Abstract
Centrosome duplication must be tightly controlled so that duplication occurs only once each cell cycle. Accumulation of multiple centrosomes can result in the assembly of a multipolar spindle and lead to chromosome mis-segregation and genomic instability. In metazoans, a centrosome-intrinsic mechanism prevents reduplication until centriole disengagement. Mitotic cyclin/cyclin-dependent kinases (CDKs) prevent reduplication of the budding yeast centrosome, called a spindle pole body (SPB), in late S-phase and G2/M, but the mechanism remains unclear. How SPB reduplication is prevented early in the cell cycle is also not understood. Here we show that, similar to metazoans, an SPB-intrinsic mechanism prevents reduplication early in the cell cycle. We also show that mitotic cyclins can inhibit SPB duplication when expressed before satellite assembly in early G1, but not later in G1, after the satellite had assembled. Moreover, electron microscopy revealed that SPBs do not assemble a satellite in cells expressing Clb2 in early G1. Finally, we demonstrate that Clb2 must localize to the cytoplasm in order to inhibit SPB duplication, suggesting the possibility for direct CDK inhibition of satellite components. These two mechanisms, intrinsic and extrinsic control by CDK, evoke two-step system that prevents SPB reduplication throughout the cell cycle.
INTRODUCTION
Maintenance of genome stability is dependent on faithful segregation of chromosomes at mitosis. The mitotic spindle mediates chromosome segregation through partitioning sister chromosomes to opposite poles. In most cell types, the mitotic spindle is organized by a pair of centrosomes present in the cell at mitosis. However, accumulation of additional centrosomes has been observed in a variety of human cancers (Lingle et al., 1998; Pihan et al., 1998). Supernumerary centrosomes have the potential to assemble a multipolar spindle, which can lead to genomic instability and aneuploidy through catastrophic errors in chromosome segregation. In fact, several studies have suggested that centrosome amplification can contribute to tumorigenesis (reviewed in D'Assoro et al., 2002).
Several circumstances could lead to generation of supernumerary centrosomes, including failed mitosis, cell fusion, de novo centrosome formation, and centrosome reduplication during a single cell cycle (Nigg, 2002). Several studies demonstrate that centrosomes can reduplicate during an extended S-phase (Sluder and Lewis, 1987; Gard et al., 1990; Balczon et al., 1995), and this reduplication has been shown to be dependent on cell cycle regulatory proteins (Hinchcliffe et al., 1999; Lacey et al., 1999; Meraldi et al., 1999). Additionally, mutations in a number of known cell cycle regulators are associated with centrosome amplification (reviewed in Nigg, 2002). These lines of evidence suggest that centrosome duplication is regulated during the cell cycle and that loss of regulation can lead to centrosome amplification.
There is evidence to suggest that the structure of the duplicating centrosome precludes reduplication. Cell fusion experiments patterned after the classic experiments of Rao and Johnson (1970) demonstrated that when cells with duplicated centrosomes were fused with cells that had yet to duplicate their centrosomes, the duplicated centrosomes did not reduplicate, even though they were placed in a cellular environment that promotes centrosome duplication (Wong and Stearns, 2003). This observation suggests that something about the constitution of the previously duplicated centrosome prevents duplication. Recently, this centrosome-intrinsic block to centrosome duplication was shown to be relieved by centriole disengagement (centriole disorientation; Tsou and Stearns, 2006). Disengagement normally occurs during late mitosis or early G1 (Kuriyama and Borisy, 1981) and appears to be driven by separase (Tsou and Stearns, 2006). These findings indicate that the structure of engaged centrioles is not permissive for centriole duplication and that disengagement exposes sites or structures necessary for centriole duplication after mitosis.
Several studies suggest a role for cyclin dependent kinases (CDKs) in regulating centrosome duplication during the cell cycle (reviewed in Delattre and Gonczy, 2004). In experimental systems that are permissive for centrosome reduplication, specific cyclin/CDK complexes promote (Cdk2, cyclins E and A; Hinchcliffe et al., 1999; Lacey et al., 1999; Meraldi et al., 1999) and restrain (cyclin B; Lacey et al., 1999) centrosome duplication. In Drosophila loss of Cdk1 leads to formation of additional centrioles, whereas expression of hyper-stable cyclin A and cyclin B may inhibit centriole duplication (Vidwans et al., 1999, 2003). These findings suggest that specific cyclin/CDK complexes are important for both promoting centrosome duplication and inhibiting centrosome reduplication during the normal cell cycle; however, it remains unclear whether CDKs control centrosome duplication directly or affect centrosome duplication indirectly by inhibiting cell cycle progression.
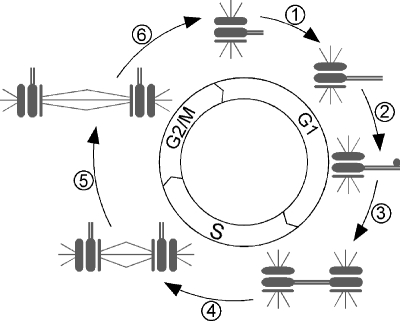
The centrosome analog in the budding yeast, Saccharomyces cerevisiae, called the spindle pole body (SPB), has been established as a model for many aspects of centrosome biology (Adams and Kilmartin, 2000). Electron microscopy has been used to describe the structure of the spindle pole body during the different stages of SPB duplication in great detail (Byers and Goetsch, 1974, 1975; Adams and Kilmartin, 1999), as summarized in Figure 1. Subsequent genetic screens identified several genes required for proper SPB duplication (reviewed in Chial and Winey, 1999; Jaspersen and Winey, 2004). By examining the structure of the unduplicated SPB in mutant cells, a detailed order of assembly for the SPB and the activities required for each step has been established (reviewed in Chial and Winey, 1999; Jaspersen and Winey, 2004). Nevertheless, relatively little is known about the regulation of SPB duplication.
Figure 1.
The SPB duplication cycle is coordinated with the cell cycle (reviewed in Chial and Winey, 1999). Steps in the SPB duplication cycle are indicated by number. Step 1, bridge elongation. The half-bridge doubles in length. Step 2, satellite assembly. An electron-dense mass of protein, thought to be a template for the new SPB, is deposited at the distal end of the half-bridge on the cytoplasmic face of the nuclear envelope. Step 3, SPB duplication. The SPB is assembled in the cytoplasm through an intermediate called the duplication plaque. The nascent SPB is then inserted into the nuclear envelope, resulting in two side-by-side SPBs connected by a full bridge. Step 4, SPB separation. The two SPBs move to opposite sides of the nucleus in a microtubule-dependent manner, splitting the full bridge into two half-bridges and creating a short spindle. Step 5, spindle elongation. At mitosis the spindle elongates, segregating chromosomes into the mother and daughter cells. Step 6, mitotic exit. At the completion of mitosis, the spindle disassembles, the cells separate, and each cell inherits a single SPB with a short half-bridge.
Several studies have demonstrated a role for CDK activity in the SPB duplication cycle (reviewed in Jaspersen and Winey, 2004). Cln1,2/Cdk1 activity has been shown to be important for SPB duplication in G1 (Figure 1, step 3) by directly phosphorylating important proteins in SPB duplication (Byers and Goetsch, 1974; Jaspersen et al., 2004). In addition, B-cyclins have been shown to be important for both promoting SPB duplication and preventing SPB reduplication (Haase et al., 2001).
