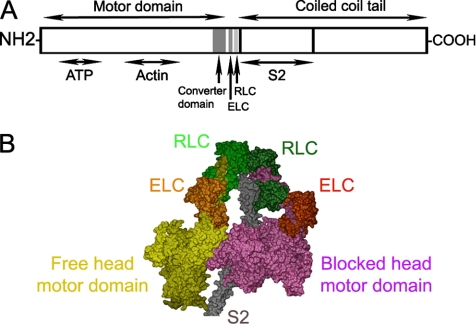
Myosin II is the motor protein that interacts with actin-containing thin filaments, using the chemical energy of ATP to produce muscle contraction and many forms of cell motility (Alberts et al., 2007). It is composed of two globular heads attached to a long, α-helical, coiled-coil tail. Each head has two functional domains (see Figure 1, A and B), a motor domain, containing actin-binding and ATP hydrolysis sites, and a regulatory domain (also called the light-chain domain or lever arm), containing two light chains (an essential light chain [ELC], and a regulatory light chain [RLC]), which amplifies structural changes in the motor domain that drive contraction (Sweeney and Houdusse, 2004; Geeves and Holmes, 2005). A property that distinguishes myosin II from other members of the myosin superfamily is its long tail, which self-associates to form myosin filaments (Craig and Woodhead, 2006). In muscle and nonmuscle cells, such myosin filaments slide past actin filaments to generate muscle shortening or cell motility (Geeves and Holmes, 2005; Alberts et al., 2007).
Figure 1.
Structural organization of myosin II. (A) Schematic layout of myosin heavy chain, consisting of: motor domain (including converter domain and ATP- and actin-binding sites), light-chain domain (containing ELC and RLC binding sites), and the coiled-coil tail domain, including S2 at its N-terminal end. (B) Proposed model for head-S2 region of smooth-muscle myosin in the off-state (Blankenfeldt et al., 2006), built from atomic models of heads (Liu et al., 2003) and S2 (Blankenfeldt et al., 2006).
In the muscles of most species, as well as in nonmuscle cells, myosin is intrinsically regulated and must be phosphorylated on its RLC (vertebrate smooth muscle, many invertebrate striated muscles, and nonmuscle cells) or bind Ca2+ via its ELC (molluscan muscle) in order to turn over ATP and interact with actin (Sellers, 1981, 1991; Szent-Gyorgyi et al., 1999; Perry, 2004). Such myosin-linked regulation is thought to be absent from vertebrate striated muscles (Lehman and Szent-Gyorgyi, 1975), although RLC phosphorylation does modulate activity (Sweeney et al., 1993).
In the inactive (relaxed) state of muscle, the heads of both regulated and unregulated myosin filaments are ordered, turn over ATP slowly, and undergo minimal interaction with actin filaments (Lymn and Taylor, 1971; Xu et al., 1996; Craig and Woodhead, 2006). Activation leads to disordering of the head array (Huxley and Brown, 1967; Zhao and Craig, 2003), increase in ATPase activity, and interaction with actin. In nonmuscle cells, inactivated myosin filaments disassemble into individual molecules, which have a folded tail (Craig et al., 1983). In this resting (or “storage”) form, ATPase and actin-binding activity are extremely low (Kendrick-Jones et al., 1987), and the compact molecules are well suited for transport to the appropriate cellular site when needed for motility. On activation by RLC phosphorylation, these molecules unfold and assemble back into filaments, which can then interact with actin.
Electron microscopic (EM) studies of vertebrate smooth-muscle myosin molecules reveal that the two myosin heads interact with each other intramolecularly in the inactive (dephosphorylated) state (Wendt et al., 2001; Burgess et al., 2007). The actin-binding region of one motor domain contacts the converter region of the other, in an asymmetric arrangement (Figure 1B). It has been suggested that in this way, actin binding is blocked on one head (the “blocked” head) by its binding to the other (“free”) head and that ATPase activity is inhibited on the free head, by binding of its converter domain to the blocked head (Wendt et al., 2001). Thus both heads would be inactivated, but by different mechanisms. Head–head interaction is also observed in native myosin filaments from tarantula striated muscle studied by cryo-electron microscopy (Woodhead et al., 2005). It is thus not an artifact of myosin isolation. The dramatic similarity between the head conformations and interactions in myosin molecules from vertebrate smooth muscle and myosin filaments from invertebrate striated muscle, separated by more than 600 million years of evolution, suggests that head–head interaction is highly conserved and may represent a common mechanism for switching off all intrinsically regulated myosin IIs, both muscle and nonmuscle (Woodhead et al., 2005; Jung et al., 2008).
Additional interactions may also contribute to the inactivated state of regulated myosin. Interaction of the blocked head and the first section of the myosin tail (myosin subfragment 2, or S2) is implied by their apparent contact in tarantula filaments (Woodhead et al., 2005; Figure 1B). Analysis of the crystal structure of S2 suggests that negatively charged residues in S2 may interact electrostatically with positively charged residues in the actin-binding “loop 2” of the blocked head motor domain, which may enhance the stability of the inactive form (Blankenfeldt et al., 2006; cf. Tama et al., 2005). Biochemical data support this view: shortening S2 so that it cannot reach the blocked head motor domain results in loss of regulation (Trybus et al., 1997).
Our goal here has been to test the hypothesis suggested by the above observations—that head-head interaction, and interaction of S2 with the blocked head, is a general mechanism for inactivating myosin II activity in both muscle and nonmuscle cells. We have approached this goal by using single molecule negative staining and image averaging to study the structure of myosin molecules isolated from tarantula and Limulus striated muscles (both phosphorylation-regulated) and from nonmuscle myosin IIA (also phosphorylation-regulated). As controls, we have isolated molecules from vertebrate striated muscles, both skeletal and cardiac, which lack an intrinsic regulatory mechanism, to determine whether this structure is specific to intrinsically regulated molecules or also occurs in unregulated myosins.
