Abstract
c-Cbl is the E3 ubiquitin ligase that ubiquitinates the epidermal growth factor (EGF) receptor (EGFR). On the basis of localization, knockdown, and in vitro activity analyses, we have identified the E2 ubiquitin-conjugating enzyme that cooperates with c-Cbl as Ubc4/5. Upon EGF stimulation, both Ubc4/5 and c-Cbl were relocated to the plasma membrane and then to Hrs-positive endosomes, strongly suggesting that EGFR continues to be ubiquitinated after internalization. Our time-course experiment showed that EGFR undergoes polyubiquitination, which seemed to be facilitated during the transport to Hrs-positive endosomes. Use of a conjugation-defective ubiquitin mutant suggested that receptor polyubiquitination is required for efficient interaction with Hrs and subsequent sorting to lysosomes. Abrupt inhibition of the EGFR kinase activity resulted in dissociation of c-Cbl from EGFR. Concomitantly, EGFR was rapidly deubiquitinated and its degradation was delayed. We propose that sustained tyrosine phosphorylation of EGFR facilitates its polyubiquitination in endosomes and counteracts rapid deubiquitination, thereby ensuring Hrs-dependent lysosomal sorting.
INTRODUCTION
Plasma membrane proteins fulfill critical cellular functions such as nutrient uptake, growth factor signaling, and cell adhesion. There are many instances that, in response to changes in extracellular inputs, they are targeted to or eliminated from the plasma membrane. Ubiquitin acts as a sorting signal to down-regulate the functions of plasma membrane proteins (Umebayashi, 2003; Mukhopadhyay and Riezman, 2007). We showed previously that the yeast tryptophan permease Tat2p is subject to ubiquitin-dependent sorting that is accurately regulated by the tryptophan concentration in the medium (Umebayashi and Nakano, 2003). It has been known that ubiquitin functions at three steps in post-Golgi trafficking: internalization from the plasma membrane, sorting into the interior of multivesicular bodies (MVBs), and Golgi-to-vacuole/lysosome transport that bypasses the plasma membrane.
In mammalian cells, endocytic trafficking of epidermal growth factor receptor (EGFR) has been extensively studied in the context of ubiquitin-dependent sorting. Upon ligand-binding, the receptor's intrinsic tyrosine kinase domain that resides on the cytoplasmic side is activated, leading to autophosphorylation at multiple tyrosine residues. The proto-oncogene product c-Cbl, a RING finger-type ubiquitin ligase (E3), is recruited to the activated receptor: the tyrosine kinase-binding domain of c-Cbl interacts with the phosphotyrosine residue of EGFR (Thien and Langdon, 2001). Receptor endocytosis by c-Cbl–mediated ubiquitination is considered to be crucial to prevent oncogenesis, because it sorts the receptor into the MVB lumen and thereby terminates the growth factor signaling. It has been suggested that the sorting step that absolutely requires EGFR ubiquitination is entry into the MVB lumen (Duan et al., 2003; Grøvdal et al., 2004; Ravid et al., 2004; Huang et al., 2006). Ubiquitination is not necessarily required for EGFR internalization from the plasma membrane, possibly because of the redundant routes (Sigismund et al., 2005). In contrast, it is indispensable for lysosomal sorting and subsequent degradation. On the endosomal membrane, the sorting machinery such as hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and the endosomal sorting complexes required for transport (ESCRTs) binds to ubiquitin and sorts ubiquitinated cargo, including EGFR, into invaginating MVB vesicles (Katzmann et al., 2002). It is envisaged that cargo has already been conjugated with ubiquitin when it first encounters the sorting machinery in endosomes and that the cargo remains ubiquitinated while it undergoes the MVB sorting. However, it is not known whether the initial ubiquitination is effective until the end of the sorting. It remains to be further elucidated where and how the cargo ubiquitination occurs.