In cells lacking the mitotic B-cyclins, SPBs will reduplicate; however, in cells lacking all six of the B-cyclin genes, SPBs duplicate but do not reduplicate (Haase et al., 2001). These observations indicate that B-cyclin/CDK activity is required to promote SPB reduplication and is likely to trigger a step(s) in the SPB duplication cycle. The B-cyclin–dependent step appears to correlate with SPB separation, which is also known to require B-cyclin/CDK activity (Fitch et al., 1992; Haase et al., 2001; Crasta et al., 2006). Although the mechanism by which mitotic cyclin/CDK complexes function to inhibit SPB reduplication is poorly understood, data suggest that mitotic cyclin/CDK acts directly, rather than through the suppression of CLN2 transcription (Haase et al., 2001). Taken together, these findings suggest a mechanism for preventing SPB reduplication reminiscent of the licensing model for the control of DNA replication (Haase et al., 2001). Evidence points to CDK regulation at multiple levels throughout the SPB duplication cycle; however, it is not yet clear how cells prevent SPB reduplication early in the cell cycle before B-cyclins are expressed, or how mitotic cyclin/CDKs inhibit reduplication when expressed later in the cell cycle.
To address the mechanisms restraining SPB duplication to once per cell cycle, we examined both CDK-dependent and -independent mechanisms that prevent SPB reduplication. Our findings reveal a role for a SPB-intrinsic block to SPB reduplication, similar to the centrosome-intrinsic block to centrosome duplication, which prevents SPB reduplication early in the cell cycle. We also demonstrate that mitotic cyclin/CDKs but not S-phase or G1 cyclin/CDKs can block SPB duplication when expressed early in G1 and that mitotic cyclin/CDK activity can inhibit early steps in the SPB duplication cycle. Our results describe distinct mechanisms that coordinate to prevent SPB reduplication throughout the yeast cell cycle.
MATERIALS AND METHODS
Plasmid and Strain Construction
All strains used in this study are derivatives of BF264-15DU (MATa; ade1; his2; leu2-3112; trp1-1; ura3Δns; Richardson et al., 1992). Relevant genotypes of strains are detailed in Table 1. SBY408 was constructed by cutting plasmid pZSPC42-GFP (Haase et al., 2001) with BglII and integrating at the TRP1 locus. SBY844 was constructed by PCR amplification of the mRFP-KanMX6 tag from the genome of ATCC201389: SPC42-mRFP-KanMX6 (Huh et al., 2003) using oligonucleotides FW (5′ATGGCCTCCTCCGAGGACGT3′) and RV (5′GTTAGTATCGAATCGACAGC3′) and cloning into pDrive (Qiagen, Valencia, CA). The sequence was reamplified using oligonucleotides FW (5′TATGTCAGAAACATTCGCAACTCCCACTCCCAATAATCGAGGAATGGCCTCCTCCGAGGACGT3′) and RV (5′TATAAAAGGCCTTTACGTTTCCGGCTTCTGTTGGAAAATAGTTAGTATCGAATCGAATCGACAGC3′) and integrated at the SPC42 locus via homologous recombination. SBY533 was constructed by cutting plasmid YIpG2-CLB2Δdb (a gift from Steve Reed) with Kpn1 and integrating at the LEU2 locus. SBY520 was made by cutting plasmid pGAL1-SIC1Δ3P (Verma et al., 1997) with EcoRV and integrating into SBY408 at the URA3 locus. SBY1353 was made by PCR amplification of CIN8 from the genome followed by ligation into pDrive (Qiagen). The cin8ts allele was made by site-directed mutagenesis using primers FW(5′GATAAAAGCGGCCATATACCTGCACGTGAATCGAAATTGACC3′) and RV (GGTCAATTTCGATTCACGTGCAGGTATATGGCCGCTTTTATC3′) to generate the temperature-sensitive F429A mutation (Gheber et al., 1999) and a unique PmlI site. A KpnI/XbaI fragment containing the cin8ts allele was then subcloned into pRS306 (Sikorski and Hieter, 1989). The resulting plasmid was cut with BglII and transformed into a version of SBY408 carrying kip1Δ::KanMX4 at the CIN8 locus. CIN8 allele replacement was achieved by growing transformants in the presence of 5′-fluoroorotic acid (5′-FOA), and confirmed by PCR followed by PmlI digestion. The resulting strain was transformed with pGAL-Sic1Δ3P (Verma et al., 1997) cut with EcoRV for integration at the URA3 locus. SBY1168 was made by amplification of the HA3 tag and KanR from plasmid pKHA3 using primers FW (5′GGTTAGAAAAAACGGCTATGATATAATGACCTTGCATGAACGCATCTTTTACCCATACG3′) and RV (5′CATACATTTTATATGGACATTTATCGATTATCGTTTTAGACATGCCATCCGTAAGATGC3′) and integrating into SBY533 at both CLB2Δdb and CLB2. SBY1099 was made by cutting CWB194 (pRS414-GAL1-CLN2-HA3; Lanker et al., 1996) with MfeI and integrating into SBY844 at the TRP1 locus. SBY1157 was made by cutting plasmid DBRI (pGAL1-HA3-CLB5Δdb; Cross et al., 1999) with EcoRV and integrating into SBY408 at the URA3 locus.
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype |
|---|---|
| SBY408 | MATa; bar1; SPC42-GFP-TRP1-ZeoR |
| SBY520 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-SIC1Δ3P-URA3 |
| SBY533 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-CLB2Δdb-LEU2 |
| SBY844 | MATa; bar1; SPC42-mRFP-KanMX6 |
| SBY916 | MATa; bar1; SPC42-mRFP-KanMX6; GAL-CLB2Δdb-GFP-TRP1 |
| SBY918 | MATa; bar1; SPC42-mRFP-KanMx6; GAL-CLB2ΔdbΔNES-GFP-TRP1 |
| SBY920 | MATa; bar1; SPC42-mRFP-KanMx6; GAL-CLB2ΔdbΔNLS-GFP-TRP1 |
| SBY1099 | MATa; bar1; SPC42-mRFP-KanMX6; GAL-CLN2-HA3-TRP1 |
| SBY1157 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-HA3-CLB5Δdb-URA3 |
| SBY1167 | MATa; bar1; CLB2-HA3-KanR |
| SBY1168 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-CLB2Δdb-HA3-LEU2-KanR; CLB2-HA3-KanR |
| SBY1171 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-HA3-CLB5Δdb-NESI-mRFP-URA3 |
| SBY1173 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; GAL-HA3-CLB5Δdb-NESA-mRFP-URA3 |
| SBY1353 | MATa; bar1; SPC42-GFP-TRP1-ZeoR; cin8ts; kip1Δ::KanMX4; GAL-SIC1Δ3P-URA3 |
The pGAL1-CLB2ΔdbΔNES-GFP and pGAL1-CLB2ΔdbΔNLS-GFP plasmids were made by swapping a BbvCI/ApaI fragment from the C-terminal half of CLB2 in pPS2191 (2μ GAL1-CLB2Δdb-GFP) with a BbvCI/ApaI fragment from either pPS2190 (2μ GAL1-CLB2(L303A)-GFP) to make 2μ GAL1-CLB2ΔdbΔNES-GFP or pPS2192 (2μ GAL1-CLB2(Δ183-200)-GFP) to make 2μ GAL1-CLB2ΔdbΔNLS-GFP (Hood et al., 2001). pRS304 GAL1-CLB2Δdb-GFP (and ΔNES and ΔNLS derivatives) were made by cutting 2μ plasmids containing the construct with EcoRI/NsiI and cloning fragments into pRS304 (Sikorski and Hieter, 1989) cut at EcoRI/PstI. SBY916, SBY918, and SBY920 were made by cutting pRS304 GAL1-CLB2Δdb-GFP constructs with Bsu36I and integrating at the TRP1 locus of SBY844.
