Abstract
Cellular stress by DNA damage induces checkpoint kinase-2 (CHK2)-mediated phosphorylation and stabilization of the E2F1 transcription factor, leading to induction of apoptosis by activation of a subset of proapoptotic E2F1 target genes, including Apaf1 and p73. This report characterizes an interaction between the aryl hydrocarbon (Ah) receptor (AHR), a ligand-activated transcription factor, and E2F1 that results in the attenuation of E2F1-mediated apoptosis. In Ahr−/− fibroblasts stably transfected with a doxycycline-regulated AHR expression vector, inhibition of AHR expression causes a significant elevation of oxidative stress, γH2A.X histone phosphorylation, and E2F1-dependent apoptosis, which can be blocked by small interfering RNA-mediated knockdown of E2F1 expression. In contrast, ligand-dependent AHR activation protects these cells from etoposide-induced cell death. In cells expressing both proteins, AHR and E2F1 interact independently of the retinoblastoma protein (RB), because AHR and E2F1 coimmunoprecipitate from extracts of RB-negative cells. Additionally, chromatin immunoprecipitation assays indicate that AHR and E2F1 bind to the Apaf1 promoter at a region containing a consensus E2F1 binding site but no AHR binding sites. AHR activation represses Apaf1 and TAp73 mRNA induction by a constitutively active CHK2 expression vector. Furthermore, AHR overexpression blocks the transcriptional induction of Apaf1 and p73 and the accumulation of sub-G0/G1 cells resulting from ectopic overexpression of E2F1. These results point to a proproliferative, antiapoptotic function of the Ah receptor that likely plays a role in tumor progression.
INTRODUCTION
Members of the E2F family of transcription factors are critical regulators of the G1/S phase transition of the cell cycle, during which their transcriptional activity is generally controlled through interaction with retinoblastoma (RB) family proteins. In addition, E2F proteins have functions beyond the G1/S phase transition that impact cell proliferation in a variety of ways (Dimova and Dyson, 2005). The E2F family consists of six extensively characterized and three less well-studied members. E2F1, -2, and -3a are potent activators of transcription, bind exclusively to RB-p105, and are cyclically expressed during the cell cycle. E2F3b and -4, which can interact with RB-p107 and -p130, and E2F5, which binds only to p130, are poor transcriptional activators, and they function mainly as repressors through their recruitment of RB proteins to E2F-regulated promoters (Dyson, 1998; Nevins, 1998; DeGregori and Johnson, 2006). In general, the E2Fs with transactivator activity promote cell cycle progression, whereas the E2Fs with transrepressor activity function in cell cycle exit and differentiation (Dimova and Dyson, 2005). E2F6-8 are distinct from the other E2F members, lacking the transactivation and RB-binding domains, and they repress transcription in an RB-independent manner (Frolov and Dyson, 2004; DeGregori and Johnson, 2006).
The best-characterized function of E2F is its ability to regulate the G1/S phase transition of the cell cycle. E2F proteins exert control over the cell cycle by modulating the transcription of a variety of essential cell cycle control genes, including cell cycle regulators, RB and related pocket proteins, enzymes for nucleotide biosynthesis, and proteins required for DNA replication (Trimarchi and Lees, 2002). RB modulates E2F-dependent transcription in at least two ways. First, the physical binding of RB to E2F directly inhibits the transactivation activity of E2F. Second, the association of RB with chromatin-modifying corepressor proteins mediates active repression of E2F-responsive genes (Dyson, 1998; Harbour and Dean, 2000; Ferreira et al., 2001). Repressor E2F/RB complexes are prevalent in G0/early G1 phases, and they are disrupted in late G1, resulting primarily from the phosphorylation of RB proteins by cyclin D-CDK4/6 and cyclin E-CDK2 complexes. Late in G1, activator E2Fs turn on the transcription of genes required for entry into S phase and DNA synthesis (Trimarchi and Lees, 2002; DeGregori, 2002; Stevaux and Dyson, 2002). However, the ability of a particular E2F protein to either induce S phase, senescence, or some other outcome is dependent on the cellular context and on the particular groups of up- or down-regulated genes (Dimova and Dyson, 2005).
In addition to its well-established proliferative function, E2F1 has also been implicated in the induction of apoptosis through p53-dependent and p53-independent mechanisms (Phillips et al., 1997; Phillips and Vousden, 2001). Several genes involved in the activation or execution of the apoptotic program are transcriptionally up-regulated by E2F1 overexpression or stabilization in response to DNA damage or to loss of RB, including the genes encoding INK4a/ARF, APAF-1, caspase-7, and TAp73, the proapoptotic p73 variant (Muller et al., 2001; Lin et al., 2001; Nahle et al., 2002; Furukawa et al., 2002; Pediconi et al., 2003; DeYoung and Ellisen, 2007). In human cells, E2F1 stabilization in response to DNA damage results from checkpoint kinase-2 (CHK2)–dependent phosphorylation at Ser364 (Stevens and La Thangue, 2004). Of all E2F family members, induction of apoptosis is predominantly a function of E2F1, with other E2Fs exhibiting a reduced propensity to mediate cell death, but the mechanisms of gene-specific regulation and proproliferative versus proapoptotic target gene selection by E2F1 are not yet fully understood (DeGregori and Johnson, 2006).
The Ah receptor (AHR) is a ligand-activated transcription factor that belongs to the basic-region helix-loop-helix (bHLH)/Per-Arnt-Sim (PAS) family of proteins. Prototypical AHR ligands include many polycyclic and halogenated aromatic compounds, such as benzo[a]pyrene (B[a]P) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). The unliganded AHR is a cytosolic protein that translocates to the nucleus upon ligand binding and dimerizes with a second bHLH/PAS protein, the AHR nuclear translocator (ARNT), to form a heterodimeric transcription factor. AHR/ARNT complexes bind to canonical DNA consensus sequences and initiate transcription of genes coding for many phase I and phase II detoxification enzymes (Hankinson, 1995). In addition to its role in the regulation of drug metabolism, evidence dating back more than 20 years shows that the AHR plays a central role in the regulation of cell proliferation. AHR activation by ligand or by deletion of the ligand-binding domain alters several cell cycle and signaling pathways, including those required for normal cell cycle regulation (Puga et al., 2002; Chang et al., 2007). Most evidence shows that AHR activation delays cell cycle progression and G1 to S phase transition, although this effect seems to be cellular context specific, because in rat oval progenitor cells the AHR promotes rather than delays cell cycle progression (Weiss et al., 2008). A canonical RB-binding cyclin D-like motif in the AHR protein sequence mediates a direct interaction between AHR and RB, and experimental work focused on the characterization of this interaction as the mediator of AHR-dependent cell cycle delay (Ge and Elferink, 1998; Puga et al., 2000; Marlowe et al., 2004; Huang and Elferink, 2005) has shown that the activated AHR cooperates with RB in its ability to repress E2F-dependent transcription and delay cell cycle progression (Puga et al., 2000; Strobeck et al., 2000).
