Abstract
Molecular chaperones prevent aggregation of denatured proteins in vitro and are thought to support folding of diverse proteins in vivo. Chaperones may have some selectivity for their substrate proteins, but knowledge of particular in vivo substrates is still poor. We here show that yeast Rot1, an essential, type-I ER membrane protein functions as a chaperone. Recombinant Rot1 exhibited antiaggregation activity in vitro, which was partly impaired by a temperature-sensitive rot1-2 mutation. In vivo, the rot1-2 mutation caused accelerated degradation of five proteins in the secretory pathway via ER-associated degradation, resulting in a decrease in their cellular levels. Furthermore, we demonstrate a physical and probably transient interaction of Rot1 with four of these proteins. Collectively, these results indicate that Rot1 functions as a chaperone in vivo supporting the folding of those proteins. Their folding also requires BiP, and one of these proteins was simultaneously associated with both Rot1 and BiP, suggesting that they can cooperate to facilitate protein folding. The Rot1-dependent proteins include a soluble, type I and II, and polytopic membrane proteins, and they do not share structural similarities. In addition, their dependency on Rot1 appeared different. We therefore propose that Rot1 is a general chaperone with some substrate specificity.
INTRODUCTION
Newly synthesized polypeptides need to be correctly folded to a proper three-dimensional structure to mature into functional proteins. In the cell, a variety of proteins assist in the folding of nascent proteins and are collectively called molecular chaperones. A molecular chaperone is basically defined as a protein that transiently associates with nascent proteins and promotes their folding and/or assembly. Based on their degree of substrate specificity, molecular chaperones can be classified from “general” ones interacting with diverse proteins to “specific” ones interacting with specific proteins or protein families. The primary function of a molecular chaperone is the stabilization of the unfolded and unstable polypeptide, and in vitro, a general chaperone is often capable of preventing aggregation of denatured proteins (Hartl, 1996). The 70-kDa heat-shock protein (Hsp70) is an abundant and the most prominent general chaperone. Hsp70 is thought to assist most nascent proteins just after and/or during their synthesis. Another abundant chaperone, Hsp90 protects several denatured proteins in vitro (Freeman and Morimoto, 1996), which suggests that Hsp90 can function as a chaperone for diverse proteins. However, in vivo, Hsp90 seems to be engaged in the folding of specific subsets of proteins such as steroid hormone receptors and some kinases (Young et al., 2004; Caplan et al., 2007). The eukaryotic chaperonin is also thought to possess some specificity for its client proteins (Spiess et al., 2004). To understand the functions of individual chaperones in a cell, it is essential to elucidate their in vivo substrate proteins, as well as to characterize the dependency of the substrates on these chaperones. Kerner et al. (2006) showed that diverse proteins are bound to the bacterial general chaperone, GroEL/ES, but suggested that only one-third of the substrates absolutely require the chaperone for folding. To date, we only have knowledge of the in vivo substrates and their dependency on general chaperones for several chaperones.
The endoplasmic reticulum (ER) is the site of protein folding in the secretory pathway. In the ER, many proteins are glycosylated and subjected to disulfide bond formation. Insertion of the membrane proteins into the membrane and the glycophosphatidylinositol (GPI) anchoring of some surface proteins also occur in the ER (Alberts et al., 2002). BiP, or Kar2 in yeast, the ER resident Hsp70, promotes translocation and folding of nascent polypeptides (Fewell et al., 2001). In addition to BiP/Kar2, various molecular chaperones and folding enzymes such as calnexin and protein disulfide isomerase fulfill many of the complex requirements for protein folding in the ER. Calnexin is a chaperone that is basically specific to N-glycosylated proteins (Williams, 2006), and protein disulfide isomerase promotes the formation and rearrangement of the disulfide bonds (Freedman, 2002; Hosoda et al., 2003). The ER also functions as the quality control compartment of folding proteins (Ellgaard et al., 1999). Proteins that fail to gain native conformation are retained in the ER and eventually degraded via a pathway called ER-associated degradation (ERAD). In the ERAD pathway, substrate proteins are retro-translocated to the cytosol, ubiquitylated and degraded by the proteasome (Carvalho et al., 2006; Denic et al., 2006). ER retention and/or ERAD of a protein strongly suggests that it is misfolded and/or unassembled.
We previously proposed that an essential, type-I ER membrane protein, Rot1 cooperates with BiP/Kar2 in the folding of nascent proteins in the ER of yeast cells (Takeuchi et al., 2006a,b). Several lines of evidences have suggested that Rot1 is involved in protein folding: ROT1 genetically interacts with genes encoding ER chaperones such as BiP/Kar2, and the unfolded protein response (Kimata et al., 2007; Kohno 2007) was induced in the temperature-sensitive rot1-2 mutant. Furthermore, folding of disulfide-reduced carboxypeptidaseY (CPY) was likely to be severely perturbed in kar2-1 rot1-2 double mutant cells (Takeuchi et al., 2006a). Rot1 does not have any known functional motif, and the details of its molecular functions have not been elucidated yet. In this report, we show that Rot1 itself functions as a general chaperone. Recombinant Rot1 protein prevented aggregation of denatured proteins in vitro. Moreover, we found several reliable or possible in vivo Rot1 substrates with various properties.
MATERIALS AND METHODS
Media, Plasmids, Yeast Strains, and Antibodies
Plasmids used in this study were constructed mainly using products of genomic PCR and are listed in Table 1. Yeast cells were grown in YPD (2% glucose, 1% yeast extract, 2% polypeptone, and 0.02% adenine sulfate) or SC (2% glucose, 0.67% yeast nitrogen base w/o amino acids; BD, Franklin Lakes, NJ) and supplemental amino acids (Kaiser et al., 1994). Standard genetic manipulations of yeast cells were performed as described (Kaiser et al., 1994). Yeast strains used in this study are listed in Table 2. For disruption of UBC7, a disruption cassette was amplified by PCR from the genome of strain W303-CQ (Hiller et al., 1996), and the resulting DNA fragment was introduced to strain YM16 (ROT1) and YM18 (rot1-2) to generate YM23 (ROT1 ubc7Δ) and YM24 (rot1-2 ubc7Δ), respectively. The kar2-1 mutation was introduced into the strains YM16 and YM18 by transformation with SpeI/SalI-digested pB-kar2-1-LEU2. LEU+, and temperature-sensitive clones were subsequently selected (examined at 39 and 32°C for YM88 [ROT1 kar2-1] and YM89 [rot1-2 kar2-1], respectively). For expression of the epitope-tagged proteins, the cells (YM16, 18, 23, 24, 88, and 89) were transformed with the corresponding plasmids digested by the indicated restrictive enzymes (see Table 1). Integration of the introduced DNA fragments into the target genes was confirmed by genomic PCR. To introduce the sec12-4 mutation, YM41 (ROT1 HA-KRE6) was transformed with pB-sec12-URA3 digested by XbaI/XhoI, and the URA+, and temperature-sensitive clones were selected. Anti-Kar2, Anti-Rot1, and anti-Sec61 antibodies were previously described (Takeuchi et al., 2006a). Anti-Mnn9 antibody was kindly provided by Dr. Koji Yoda (Tokyo University). Anti-hemagglutinin (HA; 12CA5, Roche, Basel, Switzerland) and anti-CPY (Rockland Immunochemicals, Gilbertsville, PA) were purchased.
Table 1.
Plasmids used in this study
| Name (enzyme)a | Vector | Insert |
|---|---|---|
| pB-KRE5-HA (SacI) | pBluescript II KS(+)b | KRE5(+3627c–+4095)-3HA-HDEL-stop-TRP1d,e-KRE5(+4115–+4720) |
| pB-HA-KRE6 (SacI) | pBluescript II KS(+) | KRE6(−1275–−712)-CgTRP1f-KRE6(−639–−1)-ATG-3HA-KRE6(+1–+1486) |
| pB-BIG1-HA (NotI/XhoI) | pBluescript II KS(+) | BIG1(−605–+1092)-3HA-stop-TRP1d-BIG1(+1111–+2082) |
| pB-sec12-URA3 (XbaI/XhoI) | pBluescript II KS(+) | sec12-4(−783–+1967)g-URA3h-SEC12(+1965–+2340) |
| pRS314-ATG22-HA | pRS314i (TRP1, CEN) | ATG22(−869–+1584)-3HA-stop-ADH1td,j |
| pB-DRS2-HA (KpnI/SacI) | pBluescript II KS(+) | DRS2(+3224–+4065)-3HA-stop-TRP1d-DRS2(+3657–+4065) |
| pRS424-DRS2-HA | pRS424i (TRP1, 2 μ) | DRS2(−617–+4065)-3HA-stop-ADH1t |
| pT10-GUP1-HA | pGCT10e (TRP1, CEN) | GUP1(−857–+1680)-3HA-stop-ADH1t |
| pT20-GUP1-HA | pGCT20e (TRP1, 2 μ) | GUP1(−857–+1680)-3HA-stop-ADH1t |
| pRS314-HA-OPI3 | pRS314 (TRP1, CEN) | OPI3(−643–−1)-ATG-3HA-OPI3(+1–+853) |
| pB-PMR1-HA (BamI/XhoI) | pBluescript II KS(+) | PMR1(+2046–+2850)-2HA-stop-TRP1d-PMR1(+2277–+2850) |
| pYEX-ROT1-HA-His | pYEX4Tk (leu2d, URA3, 2 μ) | TDH3(−1037–−7)-ROT1-(+1–+693)-HA-His8-stop-TDH3t(+1967–+2107) |
| pYEX-rot1-2-HA-His | pYEX4T (leu2d, URA3, 2 μ) | TDH3(−1037–−7)-rot1-2-(+1–+693)-HA-His8-stop-TDH3t(+1967–+2107) |
| pB-kar2-1-LEU2 (SpeI/SalI) | pBluescript II KS(+) | kar2-1(+1161–+2643)-LEU2-KAR2(+2080–+2990) |
a The plasmid was digested with indicated enzyme(s) and introduced to yeast cells.
b Stratagene, La Jolla, CA.
c The A residue of the initiation codon was set at +1.
d Derived from pGCT10.
f Candida glabrata TRP1; derived from pUC18-CgTRP1 (provided by Dr. Mukai, Osaka University).
g The sec12-4 mutant gene was obtained from the genome of strain MBY10-7A (Nakano et al., 1988) by PCR.
h Derived from YCp50 (Rose et al., 1987).
j Terminator.
k Clontech Lab, Mountain View, CA.