The pGAL1-HA3-CLB5Δdb-NES-mRFP constructs were made as follows. CUP1-CLB5 was amplified from pKCUP1-CLB5-HA (Haase et al., 2001) using oligonucleotides FW (5′CTTGCATGCCTGCAGGTC3′) and RV (5′GTCGACCGCGGCCGCACTTAAGATTAAATAGATTTTGAAAGTTGCTATGCATTTC3′). The resulting product was cloned into pDrive (Qiagen). A SalI fragment of pDrive-CUP1-CLB5 was cloned into SalI digested pDrive-mRFP-KanMX6 to make pDrive-CUP1-CLB5-mRFP-KanMX6 of which a SacI/BglII fragment containing CUP1-CLB5-mRFP was subcloned into YCplac33 (Gietz and Sugino, 1988) digested with SacI/BamHI. The active (NESA) and inactive (NESI) cassettes were amplified from p306-NESA and p306-NESI plasmids, respectively (Edgington and Futcher, 2001), using oligonucleotides FW (5′CGAAATGCATAGCAACTTTCAAAATCTATTTAATCTTAAGGGTTTAGCACTTAAATTAGC3′) and either NES-A RV (5′CGTCCTCGGAGGAGGCCATAATCTGAATTCGTCGACAAGCACTACCGATATCTAAACCTG3′) or NES-I RV (5′CGTCCTCGGAGGAGGCCATAATCTGAATTCGTCGACAAGCACTACCGATATCAGCACCTG3′) and cloned into SacII-digested YCplac33-CUP1-CLB5-mRFP via gap repair. GAL1-HA3-CLB5Δdb was subcloned from DBRI (Cross et al., 1999) into pRS306 (Sikorski and Hieter, 1989) on EcoRI ends. A NotI/BspEI fragment containing the C-terminus of CLB5 was replaced by NotI/BspEI fragments from YCplac33-CUP1-CLB5-NESA-mRFP or YCplac33-CUP1-CLB5-NESI-mRFP to make pRS306-GAL1-HA3-CLB5Δdb-NESA-mRFP and pRS306-GAL1-HA3-CLB5Δdb-NESI-mRFP. These constructs were digested with EcoRV and integrated into the URA3 locus of SBY408 to make SBY1173 (NES-I) and SBY1171 (NES-A).
Cell Growth and Synchronization
Yeast cultures were grown in YEP medium (1% yeast extract, 2% peptone, 0.012% adenine, 0.006% uracil supplemented with 2% sugar: dextrose, sucrose, or galactose). Cells were grown at 30°C. Mating pheromone arrest was accomplished by adding 30–60 ng/ml alpha-factor (α-factor) to the growth medium. For synchronization experiments, cells were then released into growth medium without α-factor. For mitotic arrest, cells were treated with 15 μg/ml nocodazole.
SPB reduplication experiments were performed as previously described (Haase et al., 2001). For nocodazole experiments, hydroxyurea was used at 100 mM to slow progression through S-phase, and nocodazole was used at 15 μg/ml to destabilize microtubules either at release from α-factor or at the time of induction of GAL1-SIC1Δ3P. For cin8tskip1Δ experiments, cells were incubated at either 25 or 38°C upon release from α-factor, and galactose was then added to both cultures to induce GAL1-SIC1Δ3P after the 25°C culture had become 80% budded.
Elutriation experiments were performed as follows. Cells were grown to 1.5 * 107 − 2.5 * 107 cells/ml in YEP-sucrose. The GAL1 promoter was induced by addition of 2% galactose into the media and incubated for 45 min (for CLN2 or CLB5 constructs) or 60 min (for CLB2 constructs). Cells were then put on ice, subjected to centrifugal elutriation, and small daughter cells were collected. Cells synchronized in early G1 were released into YEP-galactose at t = 0 min.
Microscopy
All samples analyzed by fluorescence microscopy were fixed in 2% paraformaldehyde for 15–30 min and then washed with PBS and stored in 30% glycerol. DNA staining with 4′,6-diamidine-2-phenylindole dihydrochloride (DAPI; Roche Diagnostics, Indianapolis, IN) was done at 1 μg/ml to visualize nuclei. Cells were viewed using a Zeiss Axio Imager widefield fluorescence microscope with a 100× objective and standard filter sets (Thornwood, NY). Foci of Spc42-GFP or Spc42-mRFP were counted for a minimum of 200 cells per sample to determine the state of SPB duplication in the sample. Images were acquired with a Hamamatsu Orca ER monochrome cooled-CCD camera with IEEE (Bridgewater, NJ) and captured using Metamorph 7.1 (Universal Imaging, Downingtown, PA). Images were merged digitally using Photoshop 7.0 (Adobe Systems, Palo Alto, CA).
Samples were prepared for electron microscopy as previously described (Byers and Goetsch, 1991). Samples were viewed using Philips CM 12 transmission electron microscope (FEI, Hillsboro, OR), and images were captured using an AMT XR100 Digital Camera (Advanced Microscopy Techniques, Danvers, MA).
Immunoblotting
Cell lysates were subjected to SDS-PAGE and immunoblotting using the following antibodies: mouse anti-HA (Roche Diagnostics), mouse anti-green fluorescent protein (GFP; Covance, Berkley, CA), mouse anti-PSTAIR (Abcam, Cambridge, MA), and HRP-conjugated goat anti-mouse (Pierce Biotechnology, Rockford, IL).
Flow Cytometry
Preparation of cells and flow cytometric analysis of DNA content was carried out as described previously (Haase and Reed, 2002).
RESULTS
SPB Separation Is Required for SPB Reduplication
Mitotic cyclin/CDKs inhibit SPB reduplication during the cell cycle (Haase et al., 2001), but mitotic cyclins are not expressed until S-phase and G2. Thus, SPB reduplication cannot be inhibited by mitotic cyclins in G1 and early S-phase, a period when SPB components are expressed and proteins are known to be important for SPB duplication are active (Jaspersen et al., 2004; Orlando et al., 2008). Previous studies indicate that B-cyclin/CDK activity is required after SPB duplication in order for SPBs to duplicate in the next cycle (Haase et al., 2001). We investigated whether B-cyclin/CDKs might function to relieve an inhibitory mechanism that normally prevents SPB reduplication in late G1 and early S-phase.
During the normal cell cycle, the SPBs separate to form a short spindle in late S-phase/G2 (Figure 1, step 4), whereas cells lacking mitotic B-cyclins, cells go on to reduplicate SPBs after separation (Haase et al., 2001). Because cells lacking all the B-cyclins neither separate, nor reduplicate their SPBs (Haase et al., 2001) and separation has been shown to be dependent on B-cyclin/CDK activity (Figure 1, step 3; Fitch et al., 1992; Crasta et al., 2006), we hypothesized that SPB separation may be required to relieve a block to SPB reduplication. To test this hypothesis, we took advantage of the finding that SPB separation is dependent on the presence of microtubules (Jacobs et al., 1988).