Paradoxically, embryo fibroblasts from Ahr gene-knockout mice also show a relative delay in cell cycle progression, which has been primarily associated with posttranscriptional stabilization of Tgfb1 mRNA (Chang et al., 2007). In these cells, several E2F transcriptional targets with AHR and E2F binding motifs in their promoters, conserved in both the human and mouse sequences (Supplemental Tables S1 and S2), were also found to be AHR targets, being repressed by both E2f1 and Ahr ablation (Supplemental Figure S1). These observations led us to hypothesize that AHR could directly interact with and modulate the transcriptional activity of E2F. We report here that inhibiting the expression of AHR triggers an increase in oxidative stress and DNA damage, leading to induction of E2F1-dependent apoptosis. Conversely, AHR activation leads to formation of AHR-E2F1 protein complexes, resulting in inhibition of E2F1-dependent gene expression and apoptosis.
MATERIALS AND METHODS
Cell Lines, Growth Conditions, and Chemical Treatments
Mouse Hepa-1c1c7 hepatoma cells and human osteosarcoma Saos-2 cells were maintained in minimal essential medium-α (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen), antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO), and 26 mM NaHCO3 at 37°C in a humidified 5% CO2 atmosphere. The stable transfectant TET-OFF cell line Off*Ahrb (Chang et al., 2007) was derived from mouse embryo fibroblasts (MEFs) of AHR knockout mice by sequential retroviral insertion of a vector expressing the regulatory tetracycline receptor protein and a second TET-OFF–regulated vector expressing the high-affinity AHR encoded by the Ahrb1 allele. The cells were grown in medium containing 600 μg/ml G418 (Invitrogen), 3 μg/ml puromycin (A.G. Scientific, San Diego, CA), and 400 μg/ml hygromycin (Calbiochem, San Diego, CA). To down-regulate AHR expression in these cells, doxycycline (Dox; Sigma-Aldrich), a tetracycline analog, was used at a final concentration of 5 μg/ml. Wild-type MEFs from C57BL/6J mice were prepared by standard techniques from 14.5-d-old fetuses and grown in α-minimal essential medium as described above. MEFs from E2f−/− mice were a gift of G. Leone (The Ohio State University). When indicated, cultures were treated with TCDD at a final concentration of 5 nM in dimethyl sulfoxide (DMSO) vehicle, never to exceed 0.1% of the final volume, and control cultures were treated with an equivalent volume of DMSO. Cell survival after etoposide treatment was determined as described previously (Chang et al., 2007). Briefly, cells were seeded in 24-well plates at concentrations ranging from 12,500 to 50,000 cells/well and exposed to the etoposide concentrations indicated in the relevant figures. After 48 h, cells were washed with phosphate-buffered saline (PBS), fixed in 70% ethanol, and incubated with 5 μg/ml Hoechst 33258 (Aldrich Chemical, Milwaukee, WI) at room temperature in the dark for 30 min. Fluorescence at 355/460 nm was measured in a Wallac Victor2 1420 plate reader (PerkinElmer Wallac, Gaithersburg, MD). Saos-2 cells do not express AHR, as determined from the lack of expression of a transfected AHR/ARNT-responsive luciferase reporter plasmid and by immunoblot analysis. Hepa-1 cells do not express RB, as determined by both immunoblot and reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Plasmid and Adenoviral Constructs
The plasmid pcDNAI/B6AHR, used to express the high-affinity murine AHR, and its truncation mutant derivatives, have been described previously (Marlowe et al., 2004). Expression of the peptides encoded by each AHR truncation mutant presented in Supplemental Figure S2 was confirmed by Western blot (data not shown). The reporter plasmid p3XE2FLuc, containing three E2F-responsive elements, has also been described previously (Whitaker et al., 1998). The kinase-dead (KD) and constitutively active (CA) CHK2 expression vectors were a gift of P. Stambrook (University of Cincinnati). The AdEasy XL Adenoviral Vector System (Stratagene, La Jolla, CA) was used to generate recombinant adenovirus for the expression of the high-affinity variant of the murine AHR. A DNA fragment containing the AHR cDNA was removed from the pcDNAI/B6AHR vector and cloned into the pShuttle-cytomegalovirus (CMV) vector to yield the pShuttle-CMV-B6AHR plasmid. This plasmid was linearized and used to transform BJ5183-AD-1 cells, containing the adenovirus plasmid pAdEasy-1, to produce the recombinant adenoviral plasmid pAdB6AHR. For control experiments, pShuttle-CMV-LacZ was similarly processed.
Adenovirus Production and Infection
AD293 cells (Stratagene) transfected with adenoviral plasmids were harvested when 80–90% of the cells detached from the growth plates. Cell pellets were resuspended in PBS and lysed by four rounds of freezing/thawing. An aliquot of the primary virus stock was used for a first round of amplification and purified virus from this first amplification was used for large-scale production of the final virus stock, which was purified by cesium chloride density gradient centrifugation. Saos-2 cells were infected with purified adenovirus containing genes for the expression of LacZ, AHR, RB, and E2F1. Infections were carried out using 100 plaque-forming units (pfu) per cell for 3 h in complete growth medium. After infection, cells were washed twice with PBS, fed fresh growth medium, and harvested after 24 h for coimmunoprecipitation assays.
Transfections and Reporter Assays
Transfection of small interfering RNAs (siRNAs) was done by procedures described previously (Schnekenburger et al., 2007a) based on the recommended manufacturer's neofection protocol (Ambion, Austin, TX). siRNA duplexes were used at a final concentration of 50 nM for predesigned SilencerR E2F1 siRNAs (ID nos. 160617 and 160618) and scrambled negative control (catalog no. 4611). At 48 h posttransfection, cells were collected and used to determine protein expression of siRNA target genes, intracellular reactive oxygen species (ROS), and caspase-3 expression.
Coimmunoprecipitation and Western Immunoblotting
For coimmunoprecipitations with nuclear and cytosolic protein extracts from Hepa-1 cells, ∼300 μg of extract was incubated with 2 μg of anti-AHR (BIOMOL Research Laboratories, Plymouth Meeting, PA) or anti-E2F1 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies covalently coupled to protein A and G beads (Millipore, Billerica, MA) overnight at 4°C. Beads were washed six times, suspended in 30 μl of electrophoresis buffer, and boiled for 5 min. Eluted proteins were analyzed in 7.5% SDS-polyacrylamide gel electrophoresis (PAGE), and AHR and E2F1 were detected by Western immunoblot after semidry electrotransfer of the proteins to polyvinylidene difluoride membranes. Antibodies against BAX and poly(ADP-ribose) polymerase (PARP) were used for detection of these proteins in Off*Ahrb fibroblasts.