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype and harboring plasmid | Source |
|---|---|---|
| FY23 | Mata leu2 trp1 ura3 | Takeuchi et al. (2006a) |
| MBY10-7A | Mata his3 his4 leu2 trp1 ura3 suc gal2 sec12-4 | Nakano et al. (1988) |
| W303CQ | Mat α ade2 his3 leu2 trp1 ura3 can1 ubc7Δ::LEU2 | Hiller et al. (1996) |
| YM16 | Mat α ade2 his3 leu2 lys2 trp1 ura3 | Takeuchi et al. (2006a) |
| YM18 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 | Takeuchi et al. (2006a) |
| YM23 | Mat α ade2 his3 leu2 lys2 trp1 ura3 ubc7Δ::LEU2 | This study |
| YM24 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 ubc7Δ::LEU2 | This study |
| YM27 | Mat α ade2 his3 leu2 lys2 trp1 ura3 KRE5-HA-HDEL::TRP1 | This study |
| YM28 | Mat α ade2 his3 leu2 lys2 trp1 ura3 KRE5-HA-HDEL::TRP1 rot1-2 | This study |
| YM41 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 | This study |
| YM42 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 rot1-2 | This study |
| YM43 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 ubc7Δ::LEU2 | This study |
| YM44 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 rot1-2 ubc7Δ::LEU2 | This study |
| YM84 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 | This study |
| YM85 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 rot1-2 | This study |
| YM86 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 ubc7Δ::LEU2 | This study |
| YM87 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 rot1-2 ubc7Δ::LEU2 | This study |
| YM88 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 | This study |
| YM89 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 rot1-2 | This study |
| YM90 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 kar2-1::LEU2 | This study |
| YM91 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 kar2-1::LEU2 rot1-2 | This study |
| YM96 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 kar2-1::LEU2 | This study |
| YM97 | Mat α ade2 his3 leu2 lys2 trp1 ura3 BIG1-HA::TRP1 kar2-1::LEU2 rot1-2 | This study |
| YM132 | Mat α ade2 his3 leu2 lys2 trp1 ura3 HA-KRE6::CgTRP1 sec12–4::URA3 | This study |
| YM173 | Mat α ade2 his3 leu2 lys2 trp1 ura3 ubc7Δ (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM174 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 ubc7Δ (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM179 | Mat α ade2 his3 leu2 lys2 trp1 ura3 (pRS314-HA-OPI3 [CEN TRP1]) | This study |
| YM180 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 (pRS314-HA-OPI3 [CEN TRP1]) | This study |
| YM181 | Mat α ade2 his3 leu2 lys2 trp1 ura3 (pT10-GUP1-HA [CEN TRP1]) | This study |
| YM182 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 (pT10-GUP1-HA [CEN TRP1]) | This study |
| YM183 | Mat α ade2 his3 leu2 lys2 trp1 ura3 DRS2-HA::TRP1 | This study |
| YM184 | Mat α ade2 his3 leu2 lys2 trp1 ura3 DRS2-HA::TRP1 rot1-2 | This study |
| YM191 | Mat α ade2 his3 leu2 lys2 trp1 ura3 (pRS314-ATG22-HA [CEN TRP1]) | This study |
| YM192 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 (pRS314-ATG22-HA [CEN TRP1]) | This study |
| YM193 | Mat α ade2 his3 leu2 lys2 trp1 ura3 (pRS424-DRS2-HA [2 μ TRP1]) | This study |
| YM194 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 (pRS424-DRS2-HA [2 μ TRP1]) | This study |
| YM195 | Mat α ade2 his3 leu2 lys2 trp1 ura3 (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM196 | Mat α ade2 his3 leu2 lys2 trp1 ura3 rot1-2 (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM197 | Mat α ade2 his3 leu2 lys2 trp1 ura3 PMR1-HA::TRP1 | This study |
| YM198 | Mat α ade2 his3 leu2 lys2 trp1 ura3 PMR1-HA::TRP1 rot1-2 | This study |
| YM199 | Mata leu2 trp1 ura3 (pYEX-ROT1-HA-His [2 μ leu2d URA3]) | This study |
| YM200 | Mata leu2 trp1 ura3 (pYEX-rot1-2-HA-His [2 μ leu2d URA3]) | This study |
| YM210 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM211 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 rot1-2 (pT20-GUP1-HA [2 μ TRP1]) | This study |
| YM212 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 (pRS424-DRS2-HA [2 μ TRP1]) | This study |
| YM213 | Mat α ade2 his3 leu2 lys2 trp1 ura3 kar2-1::LEU2 rot1-2 (pRS424-DRS2-HA [2 μ TRP1]) | This study |
| YM214 | Mat α ade2 his3 leu2 lys2 trp1 ura3 KRE5-HA-HDEL::TRP1 kar2-1::LEU2 | This study |
| YM215 | Mat α ade2 his3 leu2 lys2 trp1 ura3 KRE5-HA-HDEL::TRP1 kar2-1::LEU2 rot1-2 | This study |
Production and Purification of Recombinant Rot1 Protein
For production of Rot1HAHis8, YM199 (FY23 harboring pYEX-ROT1-HA-His) was incubated in SC not containing leucine to A600 = 1.5–1.8 at 30°C. The following procedures were performed at 4°C. The cells were lysed in L buffer (50 mM KH2PO4/K2HPO4, pH 7.4, at 4°C, 500 mM KCl, 2 mM MgCl2, 5 mM imidazole, 1 mM 2-mercaptoethanol, 1%[wt/vol] Triton X-100, 20%[vol/vol] glycerol) containing a protease inhibitor (PI) mix (1 mM PMSF, 0.2 mg/ml leupeptin and pepstatin A [Peptide Institute, Osaka, Japan], 1 mg/ml benzamidine [final concentrations]) by agitation with glass beads using Bead-beater (Biospec, Bartlesville, OK). The lysate was clarified by centrifugation at 100,000 × g for 1 h and incubated with Ni-NTA agarose resin (Qiagen, Hilden, Germany) o/n. The resin was washed with W buffer (L buffer containing 20 mM imidazole), and Rot1 was eluted with C buffer (20 mM HEPES, pH 7.4, at 4°C, 500 mM KCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol, and 20% glycerol) containing 250 mM imidazole. The eluted protein was further purified by gel filtration on Superdex75 (GE Healthcare, Amersham plc, United Kingdom) column equilibrated with C buffer. Purified Rot1 was bound to Ni-NTA agarose again, eluted, dialyzed against S buffer (C buffer containing 50% glycerol), and stored at −20°C. The rot1-2HAHis8 mutant protein was produced in YM200, purified, and stored similarly.
Aggregation Protection Assay
α-Mannosidase (Sigma, St. Louis, MO) and citrate synthase (Roche) were dialyzed against A buffer (50 mM AcOH/AcONa, pH 5.5, at 4°C, 150 mM KCl, 0.1 mM ZnSO4, and 10% glycerol) and B buffer (20 mM HEPES, pH 7.0, at 4°C, 150 mM KCl, 2 mM MgCl2, and 10% glycerol), respectively, at 4°C. The proteins were divided into aliquots, frozen with liquid N2, and stored at −80°C. For denaturation, α-mannosidase (30 μM) and citrate synthase (40 μM) were mixed with equal volumes of D buffer (R buffer [20 mM HEPES, pH 7.0, at 30°C, 50 mM KCl, and 2 mM MgCl2] containing 8 M guanidine-HCl) and incubated at RT for 1 h. Denatured α-mannosidase or citrate synthase was diluted 50-fold in R buffer to 0.3 or 0.4 μM, respectively, and aggregation was monitored by following an increase in A320. The indicated amounts of Rot1, rot1-2 mutant protein, or BSA were incubated together.
Examination of the Cellular Amount of Proteins
To examine the cellular amount of proteins by Western blotting, the KRE5-HA (YM27, 28), HA-KRE6 (YM41, 42) and BIG1-HA (YM85, 86) strains were cultured in YPD, and the strains expressing ATG22-HA (YM191, 192), DRS2-HA (YM183, 184) and GUP1-HA (YM181, 182) were grown in SC not containing tryptophan (Trp) at 23, 30, or 23°C and shifted to 37°C for 2, 4, or 6 h. The cells were harvested at A600 = 0.8–1.0 and lysed in 1% SDS-TBES (50 mM Tris, pH 7.4, at 4°C, 150 mM NaCl, and 5 mM EDTA) containing PI mix. The lysates were analyzed by Western blotting using anti-HA, anti-Rot1, or anti-Sec61 antibody. For detection of each protein, cell lysates containing the following amount of proteins were loaded: 5 μg for Kre5-HA and Atg22-HA; 0.5 μg for HA-Kre6; 10 μg for Big1-HA, Drs2-HA, and Gup1-HA; and 20 μg for Rot1 and Sec61.