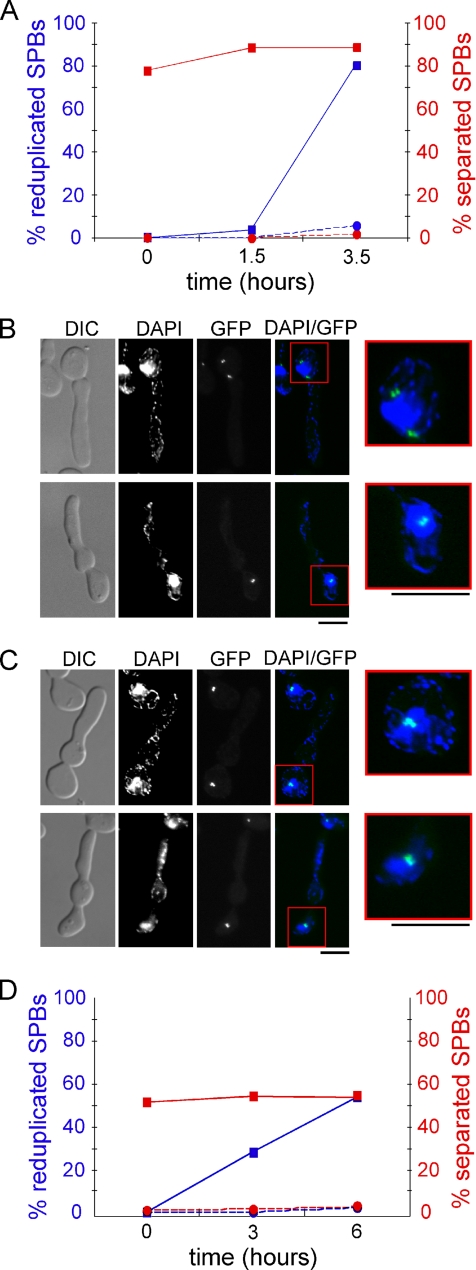
SPB separation was blocked by treating cells with the microtubule depolymerizing drug, nocodazole, and the ability of cells to reduplicate SPBs under conditions previously shown to promote SPB reduplication (Haase et al., 2001) was determined. Cells bearing a hyper-stabilized allele of SIC1 (SIC1Δ3P) controlled by the GAL1 promoter were arrested in G1 with α-factor and then released in the presence or absence of nocodazole in non-inducing medium. When cells growing in medium lacking nocodazole had separated SPBs, galactose was added to both cell cultures to induce the expression of SIC1Δ3P, thereby inhibiting B-cyclin/CDK activity and allowing SPB reduplication. SPB separation and reduplication were monitored by fluorescence microscopy of Spc42-GFP.
In cells treated with nocodazole upon release from an α-factor arrest, SPBs duplicated, but neither separated nor reduplicated, whereas untreated cells separated and reduplicated SPBs (Figure 2, A and B). To control for unanticipated effects of nocodazole treatment on SPB duplication, a third aliquot of cells was allowed to separate SPBs and form a short spindle before the addition of nocodazole, which was added concurrent with addition of galactose to stimulate expression of Sic1Δ3P. In these cells, the short spindle collapses (Jacobs et al., 1988), but some of the SPBs go on to reduplicate despite their close proximity (Figure 2C).
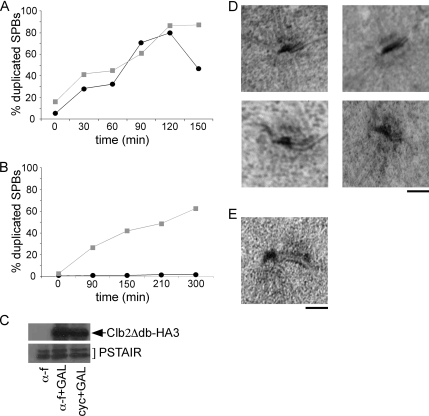
Figure 2.
SPB separation is required for SPB duplication. Cells were synchronized in G1 and cell aliquots were either allowed to separate SPBs or were prevented from separating SPBs by addition of nocodazole or shifting cin8tskip1Δ cells to 38°C. Sic1Δ3P expression was induced in all aliquots to promote SPB reduplication. SPBs were examined by fluorescent microscopy of Spc42-GFP. (A) Percentages of separated and reduplicated SPBs in cells either treated with nocodazole upon release from α-factor (●; broken line) or untreated (■; solid line) over time. (B) Top, images showing separated and reduplicated SPBs in untreated cells. Bottom, images showing unseparated and duplicated (but not reduplicated) SPBs in cells treated with nocodazole upon release from α-factor. Bar, 50 μm. (C) Images showing collapsed and reduplicated SPBs in cells released from α-factor in the absence of nocodazole to allow SPB separation and then treated with nocodazole to collapse short spindles upon expression of Sic1Δ3P. Bar, 50 μm. (D) Percentages of separated and reduplicated SPB in cin8tskip1Δ cells released at either 38°C (●; broken line) or 25°C (■; solid line) over time.
We also investigated the necessity of SPB separation before SPB reduplication using mutations in the motor proteins Cin8 and Kip1. SPB separation is defective in cin8 kip1 double mutants, and cells arrest with duplicated SPBs still attached by a full bridge structure (Hoyt et al., 1992; Roof et al., 1992). cin8tskip1Δ cells were released from α-factor arrest at either restrictive or non-restrictive temperature. Galactose was added to induce transcription of SIC1Δ3P from the GAL1 promoter in both cultures after the cells had been given sufficient time to separate their SPBs at non-restrictive temperature (25°C). SPB separation and reduplication were monitored by fluorescent microscopy of Spc42-GFP. Many of the cin8tskip1Δ cells released at 25°C separated their SPBs and by 6 h after induction of SICΔ3P most of the cells with separated SPBs had reduplicated SPBs (Figure 2D). As expected, few cin8tskip1Δ cells separated SPBs at 38°C, and only a small fraction of cells released at restrictive temperature (38°C) reduplicated SPBs after 6 h (Figure 2D). The few cells at 38°C that went on to reduplicate SPBs had also separated their SPBs.
Taken together, these results demonstrate that SPB separation is essential for SPB reduplication and suggest that duplicated side-by-side SPBs are unable to reduplicate until they have separated into two individual SPBs, each with its own short half-bridge (Figure 1, step 4).
A Mitotic Cyclin Expressed in G1 Can Inhibit SPB Duplication
After separation, it has been demonstrated that mitotic cyclin/CDKs prevent SPB reduplication (Haase et al., 2001). The structure of the SPB does not detectably change after separation until the next G1 (Figure 1, steps 5, 6, and 1; Byers and Goetsch, 1974, 1975). We posited that in the absence of mitotic cyclins, the cell cycle arrests at metaphase, but the SPB duplication cycle continues by progressing directly from the separated phase (Figure 1, after step 4) through the early steps of the duplication cycle (Figure 1, steps 1–3). The orientation of reduplicated SPBs (two sets of side-by-side SPBs separated by a short spindle; Figure 2A; Haase et al., 2001) is consistent with this hypothesis. To determine whether mitotic cyclin/CDKs normally prevent SPB reduplication by preventing the premature passage through these early steps in the SPB duplication cycle, we ectopically expressed a destruction box mutant allele of the mitotic B-cyclin Clb2, Clb2Δdb, which is stable in G1, from the GAL1 promoter in early G1 cells.