For coimmunoprecipitations from extracts of adenovirus-infected Saos-2 cells, cultures were infected with adenovirus vectors for 24 h and treated with vehicle control or 5 nM TCDD for 2 h before harvesting for whole cell extracts. Coimmunoprecipitation reactions were carried out as described above, using ∼1 mg of whole cell extract incubated with 2 μg of anti-AHR antibody. Eluted proteins were analyzed in 7.5% SDS-PAGE, and RB, AHR, and E2F1 were detected by Western immunoblotting.
Chromatin Immunoprecipitation (ChIP) Analyses
ChIP was performed with minor modifications of procedures described previously (Schnekenburger et al., 2007b). Chromatin from formaldehyde cross-linked Hepa-1 cells treated for 90 min with 5 nM TCDD or DMSO vehicle were sheared to a size range of 0.3–0.6 kb by sonication in a crushed-ice/water bath with six 30-s bursts of 200 W, with a 30-s interval between bursts using a Bioruptor (Diagenode, Sparta, NJ). Precleared chromatin was incubated overnight on a rotating platform at 4°C with rabbit polyclonal antibodies specific for AHR (SA-210; BIOMOL Research Laboratories), E2F1 (sc-193X; Santa Cruz Biotechnology), or nonspecific rabbit immunoglobulin G (IgG) (used as an immunoprecipitation control). The immunocomplexes were recovered by a 2-h incubation at 4°C with a 50% gel slurry of protein A-agarose beads (Millipore). After extensive washing, precipitated chromatin complexes were removed from the beads by incubation with elution buffer (50 mM NaHCO3 and 1% SDS), with mild vortexing. This step was repeated, and the eluates were combined. Cross-linking was reversed, and the samples were sequentially digested with RNase A and proteinase K. DNA was purified by chromatography on QIAquick columns (QIAGEN, Valencia, CA), eluted in double distilled H2O, and an aliquot was used for analysis by real-time PCR by using specific primers covering the region between −4.8 kbp and +0.1 kbp of the mouse Apaf1 gene (Supplemental Table S3). PCR products were separated by electrophoresis through 15% polyacrylamide gels and visualized after staining with ethidium bromide.
Total RNA Isolation, Reverse Transcription, and Real-Time RT-PCR
Total cellular RNA was isolated with the RNeasy Mini kit (QIAGEN). First-strand cDNAs were synthesized from 2 to 20 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and random primers. cDNAs were subjected to PCR amplification with gene-specific primer sets (Supplemental Table S4) for the various genes tested. Real-time quantitative PCR was performed using an Opticon 96-well rapid thermal cycler (MJ Research, Watertown, MA). A typical protocol included a 10-min denaturation step at 95°C followed by 40 cycles of 95°C denaturation for 30 s, annealing for 30 s at a primer-optimized temperature, and 72°C extension for 30 s. Detection of the fluorescent product was carried out during the 72°C extension period, and emission data were quantified using threshold cycle (Ct) values. Ct values for all genes analyzed were determined two to six times, averaged, and means were determined from the average Ct values for each biological duplicate. All means were then normalized to values for β-actin. The relative or -fold change from control Ct values was determined for each sample using the equation: -fold change = 2−ΔΔCt, where ΔΔCt = (CtTarget − CtActin)Test − (CtTarget − CtActin)Control. PCR product specificity from each primer pair was confirmed using melting curve analysis and subsequent gel electrophoresis.
Analysis of Reactive Oxygen Species, DNA Damage, and Apoptosis by Flow Cytometry
Relative levels of intracellular ROS were determined as described previously (Peng et al., 2007). Briefly, cells cultured on 10-cm plates were trypsinized, collected by centrifugation, and incubated in fetal bovine serum-free culture medium with 5 μM chloromethyl 2,7-dichlorofluorescein-diacetate (CM-H2DCFDA) for 30 min at 37°C. Cells were washed and suspended at 1 × 106 cells/ml in PBS and analyzed in a FACSCalibur flow cytometer (BD Biosciences Immunocytometry Systems, San Jose, CA) equipped with a 488 argon laser, for measurements of intracellular fluorescence. Mean log fluorescence intensities were determined by the CELLQUEST software program (BD Biosciences). DNA damage was evaluated using a cell-based assay system (Millipore) for the detection of histone γH2A.X phosphorylated at Ser139 in fixed cells. After fixation, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-phospho-histone H2A.X (Ser139) or negative control mouse IgG-FITC and analyzed by flow cytometry. Fluorescent signals were gated at an intensity such that <2% of the negative control cells would score as positive. For analysis of apoptotic cells, we used a flow cytometry kit to detect activated caspase-3 (BD Biosciences). Cells were harvested by trypsinization, fixed, incubated with a phycoerythrin (PE)-conjugated monoclonal rabbit anti-active caspase-3 antibody and analyzed by flow cytometry. In some cases, apoptosis was assessed from the fraction of sub-G0/G1 cells observed in cell cycle analyses of propidium iodide-stained cells.
RESULTS
Loss of AHR Increases Intracellular ROS Levels, Leading to DNA Damage and E2F1-dependent Apoptosis
AHR ablation in mice is associated with increased apoptosis in the liver by mechanisms as yet uncharacterized (Elizondo et al., 2000). Because of the known role of the AHR in cell cycle regulation, we suspected that loss of AHR might lead to loss of a cell cycle checkpoint, which in turn would drive the cells toward an apoptotic pathway. To explore this possibility, we made use of the cell line Off*Ahrb, a TET-OFF stable transfectant derived from MEFs from AHR knockout mice (Chang et al., 2007). These cells carry a TET-OFF–regulated vector expressing the high-affinity AHR receptor encoded by the Ahrb1 allele. Repression or expression of the AHR protein in these fibroblasts depends solely on the presence or absence, respectively, of Dox in the culture medium, hence ruling out genetic differences between clonally unrelated cell lines as a potential source of confounding results. Growth of these cells in the absence of Dox causes them to express the AHR protein, to proliferate at a significantly faster rate, and to deregulate expression of many cell cycle and extracellular matrix genes, compared with cells grown in the presence of Dox and hence lacking AHR expression. Furthermore, exposure to a prototypical AHR ligand or deletion of the ligand-binding domain has not effect on these AHR functions, indicating that they are truly independent of ligand (Chang et al., 2007).