MATERIALS AND METHODS
Preparation of Myosins
Muscle was dissected from scallop (Placopecten magellanicus) striated adductor muscle, tarantula (Aphonopelma spp.) leg muscle, horseshoe crab (Limulus) telson muscle, and mouse back and cardiac muscle and placed in rigor solution (0.1 M NaCl, 2 mM EGTA, 5 mM MgCl2, 1 mM DTT, 5 mM PIPES, 5 mM NaH2PO4, 1 mM NaN3, pH 7.0) on ice. Muscles (0.5–2 g tissue) were teased into thin strips and permeabilized with 0.5% saponin in rigor solution for 4 h at 4°C, followed by rinsing in rigor solution. Muscles were then finely chopped and homogenized using a Polytron (Brinkmann Instruments, Westbury, NY) homogenizer (Hidalgo et al., 2001). The homogenate was used to prepare myosin as described by Persechini and Hartshorne (1983) and modified by Ikebe and Hartshorne (1985). Crude myosin, 0.5–1 ml, at ∼1–3 mg/ml was purified on a 1 × 25 cm Sepharose 4B column equilibrated with 0.5 M KCl, 0.1 mM MgCl2, 2 mM EGTA, 0.1 mM DTT, 10 mM imidazole, and 5 mM ATP, pH 7.5, to remove contaminating actin and other proteins. Nonmuscle myosin IIA was expressed using the Baculovirus system. Myosins were used within 24 h of purification.
EM and Single-Particle Image Processing
For negative staining, ATP was added to the column-purified myosin (200–400 nM in high salt) to a final concentration of 0.1 mM. The mixture was then diluted 10-fold with “relaxing” buffer (0.15 M Na acetate, 1 mM EGTA, 2 mM MgCl2, 10 mM MOPS, and 40 μM ATP, pH 7.5). For cross-linking, the myosin was diluted 10-fold with relaxing buffer containing 0.1% glutaraldehyde and left for 1 min before grid preparation. After dilution, 5 μl of the final mixture were immediately (∼5 s) applied to a carbon-coated grid that had been glow-discharged (Harrick Plasma, Ithaca, NY) for 3 min in air, and the grid negatively stained using 1% uranyl acetate.
For metal shadowing, the column-purified myosin preparation was diluted about threefold with 1 mM EGTA, 2 mM MgCl2, 10 mM MOPS, 40 μM ATP, and 0.1% glutaraldehyde, pH 7.5, to give a final concentration of ∼100 nM. This was mixed with an equal volume of glycerol, and the resulting mixture was sprayed onto freshly cleaved mica and then rotary shadowed with platinum at an angle of 6° (Stafford et al., 2001).
Grids were examined in a Philips CM120 electron microscope (FEI, Hillsboro, OR) operated at 80 kV. Images were recorded on a 2Kx2K F224HD slow scan CCD camera (TVIPS, Gauting, Germany) at a magnification of 65,000 (0.37 nm/pixel). Single particle image processing was carried out using SPIDER (Health Research, Rensselaer, NY) and the procedures described in (Burgess et al., 2004).
UCSF Chimera was used for visualization and analysis of PDB structures (Pettersen et al., 2004). Sequence alignments were done using the web program Clustal (http://www.ebi.ac.uk/clustalw/; Chenna et al., 2003).
RESULTS
1. Head–Head Interaction in Regulated, Striated Muscle Myosin in Relaxing Conditions
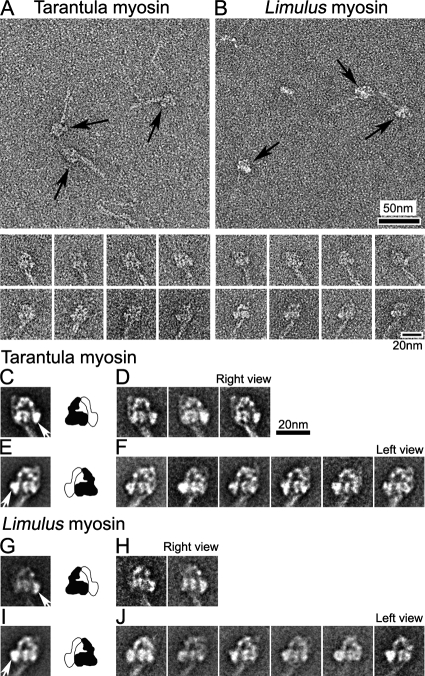
We used negative staining EM to study the structure of inactive (unphosphorylated) myosin molecules from two invertebrate species having phosphorylation-regulated myosin. For both tarantula and Limulus, almost half of the molecules appeared to show the two heads in contact with each other and had a tail that was folded twice, creating three closely apposed segments (Figure 2, A and B; Table 1). We refer to this as the “compact” conformation. These head and tail features are similar to those in vertebrate smooth-muscle myosin molecules in relaxing conditions (Trybus et al., 1982; Wendt et al., 2001; Burgess et al., 2007).
Figure 2.
Compact head conformation of inactive (unphosphorylated) myosin molecules in relaxing conditions. (A and B) Fields (top panels) and galleries (bottom panels) of negatively stained tarantula and Limulus myosins. Arrows indicate head regions of compact molecules. (C–J) Global averages and selected class averages of tarantula and Limulus myosin images (alignment based on head features). (C and E) Global averages of tarantula myosin in right and left views, produced from 122 and 214 images, respectively. (G and I) Global averages of Limulus myosin in right and left views, from 52 and 267 images, respectively. (D, F, H, and J) Class averages, each based on 20–40 individual images in each orientation. Right and left views are defined according to position of the free head motor domain (white arrows in C, E, G, and I). Diagrams show interpretation of global averages in terms of blocked (black) and free (white) heads (see text). Scale bar, 20 nm in D applies to all average images.
Table 1.
Percentage of molecules showing compact conformationa
| Native (%) | Crosslinked (%) | Native + blebbistatin (%) | Blebbistatin + crosslinking (%) | |
|---|---|---|---|---|
| Tarantula | 46 | 67 | ND | ND |
| Limulus | 40 | 68 | ND | ND |
| Vertebrate skeletal | 6 | 37 | 42 | 65 |
a The number of molecules showing the compact conformation (apposed heads) in relaxing conditions following different treatments (Figure 5 legend), were counted from five micrographs (each of a 0.75 μm x 0.75 μm region of the EM specimen), in each case. The number of compact molecules is shown as a percentage of the total number of molecules (about 100–150 total in each condition). ND, not determined.