Previous studies showed that c-Cbl is relocated to endosomes upon epidermal growth factor (EGF) stimulation, leading to the idea that EGFR ubiquitination in endosomes is important for lysosomal sorting (Levkowitz et al., 1998; Longva et al., 2002; Myromslien et al., 2006). However, it should be noted that whether the endosomal pool of c-Cbl is active in ubiquitinating EGFR is still not conclusive. An important problem is that c-Cbl is not merely an E3 but is a multifunctional adaptor protein interacting with many partners, such as signaling proteins (Schmidt and Dikic, 2005). Considering that the RING finger domain of c-Cbl interacts with an E2 ubiquitin-conjugating enzyme, the site(s) where EGFR is ubiquitinated by c-Cbl may well be identified by the localization of the E2, which was not examined previously. In this study, we focused on the E2 that is recruited to the site where c-Cbl and EGFR are colocalized. We have uncovered that Ubc4/5 is the E2 partner of c-Cbl and that Ubc4/5-c-Cbl–mediated EGFR ubiquitination continues from the plasma membrane to Hrs-positive endosomes. The ongoing ubiquitination counteracts rapid deubiquitination and seems to enhance polyubiquitination of EGFR during the transport to endosomes. The obtained results also suggest that the receptor polyubiquitination facilitates interaction with the sorting machinery and subsequent degradation in lysosomes.
MATERIALS AND METHODS
Cell Culture and Transfection
HeLa and HEK293T cells were grown in DMEM (Sigma, St. Louis, MO) supplemented with 10% FBS (Invitrogen, Carlsbad, CA) and 4 mM l-glutamine (Invitrogen). For HEK293T cells, collagen (type I)-coated dishes (Asahi Techno Glass, Chiba, Japan) were used. CHO cells were grown in F-12 nutrient mixture (Invitrogen) with 10% FBS. In the following, we refer to DMEM containing 4 mM l-glutamine or F-12 as the serum-free medium. For transfection, the cells were incubated in Opti-MEM I reduced-serum medium (Invitrogen) containing plasmid DNA, Lipofectamine (Invitrogen), and Plus reagent (Invitrogen) for 2 h. Subsequently, the medium was replaced by the serum-containing DMEM or F-12, and the cells were incubated for 12–18 h before the experiment. Overexpression of EGFR tends to significantly delay receptor degradation. Therefore, in Figure 6, Plus reagent was omitted to avoid high-level expression of the receptor.
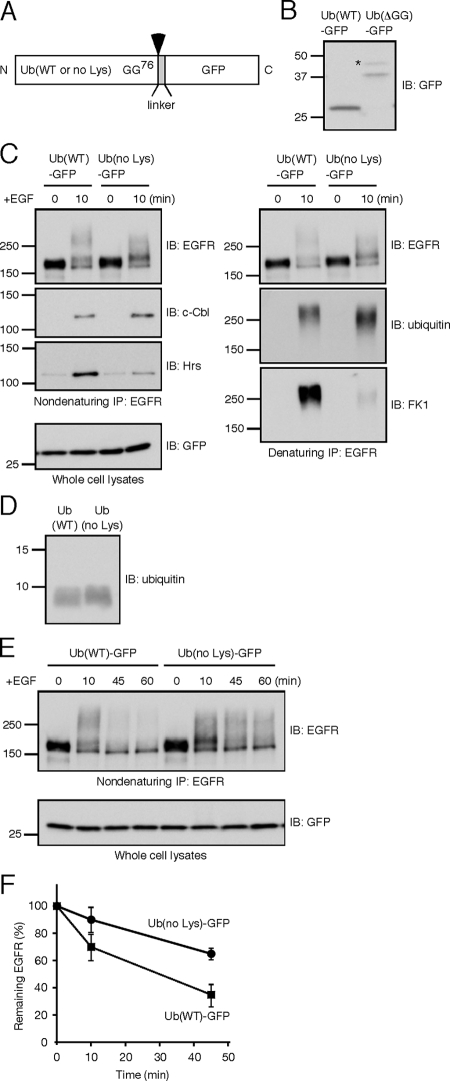
Figure 6.