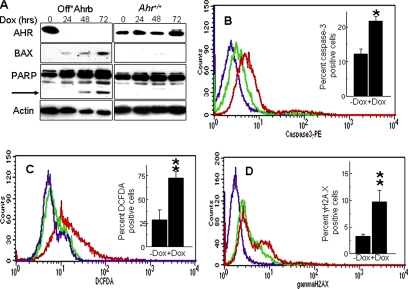
In Off*Ahrb cells, treatment with Dox for 48–72-h represses AHR expression to undetectable levels (Figure 1A). Concomitantly with the AHR repression, BAX expression and PARP cleavage, two well-characterized markers of apoptosis, are both induced (Figure 1A). Dox treatment fails to elicit similar effects from AHR-positive fibroblast cells (Figure 1A), ruling out the trivial explanation that these effects might be caused by Dox itself and thus be unrelated to AHR expression. Consistent with these data, we observe an approximate doubling of the number of cell expressing activated caspase-3 in cells treated with Dox (AHR negative) relative to untreated cells (AHR positive) (Figure 1B), thereby confirming that, as in mouse liver (Elizondo et al., 2000), the lack of AHR expression is also proapoptotic in cultured cells.
Figure 1.
Loss of the Ahr gene causes oxidative stress resulting in DNA damage and apoptosis. (A) Off*Ahrb and Ahr+/+ fibroblasts were treated with 5 μg/ml Dox for 0, 24, 48, and 72 h and analyzed by Western immunoblotting for the expression of AHR, BAX, and PARP proteins. The arrow points at the 89-kDa C-terminal PARP cleavage product resulting from caspase-3/7 activity. Actin was used as a loading control. (B) Detection of activated caspase-3 by flow cytometry. Off*Ahrb cells were treated with 5 μg/ml Dox (red trace) for 48 h or mock treated with an equivalent volume (never to exceed 0.1% of the total volume of culture) of 70% ethanol vehicle (green trace), fixed, and allowed to react with PE-labeled active caspase-3 antibodies. Control cells (purple trace) were unexposed to antibodies. The inset shows the percentage of caspase-3–positive cells, as determined by the CELLQUEST software. (C) Detection of intracellular oxidative stress by flow cytometry. Off*Ahrb cells treated as described in B were exposed to CM-H2DCFDA for 30 min at 37°C and subsequently analyzed by flow cytometry. The purple tracing corresponds to cells unexposed to DCFDA; red and green tracings are for cell treated with Dox or untreated, respectively. The inset represents the percentage of DCFDA-positive cells as determined by the CELLQUEST software. (D) Detection of γH2A.X. Off*Ahrb cells were treated with Dox (red tracing) or left untreated (green tracing) as described above, fixed, and allowed to interact with FITC-tagged antibodies to H2A.X phosphorylated at Ser139 (red and green tracings) or with a control FITC-tagged mouse IgG (purple tracing). The inset represents the percentage of γH2A.X-positive cells as determined by the CELLQUEST software. The values shown in all graphs correspond to the mean ± SD of four to six experiments. *p < 0.05 and **p < 0.01.
To determine whether the increase in apoptosis is the result of a higher level of ROS in AHR-negative cells, we labeled Dox-treated and -untreated cells with CM-H2DCFDA and identified ROS-activated fluorescence-positive cells by flow cytometry. The fluorescence intensity of Dox-treated cells was significantly higher than the intensity of Dox-untreated cells or of cells that were not exposed to CM-H2DCFDA, with about threefold more CM–H2DCFDA positive cells in the Dox-treated, AHR-negative group (Figure 1C), indicating that the absence of AHR results in a higher intrinsic oxidant level.
Persistently high ROS levels are highly reactive and interact with all cellular macromolecules, including DNA, inducing strand breaks and other forms of DNA damage. Phosphorylation of histone H2A.X is a marker of DNA damage, often used as a surrogate for direct DNA damage assays (Tanaka et al., 2007). We therefore used an antibody to phosphorylated γH2A.X to measure the extent of DNA damage in Dox-treated and -untreated cells by flow cytometry. Dox-treated, AHR-negative cells showed higher fluorescence intensities than AHR-positive cells, corresponding to ∼3 times the number of γH2A.X-positive cells and indicative of a greater extent of DNA damage in cells lacking AHR (Figure 1D).
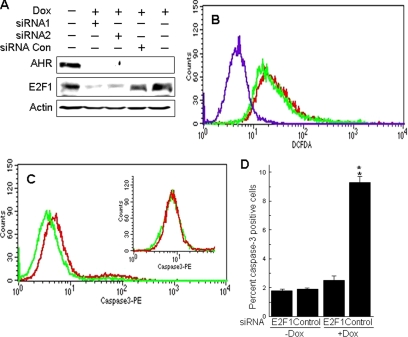
DNA damage is known to activate the ATM-CHK2-E2F1 apoptosis pathway (Stevens and La Thangue, 2004). To determine whether increases in ROS and DNA damage resulting from loss of AHR would also induce E2F1-mediated apoptosis, we assessed activated caspase-3 levels after knockdown of E2F1 expression. Forty-eight hours after transfection into Dox-treated Off*Ahrb fibroblasts, two different siRNAs blocked E2F1 expression by 70–90% in comparison with cells transfected with a scrambled RNA control or either untransfected or untreated cells (Figure 2A). Blocking E2F1 expression did not change the level of intracellular ROS in AHR-negative cells, as determined from the fluorescence intensity of Dox-treated cells exposed to CM-H2DCFDA (Figure 2B). Conversely, E2F1 siRNA significantly reduced the number of activated caspase-3 positive, Dox-treated cells compared with a scrambled RNA control, whereas it had no effect in Dox-untreated, AHR-positive cells (Figure 2, C and D). These results suggest that loss of AHR triggers a sequence of events that includes an elevated intracellular oxidant state and DNA damage, ultimately leading to E2F1-mediated induction of apoptosis.
Figure 2.
Inhibition of E2F1 with siRNA blocks apoptosis induced by loss of AHR. (A) siRNA inhibits E2F1 expression. Off*Ahrb cells grown for 48 h in the presence of 5 μg/ml Dox were transfected with two different siRNA oligonucleotides or with a negative, scrambled RNA control (for details, see Materials and Methods), and 48 h later proteins were extracted for detection of E2F1 by immunoblotting. Controls included cells not treated with Dox and cells treated with Dox but not transfected with siRNA. To ensure the loss of AHR expression in the appropriate treatments, the blots was also probed with anti-AHR antibodies. Actin was used as a loading control. We estimate that siRNA inhibited expression of E2F1 by 70–90%. (B) E2F1 inhibition did not block oxidative stress due to loss of AHR expression. Off*Ahrb cells were treated with 5 μg/ml Dox for 48 h and transfected with E2F1 siRNA (red trace) or scrambled control RNA (green trace). After 48 h in culture, transfected cells were exposed to CM-H2DCFDA for 30 min at 37°C and subsequently analyzed by flow cytometry. The purple tracing corresponds to cells unexposed to DCFDA. Note that inhibition of E2F1 expression did not change the oxidative stress levels in the cells. (C) Detection of activated caspase-3 by flow cytometry in Off*Ahrb cells treated with 5 μg/ml Dox for 48 h and transfected with E2F1 siRNA (green trace) or scrambled control RNA (red trace), fixed and allowed to react with PE-labeled active caspase-3 antibodies. The inset shows results from a similar experiment using control Off*Ahrb cells grown for 48 h in the absence of Dox and transfected with the same siRNA (green) or scrambled control (red). (D) CELLQUEST integration of the data in C, showing the percentage of caspase-3–positive cells as a function of Dox and siRNA transfection regime. The values shown are the mean ± SD of four determinations. **p < 0.01.