The molecules showed two main views, which were approximately “mirror images” of each other and were distinguishable by the orientation of the tail with respect to the heads. The orientation depended on whether the molecules adhered to the grid surface with their heads facing toward or away from the carbon substrate (Figure 2, A and B, galleries, right and left views). Within these orientations, molecules with similar appearances were grouped into classes, and each class was averaged (Burgess et al., 2004), improving the structural detail visible (Figure 2, C–J). Image averages of each class, as well as global averages containing all classes, clearly showed the disposition of the heads. One head (black, Figure 2, C–J, diagrams) curved toward and appeared to contact the other head (white), which appeared to be straighter. This appearance is essentially identical to that seen in inactive vertebrate smooth-muscle myosin molecules (Burgess et al., 2007). The black head corresponds to the “blocked” head described previously (Wendt et al., 2001; Burgess et al., 2007) and the white head to the “free” head (white arrows in global averages in Figure 2, C, E, G, and I). This asymmetric head arrangement, with apparent contact between the motor domains, is seen in all the class averages (Figure 2, D, F, H, and J).
For both tarantula and Limulus, there were fewer right than left views of the molecules (Figure 2, C–J). This suggests that the compact structure may be more stable (or bind more readily) in one orientation on the carbon film than in the other. The possibility that head structure can be perturbed by binding to substrates used in EM specimen preparation is suggested by previous EM studies of myosin (Trinick and Elliott, 1982; Craig et al., 1983; Knight and Trinick, 1984; Trybus and Lowey, 1984; Stafford et al., 2001).
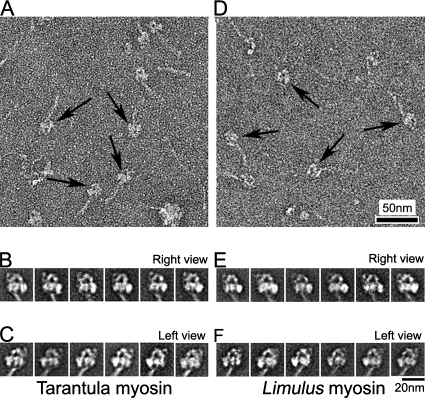
To reduce such effects, we tried cross-linking the molecules in solution to stabilize their structure before applying to the grid. After cross-linking, the total number of molecules seen was significantly greater than without cross-linking, suggesting that cross-linking at low concentration (0.1% glutaraldehyde for 1 min) was intramolecular, rather than intermolecular, and that it trapped molecules in the compact state (Figure 3, A and D), so that they were less able to form filaments on dilution with relaxing buffer (see Materials and Methods). For both myosins, the percentage of compact molecules was ∼50% greater than in unfixed specimens (Table 1). Average images showed that the detailed shape and asymmetric appearance of the two heads were well preserved (Figure 3, B, C, E, and F), and remarkably similar to unfixed molecules (cf. Figure 2). This suggests that fixation does not induce large changes in the overall structure of the compact molecules, especially the closely packed head region, but rather helps to stabilize the native compact structure present in solution. We conclude that fixation can be a useful tool for stabilizing the solution structure of single myosin molecules for EM.
Figure 3.
Myosin conformation after glutaraldehyde cross-linking. (A and D) Fields of cross-linked tarantula and Limulus myosin molecules, respectively. Arrows indicate compact molecules. (B, C, E, and F) class averages of 20–40 images each of right and left views of tarantula and Limulus myosin. Total number of images used were 284 (B), 373 (C), 240 (E), and 303 (F).
2. Head–Head Interaction in Nonmuscle Myosin II in Relaxing Conditions
Smooth muscle myosin and nonmuscle myosin II have very similar biochemical and structural properties (Craig et al., 1983; Sellers and Adelstein, 1986; Kendrick-Jones et al., 1987). Both are regulated by RLC phosphorylation, and both form a folded (10S) conformation in the dephosphorylated (switched-off) state in vitro. We therefore sought to determine whether nonmuscle myosin, like smooth-muscle myosin (Wendt et al., 2001; Burgess et al., 2007), exhibited head–head interaction in the dephosphorylated state. Figure 4, A and B, show that nonmuscle myosin IIA molecules have a compact structure similar to that seen with smooth-muscle myosin (Burgess et al., 2007), and with tarantula and Limulus myosins in this study (Figures 2 and 3). Image classification and averaging reveal clear head–head interaction, indistinguishable from that seen with the other myosins (Figure 4, C and D). We conclude that the structural mechanism that switches off nonmuscle myosin II is similar to that in smooth-muscle myosin.
Figure 4.
Compact head conformation of unphosphorylated nonmuscle myosin IIA. (A and B) Fields of non-cross-linked and cross-linked nonmuscle myosin, respectively; arrows indicate compact heads. (C and D) Global and selected class averages (left view) from head-aligned images of cross-linked molecules (359 images total). Class averages contain 20–40 images each. White arrow indicates free head.
3. Head–Head Interaction in Unregulated (Vertebrate Striated) Muscle Myosin in Relaxing Conditions
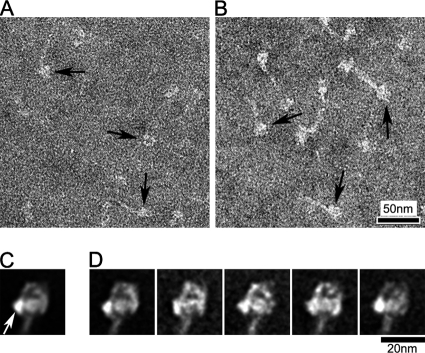
It has been suggested that the head–head interaction we have been describing is the main mechanism that inactivates the motor function of myosin II molecules regulated by phosphorylation (Wendt et al., 2001; Woodhead et al., 2005; Burgess et al., 2007), thus bringing about muscle relaxation. We tested this hypothesis by studying the structure of freshly isolated vertebrate striated muscle myosin (both skeletal and cardiac), which is thought to have no intrinsic regulation and therefore to lack an off-state (Lehman and Szent-Gyorgyi, 1975). In relaxing solution, most skeletal muscle myosin molecules showed heads with varying orientations (Figure 5A, white arrows), and only a few (∼6%) had closely apposed heads (Figure 5A, black arrow; Table 1). When the molecules were cross-linked before negative staining, as described for tarantula and Limulus, the number with closely apposed heads rose to 37% (Figure 5B, black arrows; Table 1); however, most still had separated heads, with variable orientations (Figure 5B, white arrows). These observations suggest that unregulated myosin can also adopt the interacting-head structure in relaxing conditions, but that the head interaction is weaker, and therefore less common in solution, than for regulated myosins.