Inhibition of polyubiquitination reduces EGFR-Hrs interaction and delays the receptor degradation. (A) Schematic representation of the ubiquitin–GFP fusion protein. Wild-type or lysine-less ubiquitin was fused to GFP via a 13-amino acid linker (MTVPRARDPPVAT). The possible cleavage site is between the last glycine 76 of ubiquitin and the methionine of the linker (arrowhead). (B) HEK293T cells transiently expressing Ub(WT)-GFP or Ub(ΔGG)-GFP were lysed and subject to immunoblotting with the anti-GFP antibody. An asterisk may represent monoubiquitin conjugation to the ubiquitin moiety of Ub(ΔGG)-GFP. (C) HEK293T cells transiently expressing EGFR, c-Cbl, and the indicated ubiquitin–GFP fusion were unstimulated or stimulated with EGF for 10 min. EGFR was subject to two rounds of immunoprecipitation. c-Cbl and Hrs were detected from the first, nondenaturing immunoprecipitates. Ubiquitin-conjugated EGFR was detected from the second, denaturing ones. (D) Recombinant Ub(WT) and Ub(no Lys) were detected using the anti-ubiquitin antibody. (E) HEK293T cells transiently expressing EGFR, c-Cbl, and the indicated ubiquitin–GFP fusion were stimulated with EGF for the indicated time periods. EGFR was detected after the immunoprecipitation. (F) Degradation of EGFR was monitored as shown in E, and results from three independent experiments were quantificated. In both Ub(WT) and Ub(no Lys) expressing cells, the intensities at 0 min were set as 100%. The mean + SD values are shown.
Plasmids
Plasmids expressing human EGFR, c-Cbl, and c-Cbl(C381A) were kindly provided by Minsoo Kim and Tadashi Yamamoto (The University of Tokyo, Tokyo, Japan). PCR-based mutagenesis was used to create EGFR(Y1045F). Plasmids expressing FLAG-E2s were generous gifts from Noriyuki Matsuda and Keiji Tanaka (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Plasmids containing wild-type and mutant ubiquitin sequences were kindly provided by Hiroshi Ohno (RIKEN Research Center for Allergy and Immunology, Kanagawa, Japan), and the ubiquitin sequences were inserted into pEGFP-N1 (Clontech Laboratories, Mountain View, CA) to express the ubiquitin-green fluorescent protein (GFP) fusion proteins.
Reagents and Antibodies
All Alexa fluor dyes-conjugated reagents and murine natural EGF were purchased from Invitrogen. The EGFR kinase inhibitor AG1478 was from Calbiochem (San Diego, CA). Bovine ubiquitin was from Sigma (St. Louis, MO). Recombinant human E1, UbcH5C, UbcH7, ubiquitin, and ubiquitin(no lysines) were from Boston Biochem (Cambridge, MA). For fluorescent microscopy, the following antibodies were used. Mouse anti-EEA1 and mouse anti-c-Cbl were from BD Biosciences (San Jose, CA) and rabbit anti-FLAG was from Sigma. For immunoblotting, the following antibodies were used. Goat anti-Ubc4 and rabbit anti-Cbl were from Santa Cruz Biotechnology (Santa Cruz, CA), rabbit anti-UbcH7 was from Chemicon (Temecula, CA), goat anti-GST was from GE Healthcare (Chalfont St. Giles, Buckinghamshire, United Kingdom), rabbit anti-ubiquitin was from DakoCytomation (Kyoto, Japan), HRP-conjugated mouse anti-phosphotyrosine PY20 was from BD Biosciences, FK1 and rabbit anti-GFP were from MBL (Nagoya, Japan), and mouse anti-α-tubulin was from Sigma. HRP-linked anti-mouse IgG, anti-rabbit IgG, HRP-labeled protein A, ECL, and ECL Plus and ECL Advance Western Blotting Detection System were from GE Healthcare. Affinity purified rabbit anti-Hrs (Raiborg et al., 2001) was used for fluorescent microscopy and immunoblotting. Mouse anti-EGFR Ab-11 (Lab Vision, Fremont, CA) was used for the nondenaturing immunoprecipitation and sheep anti-EGFR (Fitzgerald, Concord, MA) for fluorescent microscopy, immunoblotting, and the denaturing immunoprecipitation.
In Vitro Ubiquitination Assay
A central region of wild-type or C381A c-Cbl (amino acids 359–447) containing the RING finger domain was fused to glutathione S-transferase (GST) using pGEX-6P-1 (GE Healthcare). Expression of the GST-fusion proteins in Escherichia coli BL21 was induced with 0.4 mM IPTG at 30°C for 3 h. The cells were lysed using B-PER Bacterial Protein Extraction Reagent (Pierce, Rockford, IL), and the lysates were incubated with glutathione Sepharose 4B (GE Healthcare) at 4°C for 2 h. The Sepharose beads were washed three times with PBS containing 0.1% Triton X-100. Proteins bound to the beads were eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione), and applied to PD-10 column (GE Healthcare) for buffer exchange. Aliquots in 50 mM Tris-HCl, pH 8.0, containing 50 mM NaCl, 1 mM DTT, and 10% glycerol were frozen with liquid nitrogen and stored at −80°C.