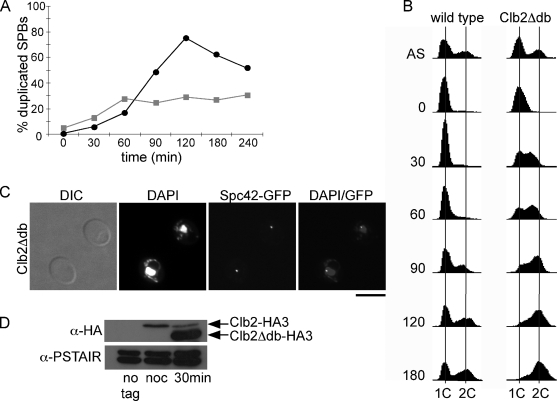
Daughter cells in early G1 were collected from an asynchronous population of cells by centrifugal elutriation 60 min after the induction of Clb2Δdb expression. SPB duplication was then monitored over time by fluorescence microscopy (Figure 3, A and C). Two hours after elutriation, cells not expressing Clb2Δdb had duplicated their SPBs and initiated mitosis; however, ∼70% of cells expressing Clb2Δdb failed to duplicate their SPBs (Figure 3A). Although early G1 cells expressing Clb2Δdb fail to bud (data not shown; Lew and Reed, 1993), they do rapidly replicate DNA and arrest with 2C DNA content (Figure 3B; Amon et al., 1994), indicating that they progress through the G1/S transition.
Figure 3.
Ectopic expression of Clb2 in early G1 inhibits SPB duplication. Clb2Δdb expression was induced from the GAL1 promoter in asynchronous populations of cells, and then small daughter cells were collected by elutriation. SPB duplication (A) and DNA content (B) were monitored over time by fluorescence microscopy and flow cytometry in cells expressing (■, gray) or not expressing (●, black ) Clb2Δdb. (C) Images of representative cells expressing Clb2Δdb 120 min after elutriation. Bar, 50 μm. (D) Western blot of Clb2 protein levels in untagged cells, cells synchronized in α-factor and released into nocodazole for 1.5 h, and in elutriated cells 30 min after elutriation. Cdk1 levels (α-PSTAIR) are shown as a loading control.
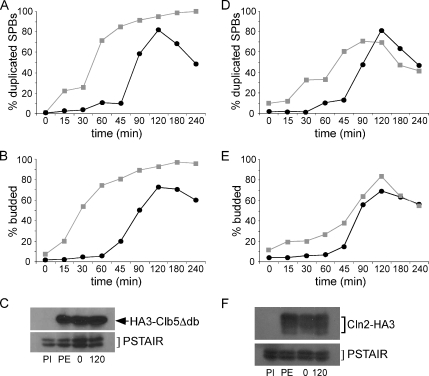
These results demonstrate that ectopic expression of Clb2 in early G1 can inhibit SPB duplication; however, there is normally very little CDK activity during early G1 (Miller and Cross, 2001). To ensure that the inhibition of SPB duplication is specific to mitotic B-cyclins and not a simply a function of excess CDK activity in early G1, we also assayed the effects of expressing the G1 cyclin, Cln2, and a destruction box mutant of the S-phase cyclin, Clb5 (Clb5Δdb; Cross et al., 1999), from the GAL1 promoter in early G1. Although Cln2 and Clb5 promote SPB duplication later in G1, it is possible that a high level of Cln2/Cdk1 or Clb5/Cdk1 activity before START would have an inhibitory role similar to a high level of Clb2/Cdk1 activity. However, expression of either Cln2 or Clb5 in early G1 did not inhibit SPB duplication, but rather accelerated the duplication process either by direct phosphorylation or by indirectly driving the cells more rapidly through G1 (as evidenced by early bud emergence in cells expressing either Cln2 or Clb5Δdb; Figure 4). Additionally, cells expressing Clb5Δdb failed to exit mitosis (Figure 4, A and B) as previously described (Jacobson et al., 2000). Our findings indicate that the inhibition of SPB duplication in early G1 is specific to mitotic cyclins and is not due to a general increase in CDK activity during this time period.
Figure 4.
Ectopic expression of G1 or S-phase cyclins in early G1 cannot inhibit SPB duplication. HA3-Clb5Δdb (A–C) or Cln2-HA3 (D–F) was expressed from the GAL1 promoter in asynchronous populations of cells, and then small daughter cells were collected by centrifugal elutriation. SPB duplication (A and D) and bud emergence (B and E) were monitored over time by fluorescence microscopy in cells expressing (■, gray) or not expressing (●, black) cyclin. (C and F) Western blot of cyclin levels before cyclin induction (PI), before elutriation (PE), after elutriation (0), and 120 min after elutriation (120). Cdk1 levels (α-PSTAIR) are shown as a loading control.
Clb2 Expression in Early G1 Inhibits Satellite Assembly
Mitotic cyclin/CDK activity could block a number of early steps in the SPB duplication cycle including bridge elongation, satellite assembly, or the assembly of a new SPB (Figure 1, steps 1–3). To determine whether Clb2 inhibits SPB duplication after satellite assembly, we asked whether Clb2 could inhibit SPB duplication in cells arrested in α-factor, when SPBs are known to have assembled a satellite (Byers and Goetsch, 1974, 1975). Cells were arrested in α-factor and then released into media containing galactose to induce expression of Clb2Δdb. Cells expressing Clb2Δdb and control cells not expressing Clb2Δdb duplicated their SPBs at a similar rate (Figure 5A). Furthermore, SPB duplication was driven by the expression of Clb2Δdb in cells that were maintained in α-factor (Figure 5B), consistent with previous observations (Amon et al., 1994). These findings suggest that Clb2 inhibits early steps in the SPB duplication cycle; however, once a satellite has formed, Clb2 no longer inhibits SPB duplication but can actually drive later steps in the SPB duplication cycle.
Figure 5.
Clb2 inhibits early steps in SPB duplication. (A) Asynchronous populations of cells were arrested in α-factor and released into media inducing expression of Clb2Δdb. (B) Cells were held in α-factor arrest as Clb2Δdb expression was induced. For both experiments, SPB duplication was monitored by fluorescence microscopy of Spc42-GFP over time for cells expressing (■, gray) and not expressing (●, black) Clb2Δdb. (C) Western blot showing Clb2Δdb-HA3 levels in arrested cells (α-f), arrested cells 30 min after Clb2Δdb induction (α-f + GAL), and in cells released from α-factor and induced to express Clb2Δdb for 30 min (cyc + GAL). Cdk1 levels (α-PSTAIR) are shown as a loading control. (D) SPB structure was analyzed by electron microscopy in elutriated cells expressing Clb2Δdb 2 h after elutriation. (E) SPB structure was analyzed by electron microscopy in cells arrested in α-factor after elutriation. Bar, 100 nm.
To investigate which early steps in the SPB duplication cycle might be inhibited by Clb2, we examined SPBs by electron microscopy in cells where SPB duplication was blocked by the expression of Clb2 in early G1. We analyzed 50 SPBs from cells expressing Clb2Δdb 2 h after elutriated cells were inoculated into fresh medium (Figure 5D). Half-bridges were observed in 33 of 50 SPBs; however, we did not observe satellite structures in any cells expressing Clb2. This result confirms that Clb2 inhibits SPB duplication before satellite assembly, although we were unable to discern whether Clb2 inhibits bridge elongation, satellite assembly, or both. Interestingly, about one-third of the SPBs were observed to be larger than 100 nm in diameter. Large SPB size has been previously observed in cells with mutations that affect SPB duplication (Byers, 1981; Winey et al., 1991; Adams and Kilmartin, 1999).