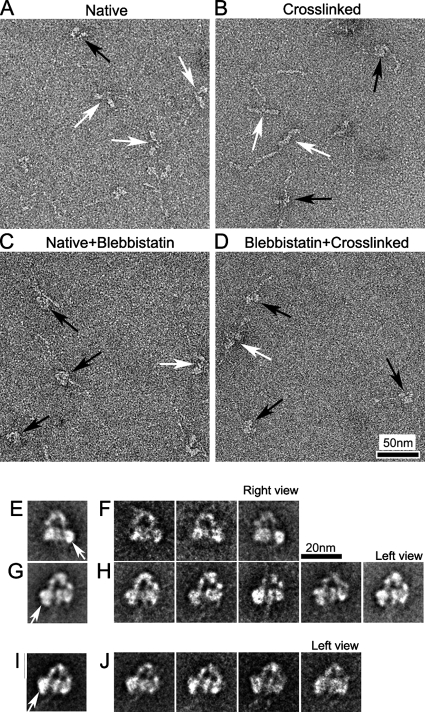
Figure 5.
Structure of vertebrate skeletal myosin molecules after different chemical treatments under relaxing conditions. (A–D) Fields of (A) Native (no treatment), (B) Cross-linking (treated with 0.1% glutaraldehyde for 1 min), (C) Native+Blebbistatin (treated with 400 nM blebbistatin), and (D) Blebbistatin+Cross-linking (treated with 400 nM blebbistatin and then cross-linked). White and black arrows in fields indicate molecules with clearly separated or closely packed heads, respectively. (E and G) Global averages of blebbistatin-treated molecules. Right and left views from 65 and 125 images, respectively. (F and H) Selected class averages of blebbistatin-treated molecules, containing 20–40 images in each. (I and J) Global and selected class averages of 113 images of blebbistatin-treated and cross-linked molecules. Each class average contains 20–40 images. (E, G, and I) White arrows indicate free head motor domain. Scale bar, 20 nm in F applies to all average images.
Regulated myosins (vertebrate smooth and scallop striated muscle) are known to “trap” ADP.Pi in the active site, thereby reducing ATPase activity to extremely low levels in the inactive state (Cross et al., 1986; Ankrett et al., 1991). Although vertebrate striated muscle myosin is also mostly in the ADP.Pi state in relaxing conditions, ADP.Pi is not trapped, and ATP turnover still occurs at an appreciable rate (Katoh et al., 1998; Takahashi et al., 1999). This may contribute to the variable conformations seen. The ADP.Pi state can be prolonged, however, by addition of the myosin inhibitor, blebbistatin, which favors closing of the “switch 2” element of the ATP-binding site, inhibiting Pi release (Kovacs et al., 2004). In this condition, the number of molecules showing closely packed heads was 42%, even without cross-linking, similar to the levels seen with untreated tarantula and Limulus myosin molecules (Figure 5C). This increased further (to 65%), when blebbistatin-treated molecules were cross-linked, similar to the cross-linked regulated myosins without blebbistatin treatment (Figure 5D; Table 1). Thus vertebrate skeletal muscle myosin can adopt the compact head conformation in the ADP.Pi state, like regulated muscle myosin under relaxing conditions.
Image classification and averaging of blebbistatin-treated skeletal myosin molecules showed that the compact structure was remarkably similar to that of regulated myosin (Figure 5, E–H), with one head (blocked) bent toward the other head (free, indicated by white arrows in Figure 5, E and G). This asymmetric interaction between the motor domains was consistently seen in class averages. The same head–head interaction was also seen in average images of cross-linked, blebbistatin-treated molecules (Figure 5, I and J).
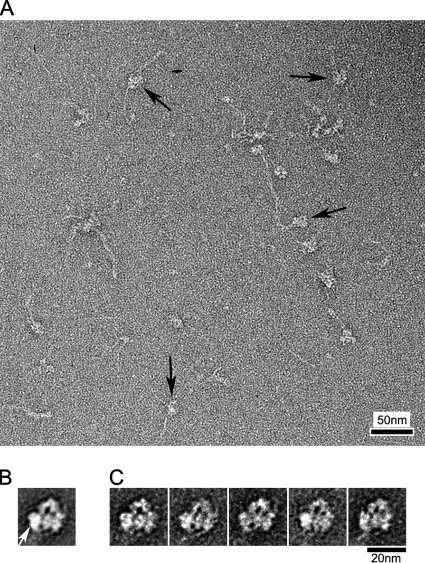
Vertebrate cardiac muscle myosin showed a similar, although weaker, tendency to form the same compact structure (Figure 6A). Although relatively few untreated molecules had this conformation, the number after blebbistatin treatment and cross-linking (63%) was similar to skeletal myosin. As with cross-linked nonmuscle and skeletal muscle myosin II (Figures 4 and 5), only left-view molecules aligned well enough to show detailed structure after averaging (Figure 6, B and C). Class averages consistently showed head–head interaction (Figure 6C), similar to that seen with relaxed regulated myosins.
Figure 6.
Head–head interaction in vertebrate cardiac muscle myosin. (A) Field of blebbistatin-treated, cross-linked molecules; arrows indicate molecules with compact head appearance. (B and C) Global (B) and class averages (C) from 144 left view images. Each class average contains 20–40 images. White arrow indicates free head motor domain.