Pulse-Chase Experiments
For [35S]methionine/cysteine (Met/Cys) pulse-chase experiments, SC medium not containing Met/Cys was used, referred to hereafter as SC. The cells were grown exponentially in SC at 23°C to A600 = 0.7–1.0, concentrated to 10 OD cells/ml, and incubated for 10 min at 37°C. The cells were labeled with [35S]Met/Cys (EXPRESS protein labeling mix; PerkinElmer, Waltham, MA) at 4 MBq/ml for 10 min at 37°C. At the beginning of the chase period, 1/50 vol of the chase solution (1% Met and 0.8% Cys in 0.1 N HCl) was added to the culture. At the indicated times, 200 μl of the culture was taken, mixed with 200 μl of YPD containing 20 mM NaN3, and placed on ice. The cells were lysed by agitation with glass beads in 100 μl of 1% SDS-TBES (1% S-TBES) containing PI mix at 4°C, and the lysate was incubated at 50°C for 5 min and centrifuged at 15,000 × g at RT for 5 min. The cleared lysate was diluted with 4 vol of 2.5% Triton X-100-TBES (2.5% T-TBES) and incubated with 1 μg of anti-HA antibody or 10 μg of anti-CPY antibody and 10 μl bed vol of protein A-Sepharose (GE Healthcare) at 4°C for 2 h or overnight. The beads were washed with 200 μl each of 1% T-TBES once, 1% T-TBES containing 0.5 M NaCl twice, and 1% T-TBES once again at RT. Recovered proteins were separated by SDS-PAGE and detected by autoradiography using the BAS2500 system (Fujifilm, Tokyo, Japan). For quantification of the radioactive signal of protein bands, Image Gauge ver. 4 software (Fujifilm) was used. For Figure 2B, cells were exponentially grown in YPD and incubated in SC for 1 h at 23°C (A600 = 0.6–1.0). The cells were then incubated for 10 min at 33°C, labeled with [35S]Met/Cys for 10 min, and chased for 30 min at 33°C. For Figure 4E, the cells were incubated for 10 min at 37°C, labeled with [35S]Met/Cys at 20 MBq/ml for 10 min, and chased for the indicated periods at 37°C. Cells (2 OD) were lysed in 100 μl 1% T-TBES containing protease inhibitors and centrifuged at 15,000 × g for 5 min at 4°C. Cleared lysates were incubated with 400 μl of 1% T-TBES, 5 μl anti-Rot1 antibody or nonimmune guinea pig serum and 10 μl bed vol of ProteinA-Sepharose for 2 h at 4°C, and the beads were washed four times with 200 μl of 1% T-TBES at 4°C. Precipitated proteins were eluted by incubation with 100 μl of 1% S-TBES for 10 min at 65°C. Immunoprecipitation of HA-Kre6 was performed as described above. Similar results were obtained in three independent experiments.
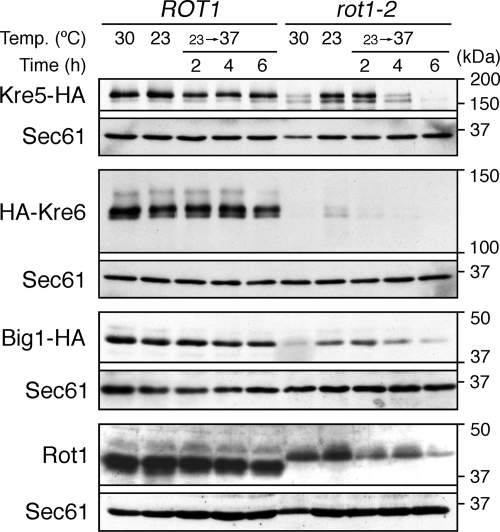
Figure 2.
Cellular levels of proteins important for cell wall 1,6-β-glucan synthesis are decreased by the rot1-2 mutation. YM27 (ROT1 KRE5-HA), YM28 (rot1-2 KRE5-HA), YM41 (ROT1 HA-KRE6), YM42 (rot1-2 HA-KRE6), YM84 (ROT1 BIG1-HA), and YM85 (rot1-2 BIG1-HA) were incubated in YPD at 23 or 30°C or shifted from 23 to 37°C for the indicated times. The cell lysates were subjected to Western blotting using anti-HA, anti-Rot1, and anti-Sec61. The cellular level of Sec61 was not affected by the rot1-2 mutation and served as a loading control.
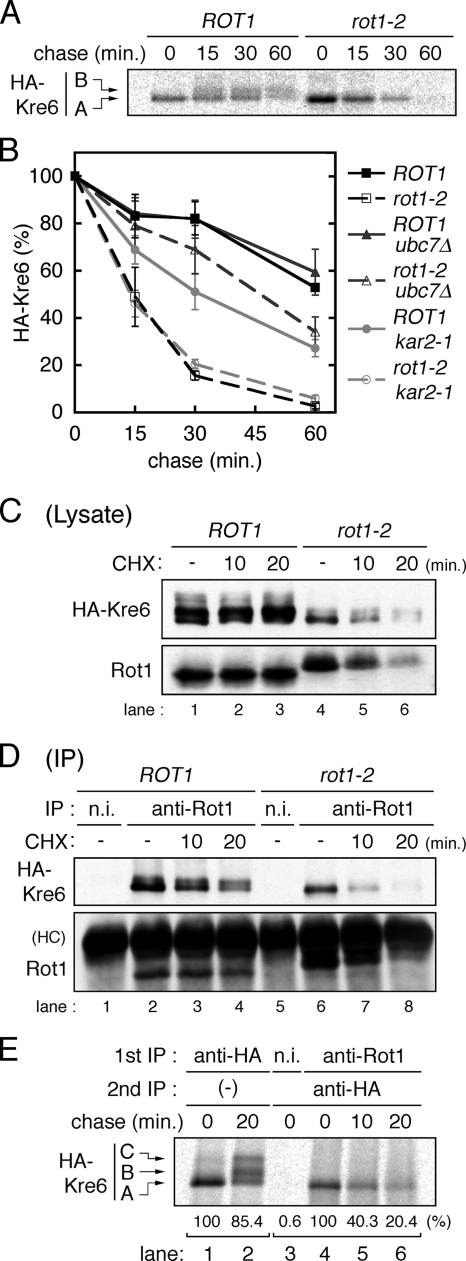
Figure 4.
Rot1 is a molecular chaperone for Kre6. (A) Conversion of HA-Kre6 does not occur in the rot1-2 mutant. ROT1 and rot1-2 cells expressing HA-Kre6 (YM41 and YM42) were incubated for 10 min at 37°C, labeled with [35S]Met/Cys for 10 min and chased for indicated periods at 37°C. Radiolabeled HA-Kre6 was recovered and detected as in Figure 3B. The positions of the A and the B forms of Kre6 are indicated in the left of the panel. The C form was barely detected under the conditions of this experiment. (B) HA-Kre6 is degraded by the ERAD in the rot1-2 cells. Pulse-chase experiments for HA-Kre6 were performed as in A, using the HA-KRE6 strains: YM41, YM42, and YM43 (ROT1 ubc7Δ); YM44 (rot1-2 ubc7Δ); YM90 (ROT1 kar2-1); and YM91 (rot1-2 kar2-1). Radioactive signals of HA-Kre6 (the A plus the B forms) were quantified, and the averages and SDs of at least five independent experiments are shown. Signal intensities of HA-Kre6 at the beginning of the chase period are set to 100%. (C and D) Rot1 interacts with HA-Kre6. Cells (YM41 and YM42) were grown in YPD at 23°C and incubated at 37°C for 10 min. Where indicated, the cells were further incubated with CHX (0.1 mg/ml) for the indicated times. Cell lysis and IP using anti-Rot1 antibody or a nonimmune serum (n.i.; as a negative control) were performed under a nondenaturing conditions at 4°C. The cell lysates (C) or the immunoprecipitants (D) were analyzed by Western blotting using anti-HA and anti-Rot1 antibodies. HC, immunoglobulin heavy chain. (E) Transient interaction of Rot1 and Kre6. Cells (YM41) were incubated, labeled with [35S]Met/Cys and chased for the indicated periods as in A. Anti-HA IP (lanes 1 and 2) or double-IP against Rot1 (or nonimmune serum) first and then HA (lanes 3–6) were performed. Relative amounts of recovered radioactive HA-Kre6 in the representative experiments are shown under the panel.
Subcellular Fractionation
YM41 (ROT1 HA-KRE6) cells were grown in 500 ml of YPD to A600 = 1.0 at 30°C. The cells were treated with 10 mM NaN3 for 5 min, collected, and incubated in 20 ml of reduction buffer (50 mM Tris, pH 9.6, at 25°C, 10 mM dithiothreitol, 10 mM NaN3) for 10 min at 30°C. The cells were spheroplasted in 10 ml spheroplasting buffer (20 mM Tris, pH 7.4, at 4°C, 150 mM NaCl, 1 M sorbitol, and 10 mM NaN3) with 2 mg of zymolyase T100 (Seikagaku Corporation, Tokyo, Japan) for 1 h at 30°C. The spheroplasts were collected, dissociated in 1.4 ml lysis buffer (50 mM Tris, pH 7.4, at 4°C, 150 mM KCl, 2 mM MgCl2, and 200 mM sucrose) containing PI mix and disrupted by a Dounce homogenizer at 4°C. The lysates were centrifuged at 600 × g, 4°C for 30 min, and 1 ml of cleared lysate was layered on the top of a 11 ml, 20–80% step sucrose gradient (from the bottom, 1 ml each of the lysis buffer containing 80, 70, 60, 55, 50, 45, 40, 35, 30, 25, and 20% sucrose was piled up). The gradient was centrifuged at 150,000 × g, 4°C for 8 h in the SW40Ti rotor (Beckman Coulter, Fullerton, CA) and 1-ml fractions were collected manually from the top of the gradient.