AHR Activation Protects Cells from Etoposide-induced Apoptosis
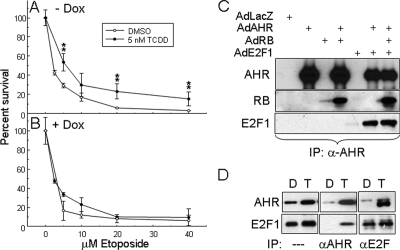
TCDD treatment inhibits p53-dependent induction of apoptosis by UV in primary rat hepatocytes in culture (Worner and Schrenk, 1996; Schrenk et al., 2004), suggesting that AHR activation might also block apoptosis induced by the DNA damage-CHK2-E2F1 pathway. To test this hypothesis, we followed the fate of TCDD-treated Off*Ahrb cells exposed to the DNA-damaging agent etoposide. Cells were grown in the presence or absence of Dox for 48 h, followed by treatment for an additional 48 h with 5 nM TCDD or DMSO vehicle and with increasing etoposide concentrations. The etoposide dose-response curve of AHR-expressing cells treated with TCDD showed a significant increase in cell survival relative to the same etoposide concentrations in cells treated with DMSO vehicle, an effect that did not take place in similarly treated cells in which AHR expression was repressed by Dox treatment (Figure 3, A and B). These results suggest that AHR activation may also protect cells from death by the E2F1-dependent apoptosis pathway.
Figure 3.
The activated AHR protects from etoposide-induced cell death and coimmunoprecipitates with E2F1. Off*Ahrb cells grown for 48 h without Dox (A) or with 5 μg/ml Dox (B) were treated with the indicated concentration of etoposide in the presence of 5 nM TCDD or DMSO vehicle. After an additional 48 h, cell numbers were determined by Hoescht 33258 staining, by using a standard curve relating Hoescht fluorescence to cell numbers (Chang et al., 2007). **p < 0.01. (C) Saos-2 cells were infected at 100 pfu/cell with purified adenoviral expression vectors for LacZ, AHR, RB, and E2F1 in the combinations indicated. After 24 h, cells were treated with 5 nM TCDD for 2 h and harvested for whole cell extracts. Coimmunoprecipitations were carried out on 1 mg of total protein extract with 2 μg of anti-AHR antibody, and eluted proteins were analyzed by Western blot. (D) AHR coimmunoprecipitates with E2F1 from Hepa-1 cell extracts. Cells were treated with 5 nM TCDD or vehicle control for 1 h. Nuclear extracts (300–500 μg of total protein) were precleared by incubation with 10 μl of a protein A/G bead mixture for 1 h at 4°C. Supernatants were transferred to tubes containing 1 μg of either an AHR or an E2F1 polyclonal antibody covalently bound to protein A/G beads, followed by overnight incubation at 4°C. After thorough washing, beads were boiled in SDS gel loading buffer, and supernatants were analyzed by SDS-PAGE and Western blot using the corresponding antibodies. An aliquot of each crude extracts was equally analyzed for comparison.
AHR Forms Complexes with E2F
AHR forms complexes with RB and enhances RB-mediated repression of E2F target genes (Puga et al., 2000). We asked whether AHR could also form complexes with E2F that were independent of the presence of RB. Transient expression of the luciferase reporter plasmid p3XE2FLuc in mouse Hepa-1 and in human Saos-2 cells, both lacking RB expression, was repressed by expression of activated AHR to the same extent as by expression of RB (Supplemental Figure S2, A and B). As reported previously (Puga et al., 2000), coexpression of AHR and RB resulted in additive repression of E2F reporter gene expression. These results suggested that AHR could not only interact with RB but also with E2F. Subsequent experiments using expression vectors bearing AHR truncation mutants (Puga et al., 2000) showed that deletion of the region between amino acid residues 322 and 495, including the ligand binding domain and part of the PAS B domain, led to loss of the ability of AHR to repress the E2F target (Supplemental Figure S2, C and D), identifying this domain as the most likely area of AHR–E2F functional interactions.
Coimmunoprecipitation experiments with crude extracts from Saos-2 cells infected with the adenoviral expression vectors Ad-AHR, Ad-E2F1, and Ad-RB showed that the AHR forms immune complexes with the other two proteins, either when only two or all three of them were overexpressed (Figure 3C), suggesting that AHR has no specific binding preference for either RB or E2F1 and that it can bind equally well with either in the presence of the other. To determine whether RB was required for the formation of AHR–E2F1 complexes, we used nuclear extracts from RB-negative Hepa-1 cells in coimmunoprecipitation experiments. Immunoblotting with anti-E2F1 showed that anti-AHR antibodies precipitated E2F1 from nuclear extracts of TCDD treated cells but not from cells treated with DMSO control. Conversely, immunoblotting with anti-AHR showed that anti-E2F1 antibodies immunoprecipitated AHR from nuclear extracts of TCDD-treated cells but not from DMSO-treated cells (Figure 3D). These data strongly indicate that AHR and E2F1 associate in a manner independent of RB and that ligand is needed to mediate AHR nuclear translocation and interaction with E2F1, which resides solely in the nucleus. We tested two other E2F family members, E2F3 and E2F4, by using the adenoviral expression system, both of which were as competent in forming complexes with AHR as E2F1 (data not shown).