The visual similarity of the interaction seen in regulated (Figures 2–4) and unregulated (Figures 5 and 6) myosins was striking. It was supported by two-dimensional (2D) fitting of the atomic model of regulated HMM (PDB 1i84; Wendt et al., 2001) to image averages of negatively stained skeletal myosin molecules (Figure 7, A–C). Although the shape of the regulatory domain, mainly associated with the regulatory light chain, does not match perfectly, the motor domain and ELC fit well. A similar 2D fit is obtained with cardiac myosin (Figure 7, D–F). We conclude that the heads of unregulated myosin (vertebrate skeletal and cardiac) in the ADP.Pi (relaxed) state can undergo head–head interactions similar to those of inactivated heads of regulated myosins.
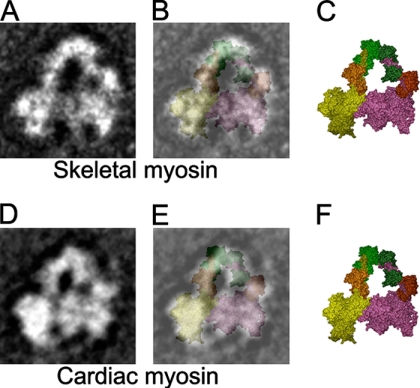
Figure 7.
Fitting of interacting-head atomic model (PDB: 1i84) to vertebrate skeletal and cardiac myosin average images. (A) Selected average of blebbistatin-treated skeletal myosin (left view, from Figure 5H). (B) Superposition of equivalent view of atomic model (C; color scheme as in Figure 1) on A. (D) Global average of blebbistatin-treated and cross-linked cardiac myosin (left view, from Figure 6B). (E) Superposition of equivalent view of atomic model (F) on D.
4. Intramolecular Interaction between the Blocked Head and S2 in Relaxed Myosins
Although the heads of regulated and unregulated myosins in the relaxed state appear similar, there are differences in the folding of the tail. The difference is especially clear in shadowed specimens (Figure 8, C and D; cf. Figure 8, A and B), in which the tails are contrasted most clearly, but is also seen in negatively stained molecules (Figure 8, E and F; cf. Figures 2–4). In the case of regulated myosins, the tail folds into three similar length segments that pack closely together, giving the appearance of a short but wide tail (Figures 2–4, 8A and B; Burgess et al., 2007). In contrast, unregulated myosin molecules fold only once (black arrows, Figure 8, C and D), at a position similar to the first fold seen in regulated molecules, creating one short and one long segment, as found previously (Katoh et al., 1998; Takahashi et al., 1999).
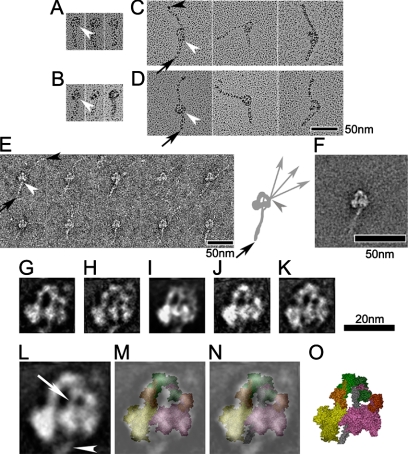
Figure 8.
Interaction between S2 and the blocked head. (A–D) Metal shadowed images of tarantula (A), Limulus (B), skeletal (C), and cardiac (D) myosins after cross-linking. (E and F) Gallery of non-cross-linked skeletal myosin molecules (E), together with their average (F). The first tail segment points down in all images (A–F). White arrowheads indicate point of emergence of S2 from the blocked head, and black arrowheads the tip of the tail. Black arrows in C–E point to the first bend in the tail. Diagram in E shows pathway of short (head to bend position) and long segments of tail. Note that long segment binds to blocked head in a position distinct from S2 (Burgess et al., 2007); C-terminal of tail is flexible beyond this point (gray arrowhead), with three different directions shown. (G–K) Average images of cross-linked myosins produced from tarantula (G; 20 images), Limulus (H; 16 images), nonmuscle (I; 20 images), skeletal (J; 36 images), and cardiac (K; 32 images). (L) Average image in the head region, produced from averaging images shown in G–K. (M) Superposition of (L) on equivalent left view of atomic model (PDB: 1i84; color scheme as in Figure 1). (N) Superposition of (L) on the atomic model with S2 (gray coiled-coil) included (O; Blankenfeldt et al., 2006). White arrow and arrowhead in L show S2 passing from head–head junction to blocked head and then emerging from blocked head, respectively. This is also seen in all individual average images (G–K).
Despite these differences in folding, the first segment, comprising most of S2, appears to follow the same path in averaged images of all the regulated and unregulated myosins that we have studied. Emanating from the junction of the two light-chain domains, the tail crosses the hole between the two heads and emerges from the middle of the far side of the blocked head (Figure 8, G–K). We interpret this segment as S2, rather than one of the other tail segments, because it has a similar appearance in heavy meromyosin (HMM; Burgess et al., 2007), which lacks the other segments, and in myosin molecules in thick filaments (Woodhead et al., 2005). Furthermore, in intact, regulated myosins, the other two segments follow a different path, distinct from S2, around the outside of the heads (Burgess et al., 2007; Jung et al., 2008). In the unregulated myosins, the tail shows significant flexibility beyond its point of emergence from the heads, demonstrated by comparing selected individual images (Figure 8E) and by the loss of tail density beyond the emergent point in image averages (Figure 8F).
The consistent path of the first segment of the tail in all the myosins suggests that, in addition to the head–head interaction, there is also interaction between the blocked head and S2. This may help to stabilize the interacting head structure in both regulated and unregulated myosins.
5. Activation Breaks the Head–Head Interaction
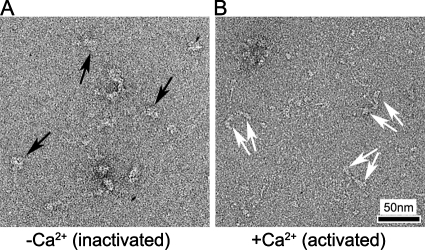
Previous studies of vertebrate smooth and invertebrate striated muscle myosin have suggested that the heads act independently of each other when activated, consistent with breaking of the head–head interaction on activation (Stafford et al., 2001; Wendt et al., 2001). However, structural observations of activation in single molecules did not clearly resolve the head–head interaction in the relaxed state (Stafford et al., 2001). We tested whether head interaction is indeed lost on activation by comparing scallop myosin in relaxing (low Ca2+) and activating (high Ca2+) conditions. In relaxing conditions, many scallop myosin molecules showed a folded tail and a compact arrangement of interacting heads (Figure 9A; Jung et al., 2008), as found in tarantula, Limulus, nonmuscle myosin IIA, and smooth-muscle myosin. In high Ca2+, the heads were generally separate from each other with varying angles between them (Figure 9B), implying breaking of the intramolecular interaction between heads on activation.