For autoubiquitination, the reaction mixture (66 μl) containing 50 mM Tris-HCl, pH 7.5, 1 mM DTT, 2.5 mM MgCl2, 4 mM ATP, 10 μg of bovine ubiquitin, 100 ng of E1, 0.75 μg of E2 (UbcH5C or UbcH7), and 1 μg of GST-RING (wild-type or C381A) was incubated at 30°C for 2 h. The reaction was terminated by adding 15 μl of 6× SDS-PAGE sample buffer (without DTT) and 9 μl of 1 M DTT and heating at 95°C for 5 min. After centrifugation, the supernatant was subject to immunoblotting.
Fluorescence Microscopy
CHO or HeLa cells grown on glass coverslips were shifted to the serum-free medium and incubated for 2 h. Then, the cells were stimulated with 100 ng/ml EGF or 1 μg/ml Alexa 647-EGF in the serum-free medium. At the indicated time periods, the cells were washed three times with ice-cold PBS, fixed with 3% paraformaldehyde in PBS for 20 min, and quenched with 50 mM NH4Cl in PBS for 10 min. The cells were permeabilized with 0.2% saponin (ICN Biomedicals, Aurora, OH) in PBS for 10 min. Alternatively, the cells were first permeabilized with 0.05% saponin in PEM buffer (80 mM PIPES-KOH, pH 6.8, 5 mM EGTA, 1 mM MgCl2) on ice for 5 min and then fixed. The permeabilized cells were incubated in blocking solution (0.1% gelatin in PBS) and then stained with primary and secondary antibodies diluted with the blocking solution. The coverslips were mounted in Vectashield Hard Set Mounting Medium (Vector Laboratories, Burlingame, CA), and the cells were observed with an Olympus FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan). Colocalization of GFP-Cbl with Hrs or EEA1 was quantitated using FV1000 analysis software FV10-ASW.
Immunoprecipitation of EGFR
HEK293T or HeLa cells grown in 10-cm dishes were starved for serum for 2 h and stimulated with 100 ng/ml EGF in the serum-free medium for the indicated time periods. The cells were washed once with ice-cold PBS plus 10 mM N-ethylmaleimide (NEM), covered with 425 μl of lysis buffer (50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1% NP-40, 1 mM EDTA, 0.02% NaN3) containing 1 mM Na3VO4, 10 mM NEM, the Complete (EDTA-free) protease inhibitor cocktail (Roche Applied Science, Basel, Switzerland), 10 μM lactacystin (Peptide Institute, Osaka, Japan), and 10 μM MG132 (Peptide Institute), scraped with a rubber policeman and lysed on ice for 30 min. The cell lysate was centrifuged at 1500 × g for 4 min, and 425 μl of the supernatant was collected. Part of the supernatant (66 μl) was withdrawn and used as the whole cell lysate, and the rest was used for nondenaturing immunoprecipitation. The lysate was mixed with 20 μl protein G Sepharose 4 Fast Flow (GE Healthcare) and rotated at 4°C for 30 min. The precleared supernatant corresponding to 1 mg (HeLa) or 2 mg (HEK293T) proteins was filled up to 300 μl, mixed with 10 μl mouse anti-EGFR Ab-11, and rotated at 4°C. After 2 h, 15 μl protein G Sepharose 4 Fast Flow was added and the microtube was rotated at 4°C for 2.5 h. The immune complexes were washed four times with the lysis buffer, resuspended in 100 μl 50 mM Tris-HCl, pH 6.8, plus 1% SDS, and heated at 95°C for 5 min. Part of the supernatant was used for immunoblotting to monitor coimmunoprecipitation. The rest, typically 55 μl, was subject to denaturing immunoprecipitation. It was diluted 10-fold with the lysis buffer containing 10 mM NEM and the protease inhibitor cocktail, mixed with 1.8 μl sheep anti-EGFR and 15 μl protein G Sepharose 4 Fast Flow, and rotated at 4°C overnight. The immune complexes were washed four times with the lysis buffer, resuspended in SDS-PAGE sample buffer, and heated at 95°C for 5 min. Immunoblotting images were acquired with the LAS-3000 imaging system (Fujifilm, Tokyo, Japan), and the band intensities were quantitated using Fujifilm Image Gauge Ver. 4.0 software.