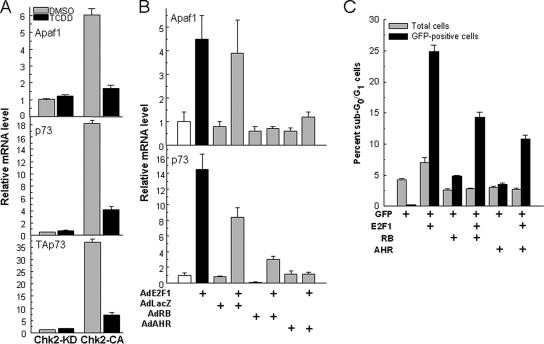
AHR Represses E2F1-dependent Transactivation of Apaf1 and p73 and Inhibits Apoptosis
The observation that the activated AHR represses E2F1-induced apoptosis and forms complexes with members of the E2F family, particularly E2F1, prompted us to test whether this effect results directly from AHR–E2F1 complex formation. We first tested the effect of AHR activation on Apaf1 and p73 induction in Hepa-1 cells transfected with a constitutively active CHK2 expression vector. Active CHK2-CA, but not kinase-dead CHK2-KD, induced Apaf1 by about sixfold, and TCDD treatment inhibited the induction (Figure 4A). Expression of p73, as measured with a set of exon 14 primers that would detect the α variants of both TAp73 and DNp73 isoforms, was induced 18-fold, but primers bracketing exons 2 and 3, specific for the proapoptotic TAp73 isoform, showed a 35-fold level of induction, almost twice as high as detected with the exon 14 primers, strongly indicating that there was little or no induction of the antiapoptotic DNp73 isoform and that C-terminal variants other than α might also be induced. As was the case for Apaf1, induction of TAp73 was significantly reduced by treatment with TCDD (Figure 4A), suggesting that the interaction of activated AHR with E2F1 suppresses the transactivation of proapoptotic E2F1 target genes.
Figure 4.
AHR-E2F1 interactions repress Apaf1 and p73 expression and inhibit apoptosis. (A) Ligand activation of the AHR represses CHK2-dependent induction of Apaf1 and p73. Mouse hepatoma Hepa-1 cells were transfected with a constitutively active CHK2 expression plasmid (CHK2-CA) or its kinase-dead counterpart (CHK2-KD). After selection for 10 d in G418, cells were treated with 5 nM TCDD or with DMSO vehicle, and 12 h later they were harvested and total RNA was extracted for determination of mRNA levels of Apaf1, p73, using both primers that detect transcripts common to TAp73 and DNp73 (middle) and primers specific for TAp73 (bottom), and β-actin control by real-time RT-PCR. The values shown are the mean ± SD of three determinations relative to β-actin. (B) AHR and RB overexpression inhibit Apaf1 and p73 induction by overexpression of E2F1. Saos-2 cells were infected with 100 pfu/cell of the adenoviral expression vectors AdE2F1, AdLacZ, AdRB, and AdAHR in the combinations indicated in the figure. RNA was extracted 24 h after infection and used to determine mRNA levels of Apaf1, p73 and β-actin control by real-time RT-PCR. The values shown are the mean ± SD of three determinations relative to β-actin. As shown previously (Puga et al., 2000; Marlowe et al., 2004) no ligand is necessary to activate the AHR in these cells. (C) AHR and RB inhibit apoptosis induction by E2F1. Saos-2 cells were transfected with plasmid vectors for expression of E2F1, GFP fused to histone H2B, RB, and AHR in the combinations indicated in the figure. Twenty-four hours after transfection, cells were analyzed by flow cytometry after propidium iodide staining. Total and GFP-positive sub-G0/G1 cells were scored separately to discriminate between the total and the transfected cell populations.
A direct test of this hypothesis made use of the Saos-2 human osteosarcoma cells, in which E2F1 overexpression and stabilization have been shown to induce apoptosis (Moroni et al., 2001; Stevens et al., 2003). Infection of these cells with the Ad-E2F1 expression vector induced accumulation of Apaf1 and TAp73 mRNA by 4.5- and 14-fold, respectively (Figure 4B). As expected, RB overexpression completely repressed the induction of both genes, as was the case when AHR was overexpressed by the corresponding adenoviral vector (Figure 4B). Neither RB, AHR, nor an adenoviral expression vector for β-galactosidase induced or repressed significantly the target genes when expressed alone.
Repression of proapoptotic E2F1 targets by AHR resulted in inhibition of apoptosis, as determined by flow cytometry analyses of sub-G0/G1 Saos-2 cells transfected with expression plasmids for E2F1, AHR, and RB. In these experiments, plasmid pH2BGFP, expressing green fluorescent protein (GFP) fused to histone H2B, was included to tag the nuclei of transfected cells, and to thereby distinguish between transfected and untransfected cells. E2F1 expression increased the fraction of sub-G0/G1 cells by five- to sixfold relative to control cells transfected only with the GFP expression vector. Neither RB nor AHR caused a significant increase over the control. However, RB, and more so AHR, significantly decreased the sub-G0/G1 fraction induced by E2F1. These effects were evident only in the transfected, GFP-positive cells, and not in the total cell population, indicating that the observations were due to the expression of the transfected genes.
AHR and E2F1 Form Complexes at E2F1 Binding Motifs on the Apaf1 Promoter
The formation of complexes between AHR and E2F1 could be unrelated to the observed effect of AHR expression on E2F1-induced transactivation and apoptosis. For example, there could be no connection between molecular interactions and biological effects, the one taking place in the nuclear matrix milieu and the other resulting from AHR binding to its consensus sites in the promoter of E2F1 target genes. Alternatively, the AHR could be recruited to canonical E2F1 binding motifs by E2F1 itself and function as an active repressor of E2F1 transactivation. To distinguish between these alternatives, we used chromatin immunoprecipitation to analyze AHR and E2F1 binding in the promoter regions of E2F1 target genes.
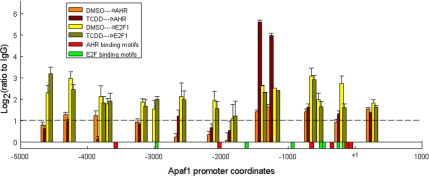
Anti-E2F and anti-AHR antibodies were used to immunoprecipitate Apaf1 promoter chromatin from Hepa-1 cells treated with 5 nM TCDD or DMSO control for 90 min. Mock immunoprecipitations were conducted as a control with nonimmune rabbit IgG. The assays were associated with a systematic mapping of the promoter, by using primer pairs for PCR amplification distributed approximately every 300 base pairs, between positions −4.8 kb and +0.1 kb from the transcription start site (Supplemental Table S3). In control and TCDD-treated cells, anti-E2F antibodies precipitated several Apaf1 promoter regions to a significantly greater extent than the control IgG. Several of these promoter regions contained consensus E2F binding sites, in particular those in the transcriptional start site-proximal domain, whereas others did not, consistent with previous observations (Wells et al., 2002). In contrast, anti-AHR antibodies precipitated a single promoter domain from TCDD-treated but not from DMSO-treated cells. This domain was localized to the −1400 region from the transcriptional start site, far from any of the canonical AHR binding sites in the Apaf1 gene but flanked by two E2F consensus sites and coinciding with an area where E2F antibodies also bound (Figure 5). These data suggest that the molecular interaction between the AHR and E2F1 proteins takes place at the promoters of proapoptotic E2F1 target genes.