Figure 9.
Organization of myosin heads on activation. (A and B) Fields of scallop striated muscle myosin molecules in low- (A) and high- (B) calcium states. In A, black arrows show compact heads, whereas in B, paired white arrows point to separated pairs of heads in different molecules.
DISCUSSION
Head–Head Interaction in Regulated Myosin
Intramolecular interaction between motor domains has been shown to occur in vertebrate smooth-muscle myosin II molecules in the inactive (dephosphorylated) state (Wendt et al., 2001; Burgess et al., 2007). This interaction suggests a simple model for inactivating the heads, by blocking the actin-binding site of one and the ATP hydrolysis site of the other. We have shown here that this structure, and therefore presumably the proposed blocking mechanism, extends beyond smooth-muscle myosin to include different striated muscle and nonmuscle myosin IIs as well. Its presence in tarantula and Limulus striated muscle myosin (in addition to smooth muscle; Figures 2 and 3) suggests that it is conserved across phosphorylation-regulated muscles. Observation of essentially the same motif in striated muscle myosin regulated by Ca2+ binding to the essential light chain implies a similar off-state structure in this distinctly different regulatory system (Jung et al., 2008). Its presence in nonmuscle myosin IIA (phosphorylation-regulated; Figure 4) suggests that this model also encompasses nonmuscle systems.
Three-dimensional (3D) reconstruction of native myosin filaments from tarantula (Woodhead et al., 2005) shows that essentially the same head–head interaction also occurs in vivo. Our observation of this structure in tarantula myosin molecules provides the first direct correlation between molecule and filament in the same species. This suggests that extrapolation of observations from molecule to filament in other species is justified. 3D reconstructions of native filaments from Limulus and scallop muscle show similar interacting head features (Zhao et al., 2008), supporting this view. We conclude that head–head interaction is a common feature of most, if not all, regulated myosin II molecules and filaments under relaxing conditions.
Head–Head Interaction in Unregulated Myosin
Our results suggest that a substantial proportion of vertebrate striated muscle myosin molecules, which are thought to lack an intrinsic regulatory mechanism (Lehman and Szent-Gyorgyi, 1975), undergo similar head–head interaction in solution, but that the interaction is weak (and/or short lived), and easily broken (see also Harris et al., 2003). The lability of this structure in unregulated myosins may explain why it has not been observed previously (Walker et al., 1985; Katoh et al., 1998; Takahashi et al., 1999).
The presence of head–head interaction in vertebrate striated muscle myosin molecules under relaxing conditions suggests that relaxed vertebrate thick filaments may also have this head arrangement. This is supported by the excellent fit of the interacting-head atomic model (PDB 1i84) to the myosin heads in 3D reconstructions of mouse cardiac thick filaments (Zoghbi et al., 2008). Thus unregulated myosin filaments and molecules from vertebrate striated muscle both exhibit head–head interaction in the relaxed state. The closing of the switch II element of the nucleotide binding site in relaxed muscle, inhibiting Pi release, may allow bending of the head at the so-called “pliant region” in the head (Houdusse et al., 2000; see Supplemental Discussion), enabling head–head interaction, which may in turn be necessary for the helical ordering of the heads that characterizes relaxed filaments. This view is supported by the findings that the closing of switch II is required for helical ordering (Zoghbi et al., 2004), and that blebbistatin, which promotes this closing, enhances both the number of interacting-head molecules (Table 1), and the ordering of myosin heads in thick filaments (Zhao et al., 2005; Zoghbi et al., 2008).
We conclude that head–head interaction is a highly conserved motif present in both regulated and unregulated myosin II systems in the relaxed state. Head–head (and other) interactions apparently serve to fully switch off activity in regulated myosins. In unregulated myosins they may serve to “park” the myosin heads in an ordered array near the thick filament surface in relaxed muscle, minimizing their interaction with thin filaments. Such parked, intramolecularly interacting heads would offer minimal resistance to re-extension of muscles after shortening, minimizing the energy cost of the contraction–relaxation cycle (Zoghbi et al., 2004). Our results on unregulated molecules and filaments imply that head–head interaction per se is insufficient to switch myosin off biochemically. In regulated myosins, additional interactions (e.g., involving the RLC or the myosin tail) may strengthen the head–head interaction, causing activity to be switched off.
Putative Ionic Interactions at the Head–Head and Head–Tail Interfaces
Scallop myosin is known to switched on by high salt in the absence of Ca2+, implying that ionic attraction plays a key role in stabilizing the off-state (Nyitrai et al., 2003). We have used comparative sequence analysis and molecular modeling to search for possible ionic interactions that may be important in stabilizing the head–head interaction (see Supplemental Discussion; Figures S1–S3). Our analysis suggests that D748 in the converter domain of the free head may interact with K368 of the blocked head and that R406 or K416 of the blocked head may interact with E169 of the free head. Mutational analysis shows that charge attraction is also critical to the switched off-state of myosin V: in this case attraction appears to occur between single charged residues in the myosin head and the globular tail domain (Li et al., 2008). Similar mutational analysis could test the significance of the charge interactions we have proposed for myosin II.
Interestingly, mutation of R406 (R403 in cardiac myosin) causes significant changes in motor activity of myosin, leading to familial hypertrophic cardiomyopathy (FHC; Palmiter et al., 2000; Yamashita et al., 2000). This mutation has been suggested to affect cardiac contractility through alteration in myosin kinetics. Our analysis suggests that changes in contractility could additionally relate to a change in the stability of the head–head interaction when cardiac muscle relaxes. Mutation of R731 (R719 of cardiac myosin), close to the putative D748/K368 charge interaction (Supplemental Figure S2B) also causes FHC. When mutated to glutamine, smooth-muscle myosin can move actin even in the unphosphorylated state (Yamashita et al., 2000; Kohler et al., 2002), consistent with breaking of the head–head interaction. Thus, although R731 may not be an integral part of the motor domain interaction, it may influence the putative nearby K368–D748 ionic interaction.