Depletion of E2s
Ubc4/5 and UbcH7 were depleted by transfection with small-interfering RNA. Among the Ubc4/5 family proteins, Ubc4, UbcH5B, and UbcH5C were simultaneously depleted using sense sequence 5′-CAGUAAUGGCAGCAUUUGUTT-3′ and antisense sequence 5′-ACAAAUGCUGCCAUUACUGTT-3′. Note that, as reported previously (Saville et al., 2004), the target sequence does not deplete UbcH5A. For UbcH7, sense sequence 5′-GGACCGUAAAAAAUUCUGUTT-3′ and antisense sequence 5′-ACAGAAUUUUUUACGGUCCTT-3′ were used according to the previous report (Verma et al., 2004). As a control, the target sequence for luciferase was used: sense sequence 5′-CAUACGCGGAAUACUUCGATT-3′ and antisense sequence 5′-UCGAAGUAUUCCGAGUACGTT-3′. HeLa cells grown in a 10-cm dish (30–50% confluent) were transfected with 600 pmol of double-strand RNA using Lipofectamine RNAiMAX (Invitrogen). After 1-d incubation, the medium was changed to the serum-containing one, and the cells were further incubated for 1 d. Then, the cells were replated to 10-cm dishes and incubated for 1.5 d. The serum starvation, EGF stimulation, and immunoprecipitation were done as described above. The goat anti-Ubc4 antibody was precleared by absorption with GST-UbcH5A (Boston Biochem), as reported previously (Saville et al., 2004), and then used for immunoblotting.
RESULTS
Ubc4/5-c-Cbl–mediated EGFR Ubiquitination Continues in Hrs-positive Endosomes
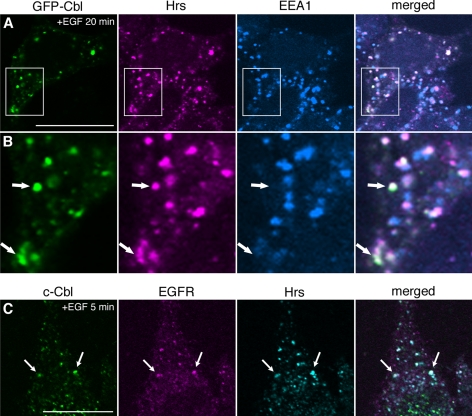
EGFR and GFP-Cbl were transiently expressed in CHO cells lacking endogenous EGFR. EGF stimulation relocated GFP-Cbl to the plasma membrane and then to endosomes, that were enlarged by the expression of the GTP-form of Rab5(Q79L) (Supplementary Figure 1). Endosomes are composed of morphologically and functionally distinct membrane domains (Gruenberg, 2001), and based on the enlargement of the GFP-Cbl–positive endosomes by Rab5(Q79L), two early endosomal markers, Hrs and EEA1, were tested. Endosomes marked by both Hrs and GFP-Cbl but not by EEA1 were often observed (Figure 1, A and B), and the quantitative colocalization analysis of 20 transfected cells indicated that 76 ± 11 and 54 ± 12% (mean ± SD) of GFP-Cbl overlapped with Hrs and EEA1, respectively. Although Hrs significantly colocalizes with EEA1 (Raiborg et al., 2001), these two proteins indeed occupy distinct early endosomal membrane domains, with Hrs being localized to the site where ubiquitinated proteins are sorted to MVB vesicles (Raiborg et al., 2002). Therefore, GFP-Cbl seems to be preferentially localized to Hrs-positive membrane domains in endosomes. We also examined the localization without overexpression, using HeLa cells that express endogenous EGFR. The cells were stimulated with EGF for 5 min, and endogenous c-Cbl, EGFR, and Hrs were stained. Note that the incubation periods with EGF were different from those in the experiments using the transfected cells, as will be also shown in the following. We confirmed that c-Cbl colocalized with EGFR as well as Hrs in endosomes (Figure 1B). These results suggest that, in endosomes, c-Cbl–mediated EGFR ubiquitination coincides with MVB sorting.