To control for our ability to identify satellite structures, we examined an isogenic strain (not expressing Clb2Δdb) that was released into medium containing α-factor to arrest the cells at START after elutriation. Cells arrested at START should contain satellite bearing SPBs as previously reported (Byers and Goetsch, 1974, 1975; Adams and Kilmartin, 1999). In the 29 control cells examined, all SPBs observed were between 75 and 100 nm in diameter, a normal size for haploid SPBs (Byers and Goetsch, 1974). Eleven SPBs had observable half-bridges and nine of those SPBs had satellite structures associated with the half-bridge (Figure 5E).
Cytoplasmic Localization of Clb2 Is Required to Inhibit SPB Duplication
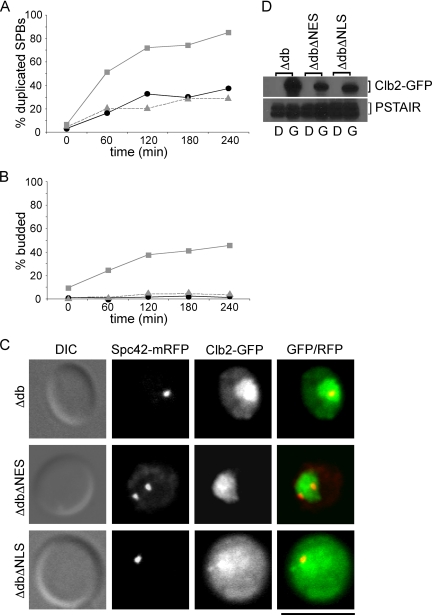
Satellite assembly is spatially confined to the cytoplasmic face of the half-bridge (Adams and Kilmartin, 1999). Clb2 has been shown to shuttle between the nucleus and cytoplasm (Hood et al., 2001) and to localize to spindle poles (Bailly et al., 2003). Thus, it is possible that Clb2 may inhibit satellite assembly by phosphorylating targets on the cytoplasmic face of the SPB. Therefore, we examined whether Clb2 localization was important for its ability to block SPB duplication in early G1 using CLB2 alleles that have mutations in either the nuclear localization sequence (NLS, Δ183-200) or the nuclear export sequence (NES, L303A; Hood et al., 2001). Clb2, Clb2ΔNES, and Clb2ΔNLS alleles with mutations in the destruction box were fused to GFP, expressed under control of the GAL1 promoter, and integrated in cells with monomeric red fluorescent protein (mRFP)-tagged Spc42 so that both Clb2 localization and SPB duplication could be monitored. We ectopically expressed these Clb2 mutants in cells synchronized in early G1 by elutriation as described above. As expected, Clb2 localized to both the nucleus and the cytoplasm, Clb2ΔNES localized to the nucleus and Clb2ΔNLS exhibited increased cytoplasmic localization but was not excluded from the nucleus (Figure 6C; Hood et al., 2001). SPB duplication was inhibited in cells expressing either Clb2 or Clb2ΔNLS, but not in cells expressing Clb2ΔNES (Figure 6A). All three constructs were expressed at comparable levels (Figure 6D). Additionally, although cells expressing Clb2 or ClbΔNLS failed to bud, some, but not all cells expressing Clb2ΔNES budded over time (Figure 6B). These findings indicate that Clb2 must localize to the cytoplasm to inhibit SPB duplication.
Figure 6.
Clb2 must be cytoplasmic in order to inhibit SPB duplication. (A–C) Asynchronous populations of cells were induced to express GFP tagged Clb2Δdb (●, black), Clb2ΔdbΔNES (■, gray), or Clb2ΔdbΔNLS (▴, gray) from the GAL1 promoter, and small daughter cells were then collected by centrifugal elutriation. SPB duplication (A) and bud emergence (B) were monitored over time by fluorescence microscopy. (C) Representative images of cells at 120 min after elutriation. In merged image, GFP is green, mRFP is red, and colocalization is yellow. Bar, 50 μm. (D) Western blot showing levels of Clb2 constructs in asynchronous cells grown in either dextrose “D” or galactose “G”. Cdk1 levels (α-PSTAIR) are shown as a loading control.
Cytoplasmic Localization Cannot Confer the Ability to Inhibit SPB Duplication to Clb5
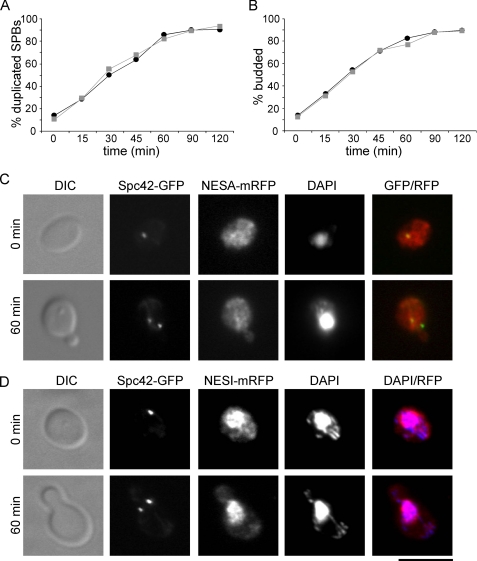
Previous studies demonstrate that the mitotic B-cyclins (Clb1, Clb2, Clb3, and Clb4) can inhibit SPB reduplication, whereas the S-phase B-cyclins, Clb5 and Clb6, cannot (Haase et al., 2001). As we have shown that Clb2 must be cytoplasmic in order in inhibit SPB duplication, it is possible that Clb5, a nuclear protein (Jacobson et al., 2000), cannot block SPB duplication because it does not localize to the cytoplasm. Thus, we asked if Clb5 could block SPB duplication if it was forced to localize to the cytoplasm. Two active leucine-rich NES sequences (NESA) or two inactive NES sequences that have key leucines mutated to alanine (NESI; Edgington and Futcher, 2001) were fused to mRFP-tagged Clb5Δdb (Cross et al., 1999) and expressed from the GAL1 promoter in early G1 cells. As expected, Clb5-NESI localized to the nucleus (Figure 7D). Clb5-NESA was observed in the nucleus and cytoplasm and was observed to colocalize with Spc42-GFP (Figure 7C), as has been shown previously for both Clb2 and Clb4 (Hood et al., 2001; Bailly et al., 2003; Maekawa and Schiebel, 2004). However, localization to the cytoplasm (and the spindle pole) did not confer the ability to inhibit SPB duplication on Clb5 (Figure 7A). Thus, Clb5/Cdk1 cannot phosphorylate the cytoplasmic Clb2/Cdk1 targets important to inhibit SPB reduplication.
Figure 7.
Cytoplasmic Clb5Δdb cannot inhibit SPB duplication. Cells were induced to express Clb5Δdb-NESA-mRFP (active NES; ■, gray) or Clb5Δdb-NESI-mRFP (inactive NES; ●, black) from the GAL1 promoter and small daughter cells were collected by centrifugal elutriation. SPB (A) duplication and bud emergence (B) were monitored over time by fluorescence microscopy. (C) Representative images showing Clb5Δdb-NESA-mRFP colocalization with Spc42-GFP as well as SPB duplication at 0 and 60 min after elutriation. In merged image GFP is green, mRFP is red, and colocalization is yellow. (D) Representative images showing Clb5Δdb-NESI-mRFP colocalization with DNA as well as SPB duplication at 0 and 60 min after elutriation. In merged image GFP is green, DAPI is blue and colocalization is magenta. Bar, 50 μm.
DISCUSSION
Ensuring that centrosomes are duplicated only once during each cell cycle is critical for assembling a proper bipolar spindle and for preventing mis-segregation events during mitosis. In budding yeast, we have now characterized both SPB-intrinsic and extrinsic control mechanisms that prevent reduplication of the SPB during distinct cell cycle intervals.