Coimmunoprecipitation
For coimmunoprecipitation of Rot1 with HA-Kre6 or Big1-HA, the cells (YM41 and 42 or YM85 and 86) were grown in YPD to A600 = 1.0 at 23°C and incubated at 37°C for 10 min. Where indicated, cycloheximide (CHX) was added to the culture to 0.1 mg/ml, and the cells were further incubated for 10 or 20 min at 37°C. For coimmunoprecipitation of Rot1 with Kre5-HA or Drs2-HA, the cells (YM27 or 194) were grown in YPD or SC not containing Trp, respectively, to A600 = 1.0 at 30°C. Then, the cells were treated with 10 mM NaN3 briefly and harvested. Cell lysis and immunoprecipitation (IP) were performed under nondenaturing conditions at 4°C as follows. Cells (2.5 OD) were disrupted by agitation with glass beads in 200 μl of 1% T-TBES containing PI mix. The lysate was cleared by the centrifugation at 15,000 × g for 5 min, and 2.5 μl of nonimmune serum and 10 μl bed vol of protein A-Sepharose were added, and the mixture was incubated for 1 h. The mixture was centrifuged at 15,000 × g for 5 min, and in the case of coIP with Kre5-HA and Drs2-HA, the supernatant was diluted twofold with 1% T-TBES. Then, 2.5 μl of anti-Rot1 antibody or nonimmune serum and 10 μl bed vol of protein A-Sepharose were added, and the mixture was rotated for 2 h. The beads were washed four times with 200 μl of 1% T-TBES, and the immunoprecipitants (one fifth of the sample for HA-Kre6, two thirds for Big1-HA; for Kre5-HA and Drs2-HA, similarly prepared two samples were combined) were analyzed by Western blotting. Similar results were obtained in at least three independent experiments.
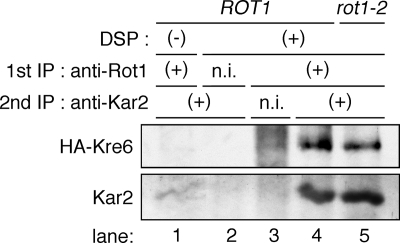
For double-immunoprecipitation in Figure 9, the cells (YM41 and YM42) were incubated, treated with NaN3, and harvested as above. Cells (5 OD) were dissociated in 400 μl of PBS and incubated with 2 mM dithiobis(succinimidyl)propionate (DSP; freshly prepared in DMSO at 200 mM) and 10 mM NaN3 for 1 h at 37°C, and Tris (pH 8.0 at RT) was added to 100 mM to quench the reaction. The cells were lysed in 200 μl of 1% S-TBES containing PI mix, incubated for 10 min at 65°C, and centrifuged at 15,000 × g for 5 min at RT. The cleared lysates were incubated with 800 μl of 2.5% T-TBES, 5 μl of nonimmune serum and 10 μl bed vol of protein A-Sepharose for 2 h at 4°C, centrifuged at 15,000 × g for 5 min at 4°C, and the supernatant was further incubated with 10 μl of anti-Rot1 or nonimmune serum and 10 μl bed vol of protein A-Sepharose overnight at 4°C. The beads were washed with 200 μl each of 1% T-TBES once, 1% T-TBES containing 0.5 M NaCl twice, and 1% T-TBES once again at RT. Recovered proteins were eluted by incubation with 100 μl of 1% S-TBES for 10 min at 65°C and incubated with 400 μl of 2.5% T-TBES, 1 μl of anti-Kar2 or nonimmune rabbit serum, and 10 μl bed vol of protein A-Sepharose for 2 h at 4°C. The beads were washed as above, and the immunoprecipitants were analyzed by Western blotting.
Figure 9.
Rot1 forms a ternary complex with BiP/Kar2 and Kre6. ROT1 and rot1-2 cells expressing HA-Kre6 (YM41 and YM42) were incubated for 10 min at 37°C, dissociated in PBS, and treated with 10 mM NaN3 and 2 mM DSP (or the vehicle, DMSO only) for 1 h at 37°C. Double immunoprecipitation was performed against Rot1 first and then Kar2. Nonimmune serums (n.i.) were used for negative controls. The second immunoprecipitants were analyzed by Western blotting using anti-HA and anti-Kar2 antibodies.
CHX Chase
The BIG1-HA cells (YM84, 85, 86, 87, 96, and 97) were grown in YPD at 23°C and incubated at 37°C for 2 h. Before the temperature shift, the density of the cultures was properly controlled so that the A600 of the culture was 0.8–1.0 after 2-h incubation at 37°C. CHX was then added to a final 0.1 mg/ml, and the cells were further incubated at 37°C. Aliquots were taken at the indicated times, and the cell lysates (equivalent to 0.05 OD cells) were subjected to Western blotting to detect Big1-HA. The protein bands on the x-ray films were scanned by ImageScanner (GE Healthcare) and quantified by Image Gauge ver. 4.
RESULTS
Recombinant Rot1 Prevents Aggregation of Denatured Proteins In Vitro
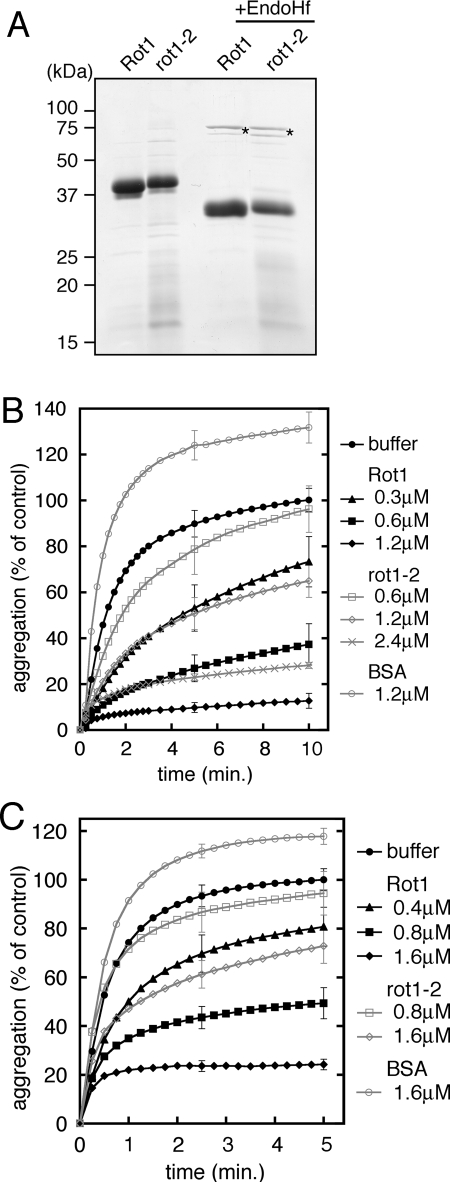
To elucidate the chaperone function of Rot1, we first generated a recombinant Rot1 and performed in vitro experiments. The recombinant Rot1 protein in which the C-terminal transmembrane region was replaced with the HA-His8 tag, and the N-terminal signal sequence was intact, was expressed in yeast cells from the strong TDH3 promoter. The protein was purified by Ni+2-chelated affinity-chromatography and gel filtration (Figure 1A). The rot1-2 mutant version was also generated and purified. Endoglycosidase H (EndoH) treatment increased the mobility of the proteins on SDS-PAGE, indicating that they were translocated into the ER and N-glycosylated as expected (Figure 1A).
Figure 1.
Recombinant Rot1 prevents aggregation of denatured proteins in vitro. (A) Purified recombinant Rot1. Wild-type Rot1 and rot1-2 proteins were expressed and purified as described in Materials and Methods. Protein, 3 μg each, was resolved by SDS-PAGE (12%), and the gel was stained with Coomasie Brilliant Blue. Where indicated, the proteins were treated with EndoHf (New England Biolabs, Beverly, MA) according to manufacturer's instructions. Asterisks, EndoHf. (B and C) Rot1 prevents aggregation of denatured α-mannosidase and citrate synthase. α-Mannosidase (B) or citrate synthase (C) was denatured with 4 M guanidine-HCl and diluted 50-fold to 0.3 μM (B) or 0.4 μM (C) at time 0. Indicated amounts of Rot1, rot1-2, or control protein BSA was incubated together, and A320 was measured at RT for the indicated periods. A320 of the buffer alone reaction at 10 min (B) or 5 min (C) is set to 100%, and the relative extent of the aggregation of denatured α-mannosidase or citrate synthase for each conditions of the reaction is shown. The averages of at least three reactions are shown. For simplicity, SDs are shown only for 5 and 10 min (B) or 2.5 and 5 min (C). Because the purity of rot1-2 was lower than Rot1, the actual concentration of rot1-2 in the reactions was estimated to be ∼75% that of Rot1.
When a protein is denatured in a chemical denaturant such as guanidine and the denaturant is rapidly diluted, the denatured protein tends to aggregate. In many cases, general chaperones bind to such denatured proteins and prevent their aggregation. We denatured α-mannosidase by guanidine and diluted it into an assay buffer. Aggregation was monitored by measuring absorbance at 320 nm, which indicates light scattering of protein aggregates. As shown in Figure 1B, Rot1 prevented aggregation of denatured α-mannosidase. The mutant rot1-2 protein also inhibited aggregation, albeit less efficiently than the wild-type Rot1 (Figure 1B). Aggregation of denatured citrate synthase was also inhibited by Rot1 (Figure 1C). These results indicate that Rot1 can directly bind to an unfolded protein, which is closely related to the in vivo functions of Rot1.