Figure 1.
Localization of c-Cbl to Hrs-positive endosomes. (A) CHO cells transiently expressing EGFR and GFP-Cbl were stimulated with EGF for 20 min. The cells were permeabilized, fixed, and stained for endogeneous Hrs and EEA1. (B) The insets in A are shown at 3× magnification. Arrows indicate endosomes marked by GFP-Cbl and Hrs, but not EEA1. (C) HeLa cells stimulated with EGF for 5 min were permeabilized, fixed, and stained for endogeneous c-Cbl, EGFR, and Hrs. Arrows indicate representative colocalizing structures among c-Cbl, EGFR, and Hrs. Bars, 20 μm.
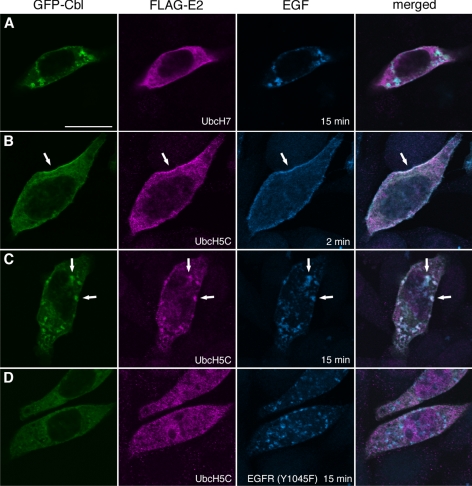
To address whether c-Cbl is active in ubiquitinating EGFR from the plasma membrane to endosomes, we examined the localization of c-Cbl's partner in the ubiquitinating reaction, i.e., the E2 enzyme. It was reported that the RING finger domain of c-Cbl interacts with UbcH7 (Yokouchi et al., 1999), and the crystal structure of a c-Cbl-UbcH7 complex was solved (Zheng et al., 2000). However, FLAG-UbcH7 was not evidently localized to the endosomes marked by both GFP-Cbl and the fluorescent EGF (Figure 2A). We further examined the localization of FLAG-tagged Ubc2, Ubc3, Ubc4, UbcH5A, UbcH5B, UbcH5C, UbcH6, UbcH8, and Ubc13 and found that the Ubc4/5 family proteins (Ubc4, UbcH5A, UbcH5B, and UbcH5C) specifically colocalized with GFP-Cbl in endosomes (Figure 2C and Supplementary Figure 2). FLAG-UbcH5C colocalized with GFP-Cbl and EGF 2 min after EGF addition in the plasma membrane (Figure 2B) and after 15 min in intracellular punctae (Figure 2C). Phosphorylated Tyr-1045 of EGFR serves as the direct c-Cbl–binding site (Levkowitz et al., 1999). In CHO cells expressing the EGFR(Y1045F) mutant, neither GFP-Cbl nor FLAG-UbcH5C was drastically relocated to the EGF-positive endosomes (Figure 2D). This suggests that the endosomal localization of FLAG-UbcH5C is dependent on the binding of GFP-Cbl to EGFR. We noticed that the endosomal relocation of GFP-Cbl was not completely abolished in cells expressing EGFR(Y1045F): in Figure 2D, faint relocation was still observed. This is probably due to another mode of binding between EGFR and c-Cbl, that is, c-Cbl binds to EGFR through Grb2 (Jiang et al., 2003). Under our experimental condition, the indirect binding mode seems to be insufficient to drastically relocate GFP-Cbl and FLAG-UbcH5C to endosomes.
Figure 2.
FLAG-UbcH5C colocalizes with GFP-Cbl and EGF in the plasma membrane and endosomes. CHO cells were transiently transfected with EGFR, GFP-Cbl, and the indicated FLAG-E2 plasmids and stimulated with Alexa 647-EGF for 2 or 15 min. In D, the mutant EGFR(Y1045F) was expressed instead of the wild-type receptor. The cells were stained with the anti-FLAG antibody. Arrows in B and C indicate representative colocalizing structures in the plasma membrane and endosomes, respectively. Bar, 20 μm.