Previous work established that SPBs reduplicate in the absence of mitotic cyclin/CDKs (Haase et al., 2001). However, we have now demonstrated that in the absence of B-cyclin/CDK activities, inhibiting SPB separation also prevents reduplication (Figure 2) before mitotic cyclin expression. As SPBs in both nocodazole-treated and cin8 kip1 cells have been shown to arrest with duplicated SPBs with an intact bridge (Jacobs et al., 1988; Hoyt et al., 1992; Roof et al., 1992), these findings suggest that duplicated, side-by-side SPBs lack the proper structure to initiate a new round of SPB duplication. The SPB duplication cycle normally begins with the elongation of the half-bridge structure and the assembly of a satellite at the distal end of the half-bridge (Figure 1, steps 1 and 2; Byers and Goetsch, 1974, 1975). It is likely that the proper cues for satellite assembly are contained in the elongated half-bridge, but not the full bridge that connects side-by-side SPBs. Thus, SPB reduplication is inhibited by an SPB-intrinsic mechanism until SPB separation exposes new half-bridge structures that can support satellite assembly. This mechanism would prevent multiple rounds of SPB duplication during G1 when B-cyclin/CDK activity is low and SPB components are expressed (Jaspersen and Winey, 2004; Orlando et al., 2008).
Similar centrosome-intrinsic mechanisms have been proposed for preventing centrosome reduplication in metazoans. Like side-by-side SPBs that are connected by a full bridge, engaged centrioles cannot initiate a new round of centriole duplication (Tsou and Stearns, 2006). Centriole disengagement is essential for a new round of centriole duplication and appears to be regulated by separase (Tsou and Stearns, 2006). Despite the similarity in mechanism, it is unlikely that separase plays a role in SPB separation, as separase mutants (esp1) do not exhibit a defect in SPB separation (Baum et al., 1988). Our work and the work of others (Fitch et al., 1992; Crasta et al., 2006) suggest that B-cyclin/CDKs trigger SPB separation, although the mechanisms have yet to be elucidated.
Mutations in the bridge protein Sfi1 have been found that block SPB separation (Anderson et al., 2007). Both of these mutations fall in a consensus CDK phosphorylation site and Sfi1 has been shown to be highly phosphorylated by Clb2/Cdk1 (Ubersax et al., 2003; Anderson et al., 2007). Additionally, these mutations are located in the C-terminus of Sfi1, where Sfi1 is proposed to bind the C-termini of other Sfi1 molecules forming the structure of the bridge (Li et al., 2006). Thus, it is possible that phosphorylation of Sfi1 by mitotic cyclin/CDKs may contribute to SPB separation by destabilization of the bridge.
A previous study demonstrated that extrinsic control by mitotic cyclin/CDK also prevents SPB reduplication (Haase et al., 2001); however, the mechanisms by which mitotic cyclins prevent SPB reduplication remained unexplored. By expressing a stabilized version of the mitotic B-cyclin (Clb2Δdb) in at different points in G1, we established that mitotic cyclin/CDKs are likely to prevent reduplication by inhibiting early events of the SPB duplication cycle (Figures 1, 3, and 5). The fact that Clb2 cannot inhibit SPB duplication after α-factor arrest suggests that SPBs are already “licensed” to duplicate at START. Previous studies dissecting SPB structure during the duplication cycle have shown that as cells transit from early G1 to START, the SPB half-bridge elongates, and a satellite is assembled at the distal end of the bridge (Byers and Goetsch, 1974, 1975). Thus mitotic cyclin/CDKs may inhibit bridge elongation or satellite assembly. Indeed, when we examined the SPB structure by EM in cells that expressed Clb2Δdb in early G1, we did not observe satellites in any of the cells analyzed, although we could not determine unambiguously if bridge elongation was inhibited. Taken together, these findings argue that the satellite-bearing SPB may be analogous to the preduplicative complex in DNA licensing models as previously hypothesized (Adams and Kilmartin, 1999; Haase et al., 2001) and that one or more steps leading up to satellite assembly are inhibited by mitotic cyclin/CDKs.
Mitotic cyclin/CDKs could inhibit satellite assembly indirectly or by direct phosphorylation of satellite or half-bridge proteins. Interestingly, previous studies have shown that mitotic cyclins Clb2 (Hood et al., 2001; Bailly et al., 2003) and Clb4 (Maekawa and Schiebel, 2004) colocalize with the SPB, suggesting that direct phosphorylation of satellite or half-bridge proteins could occur. The satellite assembles on the cytoplasmic face of the SPB (Adams and Kilmartin, 1999), suggesting that the subcellular localization of Clb2 may be important for its ability to inhibit SPB licensing. Our finding that Clb2 must be localized to the cytoplasm in order to inhibit licensing suggests that the inhibitory targets of Clb2 may indeed be associated with the half-bridge and/or satellite.
Although Clb2 can inhibit SPB duplication if expressed in early G1, neither Clb5 nor Cln2 can inhibit SPB duplication when expressed similarly, indicating that inhibition of SPB duplication in specific to Clb2 and not a function of elevated CDK activity in early G1 (Figures 3 and 4). Furthermore, we have shown that cytoplasmic localization of Clb5 is not sufficient for inhibiting SPB duplication (Figure 7), suggesting that the targets of Clb2/Cdk1 involved in inhibition of SPB duplication are not efficiently phosphorylated by Clb5/Cdk1. This finding is consistent with in vitro studies indicating that Clb5/Cdk1 may have preferences for specific substrates, whereas Clb2/Cdk1 can phosphorylate a wider range of targets (Loog and Morgan, 2005).
The results presented here suggest that at least two distinct mechanisms work together to prevent the reduplication of SPBs during the cell cycle. In early G1, when there is low mitotic cyclin/CDK activity, the preduplicative structure, the satellite bearing SPB, is allowed to assemble. After START, the activity of cyclin/CDK complexes (G1 cyclin/CDK under normal circumstances, but mitotic cyclin/CDK will suffice; Figure 5B) triggers the assembly of a new SPB, resulting in a structure with two side-by-side SPBs connected by a full bridge. This structure establishes a SPB-intrinsic block to SPB reduplication during late G1 and early S-phase when the components of the SPB are expressed (Orlando et al., 2008) and the proper activating kinases are present (Jaspersen et al., 2004). SPB separation is then triggered by the activation of mitotic B-cyclins (Fitch et al., 1992; Crasta et al., 2006) or inefficiently by S-phase cyclins (Haase et al., 2001), releasing the SPB-intrinsic block to SPB reduplication. After promoting SPB separation, mitotic cyclin/CDKs inhibit SPB reduplication for the remainder of the cycle by preventing the premature assembly of the preduplicative SPB complex. At the end of mitosis, the inhibition and destruction of mitotic cyclins releases this inhibition, and cells are able to reassemble the preduplicative SPB complex in the early G1.