Proteins Important for Cell Wall 1,6-β-Glucan Synthesis Are Decreased in the rot1-2 Mutant
ROT1 was initially reported as a gene required for the 1,6-β-glucan synthesis (Bickle et al., 1998; Machi et al., 2004). We speculated that Rot1 is required for the maturation of proteins involved in the 1,6-β-glucan synthesis, so we checked the effects of the rot1-2 mutation on the cellular amount of three proteins, Kre5, Kre6, and Big1, which are especially important for this process. Kre5 is an ER-localized soluble protein that has similarity to UDP-glucose:glycoprotein glucosyltransferase (UGGT; Levinson et al., 2002). Kre6 was initially reported to be a Golgi-localized type-II membrane protein and was predicted to be a glucosidase or transglucosylase (Montijn et al., 1999; Li et al., 2002). However, a recent report suggested that Kre6 predominantly localizes in the ER and functions with an ER membrane protein named Keg1 (Nakamata et al., 2007). Big1 is an ER-localized type-I membrane protein that does not have any known functional motif (Azuma et al., 2002). The sequence encoding the HA epitope tag was integrated into the chromosomal KRE5, KRE6, or BIG1 locus to express tagged versions of each of these proteins. As shown in Figure 2, the rot1-2 mutation caused a drastic reduction in the levels of Kre5, Kre6, and Big1, as quantitated by Western blot analysis of the tagged proteins. Kre6 was the most severely affected: in rot1-2 cells, Kre6 was detected only faintly even at 23°C (a permissive temperature for the rot1-2 mutant) and was almost undetectable at 37°C (a restrictive temperature). Kre5 and Big1 expression were considerably decreased by a temperature shift to 37°C in the rot1-2 cells but not in the ROT1 cells. The rot1-2 mutation also caused a reduction in Rot1 itself (the rot1-2 mutation changes the mobility of the protein on SDS-PAGE; Takeuchi et al., 2006a). In contrast, expression of Sec61, the α-subunit of the protein translocation channel of the ER membrane, was not affected by the rot1-2 mutation. These results suggests that severe defect in 1,6-β-glucan synthesis originally observed in rot1 mutant cells is caused by decreases in the cellular levels of Kre5, Kre6, and Big1.
Rot1 Is a Molecular Chaperone for Kre6
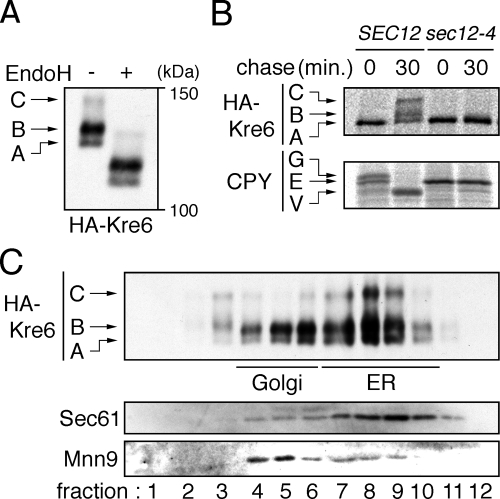
We next examined the relationship between Rot1 and Kre6 in detail. Kre6 appeared as three protein bands in cell lysate Western blots, and we designated the fastest migrating species as the A form, the middle one as the B form, and the slowest one as the C form (Figure 3A). These bands were shifted almost equally by EndoH digestion, so these differences in the gel mobility are not due to the number or modification of N-linked oligosaccharide chains. Because the predicted molecular mass of unmodified Kre6 is 80 kDa, Kre6 is likely to undergo another modification(s) such as O-glycosylation or phosphorylation (Roemer et al., 1994). To follow the modification of Kre6, we used a pulse-chase protocol in which the cells were metabolically labeled with [35S]Met/Cys and chased in the presence of excess unlabeled Met/Cys. At the beginning of the chase period, only the A form was detected, and the B and C forms appeared after 30 min (Figure 3B). This indicates that the A form was synthesized first and subsequently converted to the B or C forms, albeit not completely. When the transport from the ER to the Golgi was blocked by the sec12-4 mutation (Stevens et al., 1982), this conversion was not observed (Figure 3B), suggesting that Kre6 was modified to the B or C form in the Golgi. CPY served as a control confirming blockage of ER–Golgi transport in the sec12-4 mutant: only the ER form of CPY was detected in the mutant even after 30 min of chase, whereas the Golgi and the vacuolar forms were observed in the SEC12 cells (Figure 3B). Although a large portion of Kre6 in the cell lysate was of the B form (Figure 3A), Nakamata et al. (2007) recently showed that Kre6 is mainly localized to the ER. To address this discrepancy, we examined the intracellular distribution of Kre6 by subcellular fractionation using sucrose gradient ultracentrifugation. This analysis revealed that about two thirds of the A and B forms and most of the C form of Kre6 exist in the ER, and the rest is located in the Golgi (Figure 3C). This finding suggests that Kre6 cycles between the ER and the Golgi, which is consistent with Nakamata et al. (2007). Such cycling has also been reported for another protein, Mnn9 (Todorow et al., 2000).
Figure 3.
Modification and localization of Kre6. (A) Western blot detection of HA-Kre6. ROT1 cells expressing HA-Kre6 (YM41) was incubated in YPD at 30°C, and the cell lysates were subjected to anti-HA Western blotting. Where indicated, the lysate was treated with EndoHf. The positions of the three forms of HA-Kre6 not treated with EndoHf are indicated in the left of the panel. (B) Conversion of HA-Kre6 requires transport from the ER to Golgi. The cells (YM41 and YM132 [ROT1 HA-KRE6 sec12-4]) were incubated in SC for 1 h at 23°C, labeled with [35S]Met/Cys for 10 min at 33°C and chased for 30 min at 33°C. The cells were lysed, and HA-Kre6 or CPY was immunoprecipitated, separated by SDS-PAGE and detected by autoradiography. The positions of each forms of HA-Kre6 and CPY (E, ER; G, Golgi; V, vacuole) are indicated. (C) Intracellular distribution of HA-Kre6. YM41 was incubated in YPD at 30°C, spheroplasted and lysed gently with a Dounce homogenizer. The lysate was fractionated by a 20–80% sucrose step gradient ultracentrifugation. The fractions were subjected to Western blotting to detect HA-Kre6, Mnn9, and Sec61. Mnn9 and Sec61 were used as a Golgi or an ER marker, respectively.
To investigate the effects of the rot1-2 mutation on the modification and stability of Kre6, a pulse-chase experiment was performed at the restrictive temperature of 37°C. In the rot1-2 mutant, only the A form was detected (Figure 4A), which implies that Kre6 was retained in the ER. In addition, the rot1-2 mutation caused a rapid degradation of Kre6. Kre6 radioactivity was quantified, as shown in Figure 4B. Ubc7 is a ubiquitin-conjugating enzyme especially important for the ERAD, and ubc7Δ strongly interferes the ERAD of most known substrates (Hiller et al., 1996; Vashist and Ng, 2004; Kota et al., 2007). Kre6 degradation was inhibited in the rot1-2 ubc7Δ double mutant, indicating that Kre6 was degraded by the ERAD in the rot1-2 cells (Figure 4B). On the other hand, ubc7Δ did not affect the stability of Kre6 in a ROT1 background. These results indicate that the rot1-2 mutation causes ER retention and ERAD of Kre6, which strongly suggests that Kre6 failed to fold in the rot1-2 mutant.
One important characteristic of molecular chaperones is their transient interaction with the substrate protein. To detect the interaction between Rot1 and Kre6, Rot1 was immunoprecipitated by an anti-Rot1 antibody, and coprecipitated Kre6 was analyzed by Western blotting. As shown in Figure 4D, Kre6 was found in anti-Rot1 precipitates (lane 2), but not nonimmune serum precipitates (lane 1). Importantly, although the majority of Kre6 detected in cell lysates was of the B form (Figure 4C, lane 1), the A form of Kre6 was predominantly copurified with Rot1. This finding suggests that Rot1 selectively and transiently binds to recently synthesized Kre6. To confirm this idea, we performed double-immunoprecipitation in combination with a [35S]Met/Cys pulse-chase experiment. The cells were labeled and chased, and Rot1 was first immunoprecipitated. Then, the precipitate was subjected to a second immunoprecipitation to recover Kre6, which was separated by SDS-PAGE. Although the 35S-labeled Kre6 was stable and converted to the B and C forms during the chase period (Figure 4E, lane 1, 2), only the A form of Kre6 was found to interact with Rot1, and the amount of Rot1-bound Kre6 rapidly decreased (Figure 4E, lane 4–6). This data indicates that Rot1 transiently interacts with newly synthesized Kre6. Blockage of protein synthesis by the translation inhibitor CHX also caused a time-dependent decrease in the amount of Rot1-Kre6 complex detected (Figure 4D, lane 3 and 4), whereas the amount of cellular Kre6 was virtually unaffected (Figure 4C, lanes 1–3). An interaction between the rot1-2 mutant protein and Kre6 was also detected (Figure 4D, lane 6), but was less compared with the amount of Kre6 copurified with wild-type Rot1 (Figure 4D, compare lane 2 and 6). This result implies that the rot1-2 mutation impaired, but did not completely abolish the ability of Rot1 to bind to Kre6, as suggested by the in vitro experiments (Figure 1, B and C). However, we cannot exclude a possibility that lower abundance of Kre6 in rot1-2 cells resulted in less observed Rot1-Kre6 complex. Taken together, we conclude that Rot1 functions as a molecular chaperone for Kre6.