Figure 5.
AHR and E2F1 interact on the promoter of the Apaf1 gene. The region of the Apaf1 promoter comprised between −4.8 kbp and +0.1 kbp was scanned by chromatin immunoprecipitation analyses using antibodies to AHR, E2F1, or nonimmune IgG. Hepa-1 cells were treated with 5 nM TCDD or with DMSO vehicle, and 90 min later they were fixed with formaldehyde and processed for ChIP analyses. The values shown represent the log2 ratios of the real-time PCR amplification data of chromatin immunoprecipitated with anti-AHR, anti-E2F1, or nonimmune IgG. The values shown represent the average of two determinations ± SD. Position of canonical binding sites for AHR and E2F are denoted by the red and green bars, respectively drawn along the length of the promoter.
DISCUSSION
The results presented in this article show that deletion of the Ahr gene creates a condition of heightened cellular oxidative stress, followed by a significant level of DNA damage, quantifiable by the increase of Ser139-phosphorylated γH2A.X histone. The higher pro-oxidant status and DNA damage in AHR-negative cells promotes E2F1-mediated apoptosis, which can be inhibited by E2F1 knockdown with siRNA. These data uncover an oxidative stress pathway that connects the loss of AHR with the activation of E2F1 apoptotic functions. These functions are also induced by the DNA-damaging agent etoposide, and we find that etoposide-induced cell death in AHR-positive cells is blocked by AHR activation, which, through its interaction with E2F1, attenuates the latter's proapoptotic function. The interaction between AHR and E2F is independent of the retinoblastoma protein, because AHR coimmunoprecipitates with E2F in extracts from RB-negative cells, and, because, by blocking apoptosis, AHR–E2F1 interactions have a diametrically opposite end point, promoting, rather than inhibiting, proliferation.
Cell cycle progression, differentiation, apoptosis, and senescence are major regulatory targets of the E2F proteins, and they are also major AHR targets (Puga et al., 2005; Marlowe and Puga, 2005; Bock and Kohle, 2006). Multiple redundant pathways couple proliferation with apoptosis and protect cells when normal proliferation controls are lost (Tsantoulis and Gorgoulis, 2005). One striking example of this coupling involves the activities of the E2F1 protein, which in addition to controlling gene expression for cell cycle progression, also activates apoptosis when DNA-damaging agents activate the ATM–CHK2 signaling pathway, phosphorylating E2F1 at serine-364 (Phillips et al., 1997; Phillips and Vousden, 2001; Stevens and La Thangue, 2004). Because H2A.X phosphorylation at Ser139 is also due to activation of the ATM–CHK2 pathway (Agarwal et al., 2006), we surmise that E2F1 is also phosphorylated by CHK2 and stabilized in our cells, although the lack of antibodies that recognize mouse p-Ser-368-E2F1 (equivalent to human p-Ser-364-E2F1) precludes us from directly testing this conclusion.
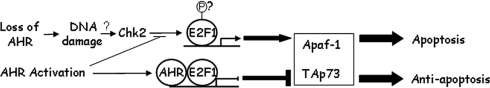
Activation by ligand translocates the AHR into the nucleus, where it physically interacts with E2F1 and causes the inhibition of E2F1-mediated apoptosis. Expression analyses and ChIP assays suggest that the mechanistic order of events leading to this outcome starts by the repression of CHK2-mediated induction of the proapoptotic E2F1 target genes, Apaf1 and TAp73 (and possibly others). Repression results from the interaction between AHR and E2F1, which, at least in the case of Apaf1, takes place at a canonical E2F1 binding motif in its promoter. The nature of the specific chromatin remodeling complexes generated by this interaction and how they function in gene repression are unexplored at this time. Figure 6 schematically represents these mechanistic events. DNA damage in general, or specifically due to loss of AHR, causes induction of the E2F1 apoptotic pathway (Stevens and La Thangue, 2004), proceeding through CHK2 activation, E2F1 phosphorylation, induction of proapoptotic genes and ultimately, cell death. At this point, we surmise that CHK2 activity and E2F1 phosphorylation also result from AHR loss. Conversely, AHR activation causes it to complex with E2F1 and block proapoptotic gene induction, presumably by blocking E2F1 phosphorylation by CHK2, and ultimately leading to an antiapoptotic endpoint.
Figure 6.
Schematic representation of the outcome of AHR–E2F1 interactions. DNA damage, in general, or specifically due to loss of AHR, causes induction of the E2F1 apoptotic pathway, which proceeds through CHK2 activation, E2F1 phosphorylation, induction of proapoptotic genes, and apoptosis. The question marks indicate that at present we do not know whether CHK2 activity and E2F1 phosphorylation also result from AHR loss. AHR activation causes it to complex with E2F1 and block proapoptotic gene induction by blocking CHK2 phosphorylation of E2F1, ultimately leading to an antiapoptotic endpoint.
Induction of apoptosis is consistent with the role of E2F1 as a tumor suppressor, a role confirmed by the finding that E2F1-deficient mice develop a range of tumor types (Field et al., 1996; Yamasaki et al., 1996). Inhibition of apoptosis, in contrast, is consistent with a mechanism of tumor promotion/progression. In diethylnitrosamine (DEN)-initiated rats, both acute and chronic treatment with TCDD results in an approximate 10-fold decrease in the rate of apoptosis in preneoplastically transformed liver foci, with no effect on the background rate of apoptosis in normal hepatocytes (Stinchcombe et al., 1995). Furthermore, AHR activation by TCDD blocks UV irradiation-induced apoptosis (Worner and Schrenk, 1996; Schrenk et al., 2004). The capacity of various Ah receptor ligands to act as tumor promoters has been attributed to their ability to inhibit the apoptotic elimination of initiated cells bearing genotoxic lesions (Schwarz et al., 2000). Our data suggest that they may do so by activating the AHR and inhibiting apoptosis through repression of E2F1 proapoptotic target genes, thus preventing the elimination of cells that have lost normal cell cycle control and promoting their proliferation. In this context, the activated AHR would effectively function as an oncogene.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Leone for a gift of E2f1−/− fibroblasts and P. Stambrook for a gift of CHK2-expressing plasmids. This research was supported by National Institute of Environmental Health Sciences (NIEHS) grants R01 ES-06273 and R01 ES-10807 and the NIEHS Center for Environmental Genetics grant P30 ES-06096.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0359) on June 4, 2008.