As has been shown for DNA replication, we suspect that mitotic cyclin/CDKs phosphorylate multiple protein targets and that any of those phosphorylations could block SPB reduplication. However, our findings here suggest that the important targets are localized to the cytoplasm and at least a subset of these proteins may be structural components of the satellite or bridge structure. In a large-scale screen for CDK targets, three of the four known components of the bridge or satellite tested were found to be phosphorylated by Clb2/Cdk1 (Ubersax et al., 2003), suggesting that many bridge and satellite proteins may, in fact, be regulated by CDK activity. One of these proteins, Spc42, a protein that has been shown to localize to the satellite (Adams and Kilmartin, 1999), is known to be phosphorylated by CDK (Donaldson and Kilmartin, 1996; Ubersax et al., 2003; Jaspersen et al., 2004). Cln1,2/Cdk1 phosphorylations have been mapped to two of the eight minimal CDK consensus sites within the Spc42. These phosphorylations likely contribute to SPB assembly (Jaspersen et al., 2004); however, it remains possible that phosphorylation of additional residues in Spc42 by B-cyclin/CDK could be important in the inhibition of SPB reduplication later in the cell cycle.
It remains unclear whether centrosome duplication in metazoans is governed by the same mechanisms we have identified in yeast, but remarkable similarities between the duplication cycles (reviewed in Adams and Kilmartin, 2000) suggest that analogous regulatory themes may exist. Centrosome-intrinsic mechanisms clearly prevent centrosome reduplication, and centriole disengagement may be an analogous process to SPB separation. However, centriole disengagement in metazoans appears to be regulated by separase (Tsou and Stearns, 2006), whereas in budding yeast, separation is regulated by B-cyclin/CDKs (Fitch et al., 1992; Crasta et al., 2006). Furthermore, centriole disengagement does not occur until mitosis, so the centrosome-intrinsic mechanism prevents reduplication for the bulk of the cell cycle. The question then arises, are additional mechanisms required to prevent centrosome reduplication during the interval between centriole disengagement in mitosis and cytokinesis, and if so, do CDKs inhibit the assembly of a preduplicative centrosome complex, perhaps by inhibiting procentriole assembly? Although several studies have suggested that cyclin B/Cdk1 inhibits centrosome duplication (Hinchcliffe et al., 1998; Lacey et al., 1999; Vidwans et al., 1999, 2003), it is not clear whether cyclin B/Cdk1 inhibits centrosome duplication directly, if it inhibits separase activation (Stemmann et al., 2001), or if it inhibits progression into cell cycle phases permissive for centrosome duplication.
ACKNOWLEDGMENTS
We thank Mark Chee, Jeffrey Kovacs (Duke University Medical Center, Durham, NC), and Daniel Lew (Duke University Medical Center) for critical reading of the manuscript. We are grateful to Mark Chee, Eric Bailly (Genome Instability and Carcinogenesis, CNRS, France), Fred Cross (The Rockefeller University, New York, NY), Steve Reed (The Scripps Research Institute, La Jolla, CA), Raphael Valdivia (Duke University Medical Center), Curt Wittenberg (The Scripps Research Institute), and Daniel Lew for providing plasmids and strains. We also thank Tom Giddings, Mark Winey, and Janet Meehl (University of Colorado, Boulder, CO) for assistance with electron microscopy. This work was funded by a grant from the National Science Foundation (MCB-0721751) to S.B.H.
Abbreviations used:
- SPB
spindle pole body
- CDK
cyclin-dependent kinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0148) on May 14, 2008.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams I. R., Kilmartin J. V. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. doi: 10.1016/s0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- Amon A., Irniger S., Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Anderson V. E., Prudden J., Prochnik S., Giddings T. H., Jr., Hardwick K. G. Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function. Mol. Biol. Cell. 2007;18:2047–2056. doi: 10.1091/mbc.E06-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M. N. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 2003;116:4119–4130. doi: 10.1242/jcs.00706. [DOI] [PubMed] [Google Scholar]
- Balczon R., Bao L., Zimmer W. E., Brown K., Zinkowski R. P., Brinkley B. R. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Yip C., Goetsch L., Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol. Cell. Biol. 1988;8:5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: Stenderup M.K.-B. A., Friis J., editors. Molecular Genetics in Yeast. vol. 16. Copenhagen: Munksgaard; 1981. pp. 119–133. [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Chial H. J., Winey M. Mechanisms of genetic instability revealed by analysis of yeast spindle pole body duplication. Biol. Cell. 1999;91:439–450. [PubMed] [Google Scholar]
- Crasta K., Huang P., Morgan G., Winey M., Surana U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006;25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Yuste-Rojas M., Gray S., Jacobson M. D. Specialization and targeting of B-type cyclins. Mol. Cell. 1999;4:11–19. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- D'Assoro A. B., Lingle W. L., Salisbury J. L. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Delattre M., Gonczy P. The arithmetic of centrosome biogenesis. J. Cell Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- Donaldson A. D., Kilmartin J. V. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington N. P., Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- Fitch I., Dahmann C., Surana U., Amon A., Nasmyth K., Goetsch L., Byers B., Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard D. L., Hafezi S., Zhang T., Doxsey S. J. Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. J. Cell Biol. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber L., Kuo S. C., Hoyt M. A. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J. Biol. Chem. 1999;274:9564–9572. doi: 10.1074/jbc.274.14.9564. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Haase S. B., Reed S. I. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Haase S. B., Winey M., Reed S. I. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Cassels G. O., Rieder C. L., Sluder G. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J. Cell Biol. 1998;140:1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Li C., Thompson E. A., Maller J. L., Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hood J. K., Hwang W. W., Silver P. A. The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci. 2001;114:589–597. doi: 10.1242/jcs.114.3.589. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K., Saunders W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. D., Gray S., Yuste-Rojas M., Cross F. R. Testing cyclin specificity in the exit from mitosis. Mol. Cell. Biol. 2000;20:4483–4493. doi: 10.1128/mcb.20.13.4483-4493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Huneycutt B. J., Giddings T. H., Jr., Resing K. A., Ahn N. G., Winey M. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev. Cell. 2004;7:263–274. doi: 10.1016/j.devcel.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G. G. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K. R., Jackson P. K., Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker S., Valdivieso M. H., Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sandercock A. M., Conduit P., Robinson C. V., Williams R. L., Kilmartin J. V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W. L., Lutz W. H., Ingle J. N., Maihle N. J., Salisbury J. L. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M., Morgan D. O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Maekawa H., Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A. M., Bartek J., Nigg E. A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Miller M. E., Cross F. R. Cyclin specificity: how many wheels do you need on a unicycle? J. Cell Sci. 2001;114:1811–1820. doi: 10.1242/jcs.114.10.1811. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Orlando D. A., Lin C. Y., Bernard A., Yang J. Y., Socolar J.E.S., Iversen E. S., Hartemink A. J., Haase S. B. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan G. A., Purohit A., Wallace J., Knecht H., Woda B., Quesenberry P., Doxsey S. J. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- Rao P. N., Johnson R. T. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Richardson H., Lew D. J., Henze M., Sugimoto K., Reed S. I. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G., Lewis K. Relationship between nuclear DNA synthesis and centrosome reproduction in sea urchin eggs. J. Exp. Zool. 1987;244:89–100. doi: 10.1002/jez.1402440111. [DOI] [PubMed] [Google Scholar]
- Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Verma R., Annan R. S., Huddleston M. J., Carr S. A., Reynard G., Deshaies R. J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Vidwans S. J., Wong M. L., O'Farrell P. H. Mitotic regulators govern progress through steps in the centrosome duplication cycle. J. Cell Biol. 1999;147:1371–1378. doi: 10.1083/jcb.147.7.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans S. J., Wong M. L., O'Farrell P. H. Anomalous centriole configurations are detected in Drosophila wing disc cells upon Cdk1 inactivation. J. Cell Sci. 2003;116:137–143. doi: 10.1242/jcs.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS 2, novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]