Rot1 Is a Molecular Chaperone for Big1
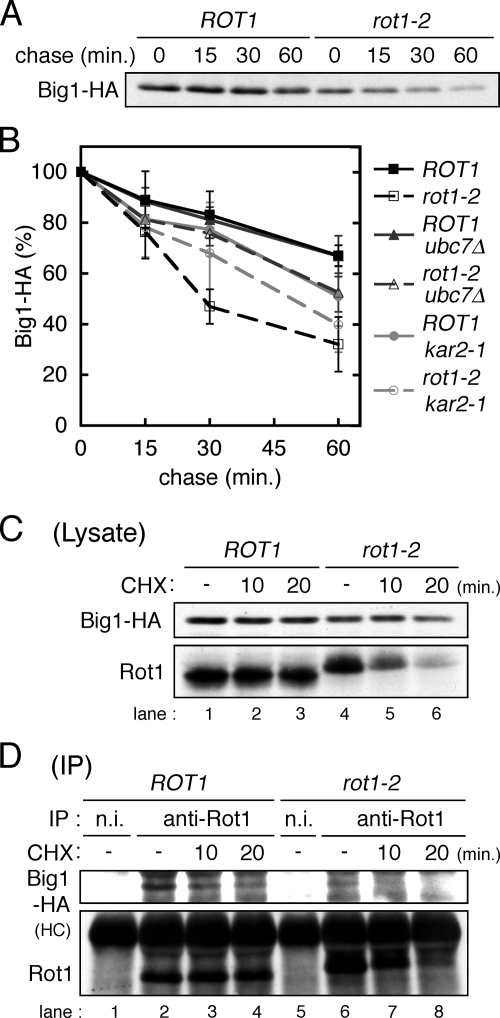
We next tested whether Rot1 also functions as a molecular chaperone for Big1. Because Big1 contains only small number of Met and Cys residues and was not labeled efficiently by [35S]Met/Cys, we used a CHX chase protocol. To follow the fate of Big1 synthesized at 37°C, the cells were incubated at 37°C for 2 h, and then CHX was added to the culture to inhibit further protein synthesis. Big1 in the cell lysates was detected by Western blotting (Figure 5A) and quantified (Figure 5B). The data clearly demonstrate that degradation of Big1 was accelerated by the rot1-2 mutation and that ubc7Δ stabilized Big1 only in the rot1-2 background. This finding indicates that the rot1-2 mutation causes the ERAD of Big1. In addition, a coIP analysis demonstrated an interaction between Rot1 and Big1, which was decreased by CHX treatment (Figure 5, C and D). This suggests that Rot1 transiently associates with Big1, probably while Big1 is still nascent. The Rot1-Big1 complex was only faintly detected in the rot1-2 cells (Figure 5D, lane 6). Taken together, these data indicate that Rot1 likely acts as a molecular chaperone for Big1.
Figure 5.
Rot1 is a molecular chaperone for Big1. (A) CHX chase of Big1-HA. ROT1 and rot1-2 cells expressing Big1-HA (YM84 and YM85) were grown in YPD at 23°C and incubated at 37°C for 2 h. The cells were further incubated with CHX (0.1 mg/ml) for indicated period at 37°C. The cell lysates were subjected to Western blotting to detect Big1-HA. (B) Big1-HA is degraded by the ERAD in the rot1-2 cells. CHX chase experiments were performed as in A using the BIG1-HA strains: YM84, YM85, and YM86 (ROT1 ubc7Δ); YM87 (rot1-2 ubc7Δ); YM96 (ROT1 kar2-1); and YM97 (rot1-2 kar2-1). The signal intensity of Big1-HA was quantified, and the averages and SDs of at least four independent experiments are shown. (C and D) Rot1 interacts with Big1-HA. Lysis of YM84 and YM85 cells and IP were performed, and results are presented as Figure 4, C and D.
Rot1 May Chaperone Kre5 and Atg22
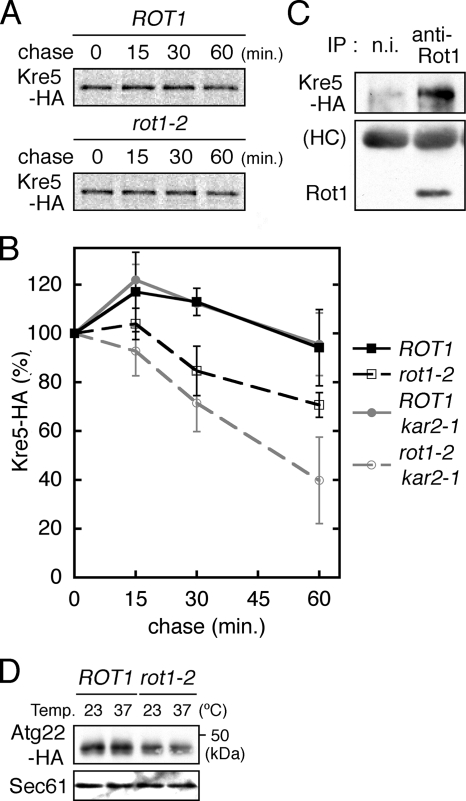
The relationship of Rot1 with maturation of Kre5 was also examined. Kre5 stability was examined by the [35S]Met/Cys pulse-chase experiment. As shown in Figure 6A and B, the rot1-2 mutation caused a weak but significant acceleration of Kre5 degradation. IP of Rot1 coprecipitated Kre5 (Figure 6C), indicating an interaction between Rot1 and Kre5. We suggest that Rot1 is involved in the folding and/or stabilization of Kre5, although the drastic reduction of Kre5 observed in the rot1-2 cells (Figure 2) could also be a separate and indirect consequence of prolonged incubation at 37°C.
Figure 6.
The relationship of Rot1 with Kre5 and Atg22 (A and B) The stability of Kre5-HA in YM27 (ROT1 KRE5-HA), YM28 (rot1-2 KRE5-HA), YM214 (ROT1 kar2-1), and YM215 (rot1-2 kar2-1) was examined by [35S]Met/Cys pulse-chase experiments as described for HA-Kre6 (Figure 4, A and B). A representative autoradiograph is shown in A. Kre5-HA radioactivity was quantified, and the averages and SDs of at least four independent experiments are shown in B. (C) Interaction between Rot1 and Kre5-HA. YM27 was incubated in YPD at 30°C and lysed, and IP using anti-Rot1 antibody or nonimmune serum (n.i.) was performed under nondenaturing conditions at 4°C. The immunoprecipitants were subjected to Western blotting to detect Kre5-HA and Rot1. (D) Cellular level of Atg22-HA is decreased by the rot1-2 mutation. ROT1 and rot1-2 cells expressing ATG22-HA from a centromeric plasmid (YM191 and YM192 [pRS314-ATG22-HA]) were incubated in SC at 23°C or shifted to 37°C for 6 h. The cells were lysed, and Atg22-HA and Sec61 were detected by Western blotting.
We previously showed that the lysis of the autophagic bodies is perturbed in the rot1-2 mutant (Takeuchi et al., 2006a). We thus asked if Rot1 chaperones proteins that are involved in this cellular event. Atg22 is a polytopic membrane protein and probably acts as an amino acid permease in the vacuolar membrane (Yang et al., 2006). We found that the cellular level of Atg22 was decreased by the rot1-2 mutation at 37°C (Figure 6D). Because of some technical reasons, neither 35S-metabolic labeling of Atg22 nor the CHX chase protocol was successful for checking stability of Atg22.
Finding Rot1-dependent Proteins from the E-MAP
To find other Rot1-dependent proteins, we utilized an epistatic miniarray profile (E-MAP) of yeast genes reported by Schuldiner et al. (2005), which illustrates the genetic relationship among mutant alleles of 424 genes related to the secretory pathway. In their study, 57 essential genes, including ROT1, were mutagenized by insertion of a nucleotide sequence to destabilize the product mRNAs, whereas another 367 genes were deleted. Next, the interactions of most of the gene pairs were estimated by checking the growth phenotype of the double mutant of the two genes. The combinations of the 424 mutations were examined, except for pairs of essential genes, and this comprehensive data set comprises the E-MAP.
Here we assumed the following scenario. If the product of gene S is a substrate protein of Rot1, its cellular level would decrease upon introduction of the rot1 mutation. On the other hand, the mutation in S also causes loss or reduction of its own product. Therefore, the mutations in ROT1 and S would show similar phenotypes, which in this case, is a genetic interaction(s) with another gene(s). On the basis of this model, we chose four genes, DRS2, GUP1, OPI3, and PMR1, as candidates for gene S from the 424 genes.
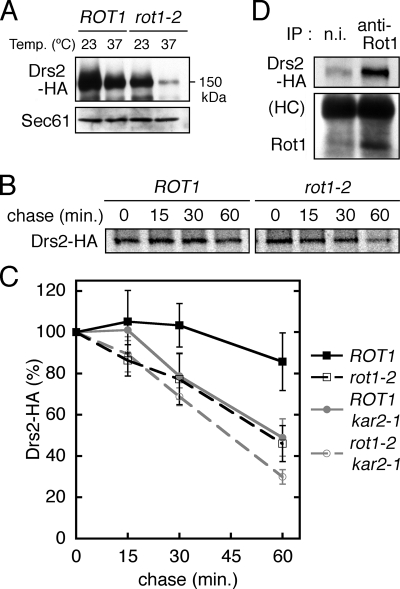
Drs2 is a polytopic membrane protein belonging to the P-type ATPases and cycles between the trans-Golgi network and the plasma membrane (Saito et al., 2004). The HA-tagging sequence was integrated into the chromosomal DRS2 locus to generate Drs2-HA. Western blot analysis of cell lysates revealed that, compared with ROT1 cells, Drs2-HA protein levels were lower in rot1-2 mutants incubated at 23°C and drastically lower in rot1-2 mutants incubated at 37°C (Figure 7A). We next examined the stability of Drs2 by [35S]Met/Cys pulse-chase. In this experiment, Drs2-HA was expressed from a multicopy plasmid for efficient detection. As shown in Figure 7, B and C, Drs2 was degraded more quickly in the rot1-2 cells than in the ROT1 cells. In addition, we detected an association between Rot1 and Drs2 by coIP (Figure 7D). These findings suggest that Rot1 functions as a chaperone for Drs2. We could not determine if the ubc7Δ mutation stabilizes Drs2 in the rot1-2 cells, because it unexpectedly induced degradation of Drs2 even in the ROT1 cells (data not shown).
Figure 7.
The relationship between Rot1 and Drs2. (A) Cellular amount of Drs2-HA is decreased by the rot1-2 mutation. The cells (YM183 [ROT1 DRS2-HA] and YM184 [rot1-2 DRS2-HA]) were incubated in SC at 23°C or shifted to 37°C for 6 h, lysed, and analyzed by anti-HA and anti-Sec61 Western blotting. (B and C) Accelerated degradation of Drs2-HA in the rot1-2 cells. ROT1 and rot1-2 cells expressing Drs2-HA from a multicopy plasmid (YM193 [ROT1], YM194 [rot1-2], YM212 [ROT1 kar2-1], and YM213 [rot1-2 kar2-1]; pRS424-DRS2-HA) were subjected to the [35S]-Met/Cys pulse-chase experiments to examine stability of Drs2-HA as described for HA-Kre6 (Figure 4, A and B). Representative autoradiograph is shown in B. The amount of radiolabeled Drs2-HA was quantified, and the averages and SDs of at least four independent experiments are shown in C. (D) Interaction between Rot1 and Drs2-HA. YM193 was incubated in SC at 30°C and lysed, and IP using anti-Rot1 antibody or nonimmune serum (n.i.) was performed under a nondenaturing condition. Then the immunoprecipitants were analyzed by anti-HA and anti-Rot1 Western blotting.