The possibility that interaction of the blocked head with S2 was involved in the off-state was first suggested in studies of tarantula filaments (Woodhead et al., 2005) and is supported by images of smooth and scallop HMM molecules (Jung et al., 2008). Our observations add further weight to this concept (Figure 8 and Supplemental Discussion) and demonstrate that it also applies to unregulated myosins. The physiological importance of S2–head interaction is implied by the loss of regulation that occurs when smooth-muscle myosin S2 is truncated so that it can no longer reach the blocked head (Trybus et al., 1997). We conclude that interaction of S2 with the blocked head is an important contributor to the relaxed state of myosin. It has been suggested that this interaction occurs between a patch of negative charge on S2 (E894-D906) and a conserved patch of positively charged residues in the actin-binding “loop 2” (residues 627–646) of the blocked head (Blankenfeldt et al., 2006). The point mutation L908V in cardiac S2 alters the kinetics of actin-myosin interaction, and results in FHC (Palmiter et al., 2000; Yamashita et al., 2000). One interpretation of this effect is that L908, only two residues from D906, in some way influences the interaction between S2 and the blocked head and that this interaction occurs transiently during the cross-bridge cycle, not only in the relaxed state.
Intramolecular Interaction: A Common Theme for Switching Off Molecular Motors
Intramolecular interaction appears to be a common mechanism for switching off other motor proteins as well as myosin II. Kinesin is switched off by interaction of the heavy chain tail with the motor domains (Friedman and Vale, 1999). Similarly, myosin V is inactivated by intramolecular interaction between the globular tail domain and the heads (Li et al., 2006; Thirumurugan et al., 2006). In both cases, activity is switched on by binding of cargo to the tail domain, displacing it from the heads. In the case of regulated myosin II, switching on breaks the head–head interaction, switching on ATPase activity and enabling the heads to extend from the filament backbone and bind to actin. Although vertebrate striated muscle myosin is not regulated, we have shown that it can still form head–head interactions (probably weak), which may be required for “parking” the heads in their ordered array on relaxed thick filaments. Phosphorylation in this case enhances muscle contractility (Sweeney et al., 1993), presumably by further weakening the head–head interaction, facilitating extension of the heads from the thick filament and thus binding to actin (Levine et al., 1996).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Wulf Blankenfeldt (Max Planck Institute of Molecular Physiology, Dortmund, Germany) for the PDB file of smooth-muscle myosin heads with attached S2, and Dr. Dong Young Kim for help with modeling flexible loops at the head–head junction. We are grateful to Dr. Maria-Elena Zoghbi for help with preparing mouse striated muscles and to her and Dr. John Woodhead for discussions and comments on the manuscript. This work was supported by National Institutes of Health Grants AR34711 (to RC) and DC006103, AR048526, and AR048898 (to MI). Electron microscopy was carried out in the Core Electron Microscopy Facility of the University of Massachusetts Medical School, supported in part by Diabetes Endocrinology Research Center Grant DK32520. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
Abbreviations used:
- ELC
essential light chain
- RLC
regulatory light chain
- HMM
heavy meromyosin
- S2
subfragment 2 of myosin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0206) on May 21, 2008.
REFERENCES
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. New York: Garland Science; 2007. [Google Scholar]
- Ankrett R. J., Rowe A. J., Cross R. A., Kendrick-Jones J., Bagshaw C. R. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J. Mol. Biol. 1991;217:323–335. doi: 10.1016/0022-2836(91)90546-i. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt W., Thoma N. H., Wray J. S., Gautel M., Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc. Natl. Acad. Sci. USA. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S. A., Walker M. L., Thirumurugan K., Trinick J., Knight P. J. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Burgess S. A., Yu S., Walker M. L., Hawkins R. J., Chalovich J. M., Knight P. J. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Smith R., Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- Craig R., Woodhead J. L. Structure and function of myosin filaments. Curr. Opin. Struct. Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Cross R. A., Cross K. E., Sobieszek A. ATP-linked monomer-polymer equilibrium of smooth muscle myosin: the free folded monomer traps ADP.Pi. EMBO J. 1986;5:2637–2641. doi: 10.1002/j.1460-2075.1986.tb04545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. S., Vale R. D. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Geeves M. A., Holmes K. C. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- Harris S. P., Heller W. T., Greaser M. L., Moss R. L., Trewhella J. Solution structure of heavy meromyosin by small-angle scattering. J. Biol. Chem. 2003;278:6034–6040. doi: 10.1074/jbc.M210558200. [DOI] [PubMed] [Google Scholar]
- Hidalgo C., Padron R., Horowitz R., Zhao F. Q., Craig R. Purification of native myosin filaments from muscle. Biophys. J. 2001;81:2817–2826. doi: 10.1016/S0006-3495(01)75923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A., Szent-Gyorgyi A. G., Cohen C. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley H. E., Brown W. The low-angle X-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- Ikebe M., Hartshorne D. J. Effects of Ca2+ on the conformation and enzymatic activity of smooth muscle myosin. J. Biol. Chem. 1985;260:13146–13153. [PubMed] [Google Scholar]
- Jung H. S., Burgess S. A., Billington N., Colegrave M., Patel H., Chalovich J. M., Chantler P. D., Knight P. J. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. USA. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T., Konishi K., Yazawa M. Skeletal muscle myosin monomer in equilibrium with filaments forms a folded conformation. J. Biol. Chem. 1998;273:11436–11439. doi: 10.1074/jbc.273.19.11436. [DOI] [PubMed] [Google Scholar]
- Kendrick-Jones J., Smith R. C., Craig R., Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J. Mol. Biol. 1987;198:241–252. doi: 10.1016/0022-2836(87)90310-x. [DOI] [PubMed] [Google Scholar]