REFERENCES
- Agarwal C., Tyagi A., Agarwal R. Gallic acid causes inactivating phosphorylation of cdc25A/cdc25C-cdc2 via ATM-Chk2 activation, leading to cell cycle arrest, and induces apoptosis in human prostate carcinoma DU145 cells. Mol. Cancer Ther. 2006;5:3294–3302. doi: 10.1158/1535-7163.MCT-06-0483. [DOI] [PubMed] [Google Scholar]
- Bock K. W., Kohle C. Ah receptor: dioxin-mediated toxic responses as hints to deregulated physiologic functions. Biochem. Pharmacol. 2006;72:393–404. doi: 10.1016/j.bcp.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Chang X., Fan Y., Karyala S., Schwemberger S., Tomlinson C. R., Sartor M. A., Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol. Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Johnson D. G. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- DeYoung M. P., Ellisen L. W. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- Dimova D. K., Dyson N. J. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Elizondo G., Fernandez-Salguero P., Sheikh M. S., Kim G. Y., Fornace A. J., Lee K. S., Gonzalez F. J. Altered cell cycle control at the G(2)/M phases in aryl hydrocarbon receptor-null embryo fibroblast. Mol. Pharmacol. 2000;57:1056–1063. [PubMed] [Google Scholar]
- Ferreira R., Naguibneva I., Pritchard L. L., It-Si-Ali S., Harel-Bellan A. The Rb/chromatin connection and epigenetic control: opinion. Oncogene. 2001;20:3128–3133. doi: 10.1038/sj.onc.1204337. [DOI] [PubMed] [Google Scholar]
- Field S. J., Tsai F. Y., Kuo F., Zubiaga A. M., Kaelin W. G., Jr, Livingston D. M., Orkin S. H., Greenberg M. E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Frolov M. V., Dyson N. J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Nishimura N., Furukawa Y., Satoh M., Endo H., Iwase S., Yamada H., Matsuda M., Kano Y., Nakamura M. Apaf-1 is a mediator of E2F-1-induced apoptosis. J. Biol. Chem. 2002;277:39760–39768. doi: 10.1074/jbc.M200805200. [DOI] [PubMed] [Google Scholar]
- Ge N.-L., Elferink C. J. A direct interaction between the aryl hydrocarbon receptor and retinoblatoma protein. J. Biol. Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Harbour J. W., Dean D. C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Huang G., Elferink C. J. Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol. Pharmacol. 2005;67:88–96. doi: 10.1124/mol.104.002410. [DOI] [PubMed] [Google Scholar]
- Lin W. C., Lin F. T., Nevins J. R. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Marlowe J. L., Knudsen E. S., Schwemberger S., Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S-phase specific gene expression. J. Biol. Chem. 2004;279:29013–29022. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- Marlowe J. L., Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell Biochem. 2005;96:1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- Moroni M. C., Hickman E. S., Lazzerini D. E., Caprara G., Colli E., Cecconi F., Muller H., Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- Muller H., Bracken A. P., Vernell R., Moroni M. C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J. D., Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. Direct coupling of the cell cycle and cell death machinery by E2F1. Nat. Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- Pediconi N., et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- Peng Z., Peng L., Fan Y., Zandi E., Shertzer H. G., Xia Y. A critical role for IkappaB kinase beta in metallothionein-1 expression and protection against arsenic toxicity. J. Biol. Chem. 2007;282:21487–21496. doi: 10.1074/jbc.M702510200. [DOI] [PubMed] [Google Scholar]
- Phillips A. C., Bates S., Ryan K. M., Helin K., Vousden K. H. Induction of DNA synthesis and apoptosis are separable functions of E2F–1. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- Phillips A. C., Vousden K. H. E2F-1 induced apoptosis. Apoptosis. 2001;6:173–182. doi: 10.1023/a:1011332625740. [DOI] [PubMed] [Google Scholar]
- Puga A., Barnes S. J., Dalton T. P., Chang C., Knudsen E. S., Maier M. A. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 2000;275:2943–2950. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- Puga A., Tomlinson C. R., Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Puga A., Xia Y., Elferink C. Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem. Biol. Interact. 2002;141:117–130. doi: 10.1016/s0009-2797(02)00069-8. [DOI] [PubMed] [Google Scholar]
- Schnekenburger M., Peng L., Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim. Biophys. Acta. 2007a;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M., Talaska G., Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol. Cell Biol. 2007b;27:7089–7101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk D., Schmitz H. J., Bohnenberger S., Wagner B., Worner W. Tumor promoters as inhibitors of apoptosis in rat hepatocytes. Toxicol. Lett. 2004;149:43–50. doi: 10.1016/j.toxlet.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Schwarz M., Buchmann A., Stinchcombe S., Kalkuhl A., Bock K. Ah receptor ligands and tumor promotion: survival of neoplastic cells. Toxicol. Lett. 2000:112–113. 69–77. doi: 10.1016/s0378-4274(99)00247-7. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dyson N. J. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- Stevens C., La Thangue N. B. The emerging role of E2F-1 in the DNA damage response and checkpoint control. DNA Repair. 2004;3:1071–1079. doi: 10.1016/j.dnarep.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Stevens C., Smith L., La Thangue N. B. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- Stinchcombe S., Buchmann A., Bock K. W., Schwarz M. Inhibition of apoptosis during 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated tumour promotion in rat liver. Carcinogenesis. 1995;16:1271–1275. doi: 10.1093/carcin/16.6.1271. [DOI] [PubMed] [Google Scholar]
- Strobeck M. W., Fribourg A. F., Puga A., Knudsen E. S. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene. 2000;19:1857–1867. doi: 10.1038/sj.onc.1203510. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Huang X., Halicka H. D., Zhao H., Traganos F., Albino A. P., Dai W., Darzynkiewicz Z. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry A. 2007;71:648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J. M., Lees J. A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Tsantoulis P. K., Gorgoulis V. G. Involvement of E2F transcription factor family in cancer. Eur. J. Cancer. 2005;41:2403–2414. doi: 10.1016/j.ejca.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Weiss C., et al. TCDD deregulates contact inhibition in rat liver oval cells via Ah receptor, JunD and cyclin A. Oncogene. 2008;27:2198–2207. doi: 10.1038/sj.onc.1210859. [DOI] [PubMed] [Google Scholar]
- Wells J., Graveel C. R., Bartley S. M., Madore S. J., Farnham P. J. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker L. L., Su H., Baskaran R., Knudsen E. S., Wang J. Y. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol. Cell Biol. 1998;18:4032–4042. doi: 10.1128/mcb.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worner W., Schrenk D. Influence of liver tumor promoters on apoptosis in rat hepatocytes induced by 2-acetylaminofluorene, ultraviolet light, or transforming growth factor beta 1. Cancer Res. 1996;56:1272–1278. [PubMed] [Google Scholar]
- Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N. J. Tumor induction and tissue atrophy in mice lacking E2F–1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.