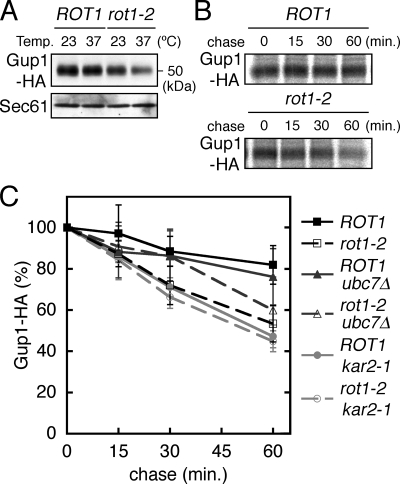
Gup1 is a multispanning membrane protein that is located in the ER and is involved in remodeling of the GPI anchor (Bosson et al., 2006). HA-tagged Gup1 was expressed under the control of its native promoter from a centromeric plasmid. Western blot analysis revealed that the cellular level of Gup1 was reduced by the rot1-2 mutation, especially at 37°C (Figure 8A). By the [35S]Met/Cys pulse-chase protocol, stability of Gup1 expressed from a multicopy plasmid was examined. The rot1-2 mutation caused accelerated degradation of Gup1, an effect that was weakly but significantly alleviated by the ubc7Δ mutation (Figure 8, B and C). Comigration of Gup1 with the immunoglobulin heavy chain on SDS-PAGE prevented us from detecting an interaction of Rot1 and Gup1 by coIP.
Figure 8.
The relationship between Rot1 and Gup1. (A) Cellular level of Gup1-HA was decreased by the rot1-2 mutation. ROT1 and rot1-2 cells expressing Gup1-HA from a centromeric plasmid (YM181 and YM182 [pT10-GUP1-HA]) were incubated in SC at 23°C or shifted to 37°C for 6 h. Gup1-HA and Sec61 in the cell lysates were detected by Western blotting. (B and C) Accelerated degradation of Gup1-HA in the rot1-2 cells. Stability of Gup1-HA was examined by [35S]Met/Cys pulse-chase experiments as described for HA-Kre6 (Figure 4, A and B) using the strains (YM195 [ROT1], YM196 [rot1-2], YM173 [ROT1 ubc7Δ], YM174 [rot1-2 ubc7Δ], YM210 [ROT1 kar2-1], and YM211 [rot1-2 kar2-1]) harboring a multicopy plasmid for expression of Gup1-HA (pT20-GUP1-HA). Representative autoradiograph is shown in B. Radioactive signals of Gup1-HA were quantified, and the averages and SDs of at least four independent experiments are shown in C.
Our analysis of Opi3 and Pmr1 showed little or no effect of the rot-2 mutation on their cellular amount or stability (Supplemental Figure 1), eliminating these as potential substrates. Taken together, these data suggest that Rot1 is a chaperone for Drs2 and Gup1, but not for Opi3 or Pmr1.
Rot1 May Cooperate with BiP/Kar2 in the Folding of Kre6
We previously proposed that Rot1 cooperates with BiP/Kar2 in nascent protein folding (Takeuchi et al., 2006a,b), and we examined this possibility here. The kar2-1 mutation causes an amino acid substitution in the peptide binding region of BiP/Kar2, and kar2-1 cells are likely to be defective in protein folding and ERAD (Brodsky et al., 1999; Kimata et al., 2003). We found that the kar2-1 mutation accelerated degradation of most of the Rot1-substrate proteins, viz. Kre6, Big1, Drs2, and Gup1 (Figures 4B, 5B, 7C, and 8C), which suggests that BiP/Kar2 is also involved in their folding. Although the kar2-1 mutation alone did not seem to affect the stability of Kre5, the rot1-2 kar2-1 double mutation caused a rapid degradation of Kre5 (Figure 6B). The combination of rot1-2 and kar2-1 also enhanced degradation of Drs2 (Figure 7C). In the case of Big1, kar2-1 apparently stabilized this protein in a rot1-2 background, perhaps due to inhibition of ERAD (Figure 5B).
If Rot1 and BiP/Kar2 cooperate in protein folding, we would expect them to simultaneously associate with the substrate proteins. To detect this potential ternary complex, cells were treated with a membrane-permeable and thiol-cleavable cross-linker, DSP, lysed, and double-immunoprecipitation was performed against Rot1 first and then BiP/Kar2. Kre6 was detected in the second immunoprecipitant (Figure 9, lane 4), indicating that Rot1, BiP/Kar2, and Kre6 form a ternary complex. The ternary complex was also detected in the rot1-2 cells (Figure 9, lane 5). These results strongly suggest that Rot1 cooperates with BiP/Kar2 in the folding of Kre6.
DISCUSSION
In this report, we provided evidence showing that Rot1 functions as a molecular chaperone in the ER. A molecular chaperone transiently interacts with unfolded proteins to inhibit their aggregation and support their folding and/or assembly. The recombinant Rot1 protein prevented aggregation of denatured proteins in vitro (Figure 1, B and C), indicating that Rot1 can bind directly to unfolded proteins. The rot1-2 mutation partially impaired this function in vitro (Figure 1, B and C), and observations in vivo also implied it (Figures 4D and 5D). Therefore, interaction with unfolded proteins probably underlies the function of Rot1 in cells. Moreover, cellular levels of six proteins in the secretory pathway were lower in the rot1-2 mutation (Figures 2, 6D, 7A, and 8A). As for five of these proteins, we demonstrated that the rot1-2 mutation accelerates degradation, which at least for three of them depends on the Ubc7-mediated ERAD pathway (Figures 4B, 5B, 6B, 7C, and 8C). These findings strongly suggest that Rot1 stabilizes these proteins by supporting their folding and/or assembly. In addition, we demonstrated a physical interaction of Rot1 with four of those Rot1-dependent proteins (Figures 4D, 5D, 6C, and 7D). In particular, Rot1 was found to transiently associate with newly synthesized Kre6 (Figure 4E). Taken together, we propose that Rot1 functions as a molecular chaperone that directly and probably transiently interacts with nascent unfolded proteins and assists in their folding and/or assembly.
The Rot1-dependent proteins presented in this study are listed in Table 3. It should be noted that they display different properties. They include a soluble protein, and type I and II and polytopic membrane proteins. Their subcellular localizations and functions are diverse, and each of the proteins is categorized into different families. Considering together that Rot1 directly interacts with two model denatured proteins in vitro, we propose that Rot1 is a general chaperone. As described above, information about the dependency of the substrate proteins on individual chaperones is also important to understanding protein folding in a cell. In our study, Kre6 stability was severely affected by the rot1-2 mutation, suggesting that Kre6 is an obligate substrate of Rot1. On the other hand, Big1, Kre5, Drs2, and Gup1 each exhibited different dependencies on Rot1. We also found that some proteins may be folded independent of Rot1. Although Drs2 and Pmr1 are both P-type ATPases, only Drs2 was dependent on Rot1 (Figure 7 and Supplemental Figure 1). Moreover, we previously reported that CPY was normally folded in rot1-2 single mutant cells (Takeuchi et al., 2006a). Therefore, we suggest that Rot1 functions with some specificity in the cells, as do Hsp90 and the eukaryotic chaperonin. It is unlikely that all of the Rot1-client proteins inevitably require Rot1 for their folding. Nevertheless, Rot1 is an essential protein, and we believe that Rot1 plays a unique role(s) in protein folding in the ER. To illustrate Rot1-dependent protein folding in vivo more comprehensively, large-scale identification and examination of the substrate proteins using a proteomic approach such that of Kerner et al. (2005) and Jessop et al. (2007) would be desirable.
Table 3.
Rot1-depending proteins examined in this study
| Protein | Property | Localization | Cellular function | Destabilization by rot1-2 | Physical interaction with Rot1 | Evolutional conservation |
|---|---|---|---|---|---|---|
| Kre5 | Soluble protein | ER | 1,6-β-glucan synthesis | Smalla | Yes | Conserved?b |
| Kre6 | Type II m.p.c | ER∼Golgi | 1,6-β-glucan synthesis | Higha | Yes | Yeast specific |
| Big1 | Type I m.p. | ER | 1,6-β-glucan synthesis | Mediumd | Yes | Yeast specific |
| Atg22 | m.p. (10 TMs)e | Vacuole | Amino acid permease? | Mediumf | Not determined | Yeast, bacteria |
| Drs2 | m.p. (8 TMs)e | Golgi∼PMg | Phospholipid traslocation? | Mediuma | Yes | Conservedh |
| Gup1 | m.p. (11 TMs)e | ER | GPI anchor remodeling | Mediuma | Not detected | Conservedh |
a Estimated by [35S]Met/Cys pulse-chase experiments.
b Function(s) of Kre5 may be different from that of UGGT (Levinson et al., 2002).
c Membrane protein.
d Estimated by cycloheximide-chase experiments.
e Number of the transmembrane regions predicted by SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/).
f Estimated by Western blotting.
g Plasma membrane.
h Homologous protein(s) is registered in SWISS-PLOT, but its function is unknown.