- Knight P., Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J. Mol. Biol. 1984;177:461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- Kohler J., Winkler G., Schulte I., Scholz T., McKenna W., Brenner B., Kraft T. Mutation of the myosin converter domain alters cross-bridge elasticity. Proc. Natl. Acad. Sci. USA. 2002;99:3557–3562. doi: 10.1073/pnas.062415899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs M., Toth J., Hetenyi C., Malnasi-Csizmadia A., Sellers J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Lehman W., Szent-Gyorgyi A. G. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J. Gen. Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. J., Kensler R. W., Yang Z., Stull J. T., Sweeney H. L. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys. J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. D., Jung H. S., Mabuchi K., Craig R., Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J. Biol. Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- Li X. D., Jung H. S., Wang Q., Ikebe R., Craig R., Ikebe M. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc. Natl. Acad. Sci. USA. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wendt T., Taylor D., Taylor K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- Lymn R. W., Taylor E. W. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- Nyitrai M., Stafford W. F., Szent-Gyorgyi A. G., Geeves M. A. Ionic interactions play a role in the regulatory mechanism of scallop heavy meromyosin. Biophys. J. 2003;85:1053–1062. doi: 10.1016/S0006-3495(03)74544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter K. A., Tyska M. J., Haeberle J. R., Alpert N. R., Fananapazir L., Warshaw D. M. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J. Muscle Res. Cell Motil. 2000;21:609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- Perry S. V. Activation of the contractile mechanism by calcium. In: Engel A. G., Franzini-Armstrong C., editors. Myology. New York: McGraw-Hill; 2004. pp. 281–306. [Google Scholar]
- Persechini A., Hartshorne D. J. Ordered phosphorylation of the two 20 000 molecular weight light chains of smooth muscle myosin. Biochemistry. 1983;22:470–476. doi: 10.1021/bi00271a033. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Sellers J. R. Phosphorylation-dependent regulation of Limulus myosin. J. Biol. Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- Sellers J. R. Regulation of cytoplasmic and smooth muscle myosin. Curr. Opin. Cell Biol. 1991;3:98–104. doi: 10.1016/0955-0674(91)90171-t. [DOI] [PubMed] [Google Scholar]
- Sellers J. R., Adelstein R. S. Regulation of contractile activity. In: Boyer P. D., Krebs E. G., editors. The Enzymes. Orlando: Academic Press; 1986. pp. 381–418. [Google Scholar]
- Stafford W. F., Jacobsen M. P., Woodhead J., Craig R., O'Neall-Hennessey E., Szent-Gyorgyi A. G. Calcium-dependent structural changes in scallop heavy meromyosin. J. Mol. Biol. 2001;307:137–147. doi: 10.1006/jmbi.2000.4490. [DOI] [PubMed] [Google Scholar]
- Sweeney H. L., Bowman B. F., Stull J. T. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am. J. Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- Sweeney H. L., Houdusse A. Mammalian muscle myosin. In: Engel A. G., Franzini-Armstrong C., editors. Myology. New York: McGraw-Hill; 2004. pp. 167–186. [Google Scholar]
- Szent-Gyorgyi A. G., Kalabokis V. N., Perreault-Micale C. L. Regulation by molluscan myosins. Mol. Cell Biochem. 1999;190:55–62. [PubMed] [Google Scholar]
- Takahashi T., Fukukawa C., Naraoka C., Katoh T., Yazawa M. Conformations of vertebrate striated muscle myosin monomers in equilibrium with filaments. J. Biochem. 1999;126:34–40. doi: 10.1093/oxfordjournals.jbchem.a022433. [DOI] [PubMed] [Google Scholar]
- Tama F., Feig M., Liu J., Brooks C. L., III, Taylor K. A. The requirement for mechanical coupling between head and S2 domains in smooth muscle myosin ATPase regulation and its implications for dimeric motor function. J. Mol. Biol. 2005;345:837–854. doi: 10.1016/j.jmb.2004.10.084. [DOI] [PubMed] [Google Scholar]
- Thirumurugan K., Sakamoto T., Hammer J. A., III, Sellers J. R., Knight P. J. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick J., Elliott A. Effect of substrate on freeze-dried and shadowed protein structures. J. Microsc. 1982;126:151–156. doi: 10.1111/j.1365-2818.1982.tb00364.x. [DOI] [PubMed] [Google Scholar]
- Trybus K. M., Freyzon Y., Faust L. Z., Sweeney H. L. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc. Natl. Acad. Sci. USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K. M., Huiatt T. W., Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K. M., Lowey S. Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J. Biol. Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- Walker M., Knight P., Trinick J. Negative staining of myosin molecules. J. Mol. Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- Wendt T., Taylor D., Trybus K. M., Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Zhao F. Q., Craig R., Egelman E. H., Alamo L., Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- Xu J. Q., Harder B. A., Uman P., Craig R. Myosin filament structure in vertebrate smooth muscle. J. Cell Biol. 1996;134:53–66. doi: 10.1083/jcb.134.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H., Tyska M. J., Warshaw D. M., Lowey S., Trybus K. M. Functional consequences of mutations in the smooth muscle myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. J. Biol. Chem. 2000;275:28045–28052. doi: 10.1074/jbc.M005485200. [DOI] [PubMed] [Google Scholar]
- Zhao F. Q., Craig R. Ca2+ causes release of myosin heads from the thick filament surface on the milliseconds time scale. J. Mol. Biol. 2003;327:145–158. doi: 10.1016/s0022-2836(03)00098-6. [DOI] [PubMed] [Google Scholar]
- Zhao F. Q., Craig R., Padron R. Structural effects of blebbistatin on muscle thick filaments. Biophysical Society Meeting Abstracts. Biophys. J. 2005;(Supplement) Abstract, 632-Pos. [Google Scholar]
- Zhao F. Q., Woodhead J. L., Craig R. Head-head interaction characterizes the relaxed state of scallop and Limulus muscle myosin filaments. Biophysical Society Meeting Abstracts. Biophys. J. 2008;(Supplement) doi: 10.1016/j.jmb.2008.10.038. Abstract, 630-Pos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi M. E., Woodhead J. L., Craig R., Padron R. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J. Mol. Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Zoghbi M. E., Woodhead J. L., Moss R. L., Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.