Although the mutant rot1-2 protein exhibited impaired substrate binding, this defect was not complete, and the rot1-2 protein may be partially active. Therefore, the dependency of proteins such as Big1, Kre5, Drs2, and Gup1 on Rot1 may be underestimated in this study comparing rot1-2 to wild-type cells. It is also possible that there are other proteins with functions redundant to Rot1 caused. We previously demonstrated that deletion of CNE1, LHS1, or SCJ1 aggravates the defective growth phenotype of rot1-2 cells, and conversely, that overexpression of each of them partially rescues it (Takeuchi et al., 2006a). Proteins encoded by these genes are ER-resident chaperones and may share partially redundant function(s) with Rot1. We have also reported that the rot1-2 kar2-1 double mutation causes temperature-dependent synthetic lethality (Takeuchi et al., 2006a). In this study, we showed that BiP/Kar2 is also required for the folding of Rot1-dependent proteins, except for Kre5 (Figures 4B, 5B, 6B, 7C, and 8C) and that Rot1 and BiP/Kar2 interact with Kre6 simultaneously (Figure 9). These findings suggest that Rot1 and BiP/Kar2 play distinct roles and cooperate in the folding of Kre6 and possibly other proteins. On the other hand, the additive effects of the rot1-2 and kar2-1 mutations on destabilization of Kre5 and Drs2 imply partial functional redundancy of Rot1 and BiP/Kar2.
Recently, three reports from other groups have reported possible functions of Rot1. Orlowski et al. (2007) reported that multicopy expression of ROT1 suppressed the sec59-1 mutation. Sec59 is an ER-localized polytopic membrane protein that functions as a dolichol kinase. We speculate that Rot1 can support the correct folding of the mutant sec59-1 protein. Second, Juanes et al. (2007) reported that in a Rot1-depleted cells, apical growth of bud and septum formation were perturbed. It is likely that these phenotypes are the secondary effects of misfolding and/or destabilization of Rot1-client proteins, because in their experiments, the cells were incubated for more than 6 h under conditions where Rot1 was depleted. Drs2 and its family members have been hypothesized to generate a biased distribution of aminophospholipids between the two leaflets of the lipid bilayer, and this may be important for assembly of the cortical actin patch required for apical bud growth (Kishimoto et al., 2005). In addition, Kre6 was shown to interact with the actin patch assembly proteins, Las21 and Sla1, through its cytoplasmic tail (Li et al., 2002). We speculate that a decrease in Drs2, Kre6, and other Rot1-dependent proteins collectively could cause abnormalities in actin patch assembly and septum formation. Finally, Nakamata et al. (2007) reported that an essential, ER membrane protein, Keg1, interacts with Kre6 and that overexpression of ROT1 suppresses the temperature-sensitive growth defect of the keg1-1 mutant. As they suggested, it is possible that Rot1 chaperones the mutant keg1-1 protein and/or indirectly supports its function by chaperoning Kre6. As described above, Rot1 was initially reported as a protein involved in cell wall 1,6-β-glucan synthesis, but importantly, these pleiotropic phenotypes of the rot1 mutation and overexpression of ROT1 are consistent with our idea that Rot1 is a general chaperone.
We have shown here that Rot1 functions as a molecular chaperone and acts together with BiP/Kar2 in some cases. Hsp70 cofactors such as Hsp40 possess chaperone activity and bind to substrate proteins together with Hsp70. At the same time, Hsp70 cofactors generally regulate the ATPase activity of Hsp70. We speculate that Rot1 cooperates with BiP/Kar2 in a similar manner. Can Rot1 bind BiP/Kar2 directly and regulate its ATPase activity? This is an important question to be addressed in future studies. Although Rot1 homologues are found only in yeast species, there may exist a protein(s) having functions similar to Rot1 in the ER of higher eukaryotes, because some of the Rot1-client proteins are evolutionally conserved. Alternatively, Rot1 may meet the requirements specific to the yeast ER. Large-scale identification of Rot1 substrates and a detailed analysis of Rot1 functions, including possible cooperation with BiP/Kar2, will deepen our understanding of protein folding in the ER.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Akihiko Nakano (The University of Tokyo, Riken), Satoshi Harashima (Osaka University), and Koji Yoda (The University of Tokyo) for the yeast strain, plasmid and antibody, respectively. We are grateful to Junko Iida, Hisayo Masuda, and Miki Matsumura for technical assistance. M.T. thanks all members of Kohno's laboratory for helpful discussions and everyday conversation. This work was supported by Grants-in-Aids for Scientific Research on Priority Areas (14037240, 19058010 to K. K., 18050024 to Y. K.) and for 21st Century COE Program from MEXT, and JSPS.KAKENHI (18570179 to Y.K.).
Abbreviations used:
- CHX
cycloheximide
- CPY
carboxypeptidase Y
- DSP
dithiobis(succinimidyl)propionate
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- GPI
glycophosphatidylinositol
- Hsp
heat-shock protein
- HC
immunoglobulin heavy chain
- IP
immunoprecipitation
- n.i.
nonimmune serum.
- PI
protease inhibitor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/italic) on May 28, 2008.
REFERENCES
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- Azuma M., Levinson J. N., Page N., Bussey H. Saccharomyces cerevisiae Big1p, a putative endoplasmic reticulum membrane protein required for normal levels of cell wall β-1,6-glucan. Yeast. 2002;19:783–793. doi: 10.1002/yea.873. [DOI] [PubMed] [Google Scholar]
- Bickle M., Delley P. A., Schmidt A., Hall M. N. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell. 2006;17:2636–2645. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Caplan A. J., Mandal A. K., Theodoraki M. A. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Molinari M., Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Fewell S. W., Travers K. J., Weissman J. S., Brodsky J. L. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Freedman R. B. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. C., Morimoto R. I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hosoda A., Kimata Y., Tsuru A., Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 2003;278:2669–2676. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- Iha H., Tsurugi K. Shuttle-vector system for Saccharomyces cerevisiae designed to produce C-terminal-Myc-tagged fusion proteins. Biotechniques. 1998;25:936–938. doi: 10.2144/98256bm01. [DOI] [PubMed] [Google Scholar]
- Jessop C. E., Chakravarthi S., Garbi N., Hammerling G. J., Lovell S., Bulleid N. J. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanes M. A., Queralt E., Bano M. C., Igual J. C. Rot1 plays an antagonistic role to Clb2 in actin cytoskeleton dynamics throughout the cell cycle. J. Cell Sci. 2007;120:2390–2401. doi: 10.1242/jcs.002758. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kerner M. J., et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Kimata Y., Ishiwata-Kimata Y., Shimizu Y., Abe H., Farcasanu I. C., Takeuchi M., Rose M. D., Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Ishiwata-Kimata Y., Ito T., Hirata A., Suzuki T., Oikawa D., Takeuchi M., Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., Yamamoto T., Tanaka K. Defects in structural integrity of ergosterol and the Cdc50p-Drs2p putative phospholipid translocase cause accumulation of endocytic membranes, onto which actin patches are assembled in yeast. Mol. Biol. Cell. 2005;16:5592–5609. doi: 10.1091/mbc.E05-05-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K. How transmembrane proteins sense endoplasmic reticulum stress. Antioxid. Redox. Signal. 2007;9:2295–2303. doi: 10.1089/ars.2007.1819. [DOI] [PubMed] [Google Scholar]
- Kota J., Gilstring C. F., Ljungdahl P. O. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J. Cell Biol. 2007;176:617–628. doi: 10.1083/jcb.200612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson J. N., Shahinian S., Sdicu A. M., Tessier D. C., Bussey H. Functional, comparative and cell biological analysis of Saccharomyces cerevisiae Kre5p. Yeast. 2002;19:1243–1259. doi: 10.1002/yea.908. [DOI] [PubMed] [Google Scholar]
- Li H., Page N., Bussey H. Actin patch assembly proteins Las17p and Sla1p restrict cell wall growth to daughter cells and interact with cis-Golgi protein Kre6p. Yeast. 2002;19:1097–1112. doi: 10.1002/yea.904. [DOI] [PubMed] [Google Scholar]
- Machi K., Azuma J., Igarashi K., Matsumoto T., Fukuda H., Kondo A., Ooshima H. Rot1p of Saccharomyces cerevisiae is a putative membrane protein required for normal levels of the cell wall 1, 6-β-glucan. Microbiol. 2004;150:3163–3173. doi: 10.1099/mic.0.27292-0. [DOI] [PubMed] [Google Scholar]
- Montijn R. C., Vink E., Muller W. H., Verkleij A. J., Van Den Ende H., Henrissat B., Klis F. M. Localization of synthesis of β-1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 1999;181:7414–7420. doi: 10.1128/jb.181.24.7414-7420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamata K., Kurita T., Bhuiyan M. S., Sato K., Noda Y., Yoda K. KEG1/YFR042w encodes a novel Kre6-binding ER membrane protein responsible for β-1,6-glucan synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:34315–34324. doi: 10.1074/jbc.M706486200. [DOI] [PubMed] [Google Scholar]
- Nakano A., Brada D., Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J., Machula K., Janik A., Zdebska E., Palamarczyk G. Dissecting the role of dolichol in cell wall assembly in the yeast mutants impaired in early glycosylation reactions. Yeast. 2007;24:239–252. doi: 10.1002/yea.1479. [DOI] [PubMed] [Google Scholar]
- Roemer T., Paravicini G., Payton M. A., Bussey H. Characterization of the yeast (1→6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C., Meyer A. S., Reissmann S., Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Kimata Y., Hirata A., Oka M., Kohno K. Saccharomyces cerevisiae Rot1p is an ER-localized membrane protein that may function with BiP/Kar2p in protein folding. J. Biochem. 2006a;139:597–605. doi: 10.1093/jb/mvj063. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Kimata Y., Kohno K. Causal links between protein folding in the ER and events along the secretory pathway. Autophagy. 2006b;2:323–324. doi: 10.4161/auto.3089. [DOI] [PubMed] [Google Scholar]
- Todorow Z., Spang A., Carmack E., Yates J., Schekman R. Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2000;97:13643–13648. doi: 10.1073/pnas.250472397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. B. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D. J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.