The localization results raise the possibility that c-Cbl, in cooperation with Ubc4/5, ubiquitinates EGFR from the plasma membrane to Hrs-positive endosomes. However, this is apparently inconsistent with the previous reports that c-Cbl interacts with UbcH7 (Yokouchi et al., 1999; Zheng et al., 2000). We therefore examined the effects of the E2s' knockdown on the receptor ubiquitination and degradation. To deplete Ubc4/5 proteins simultaneously, an siRNA target sequence that matches those in Ubc4 and UbcH5B and -C was selected according to the previous report (Saville et al., 2004). The results are shown in Figure 3. First, specific depletion of Ubc4/5 and UbcH7 was confirmed. EGFR was immunoprecipitated under the denaturing condition (see below in Figure 5), and the precipitates were blotted with the anti-ubiquitin antibody. Strikingly, the receptor ubiquitination was severely inhibited by knockdown of Ubc4/5 but not UbcH7. The anti-EGFR immunoblotting showed comparable degradation rates in the control and UbcH7-depleted cells. In contrast, the depletion of Ubc4/5 retarded the degradation. The mean ± SD values from three independent experiments showed that 34 ± 2.3, 77 ± 10, and 31 ± 3.8% of EGFR were remaining at 60 min in control, Ubc4/5- and UbcH7-depleted cells, respectively. Thus, the depletion of Ubc4/5, but not UbcH7, inhibited both the ubiquitination and degradation of EGFR. We examined the degradation at 30 min and obtained the same result (Supplementary Figure 3A). Also, as a control, another target sequence was tested (Supplementary Figure 3B). It was recently reported that the internalization routes of EGFR is affected by EGF concentrations (Sigismund et al., 2005). We tested at a lower EGF concentration (20 ng/ml), and found that the dependence of the receptor ubiquitination on Ubc4/5 was not influenced (Supplementary Figure 3C).
Figure 3.
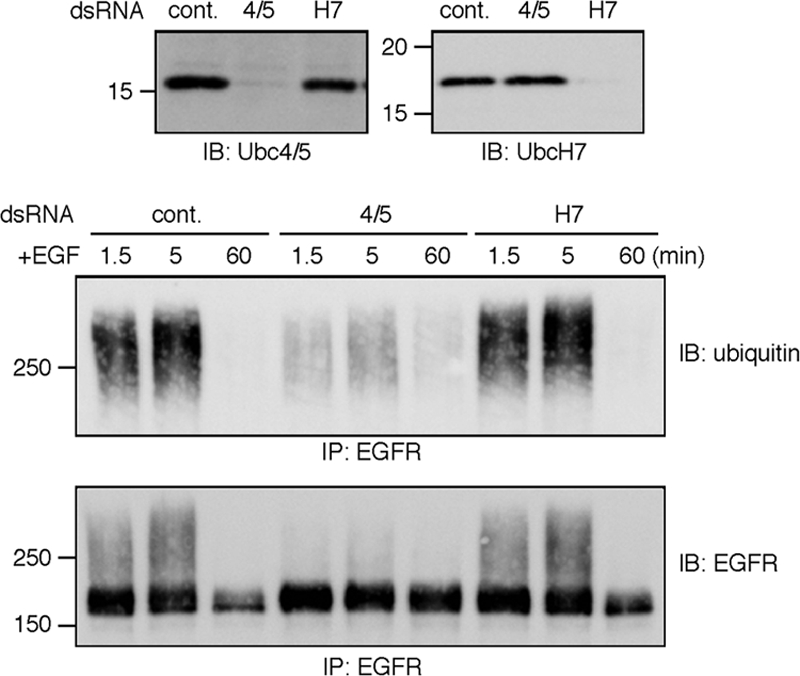
Ubc4/5, but not UbcH7, is required for ubiquitination and degradation of EGFR. HeLa cells were depleted of Ubc4/5 or UbcH7 using the corresponding double-stranded RNA. As a control, the luciferase sequence was used. The cells were stimulated with EGF for the indicated time periods. The depletion was confirmed by the immunoblotting of the whole cell lysates. To detect ubiquitin conjugates of EGFR, immunoprecipitation was done under the denaturing condition.
Figure 5.
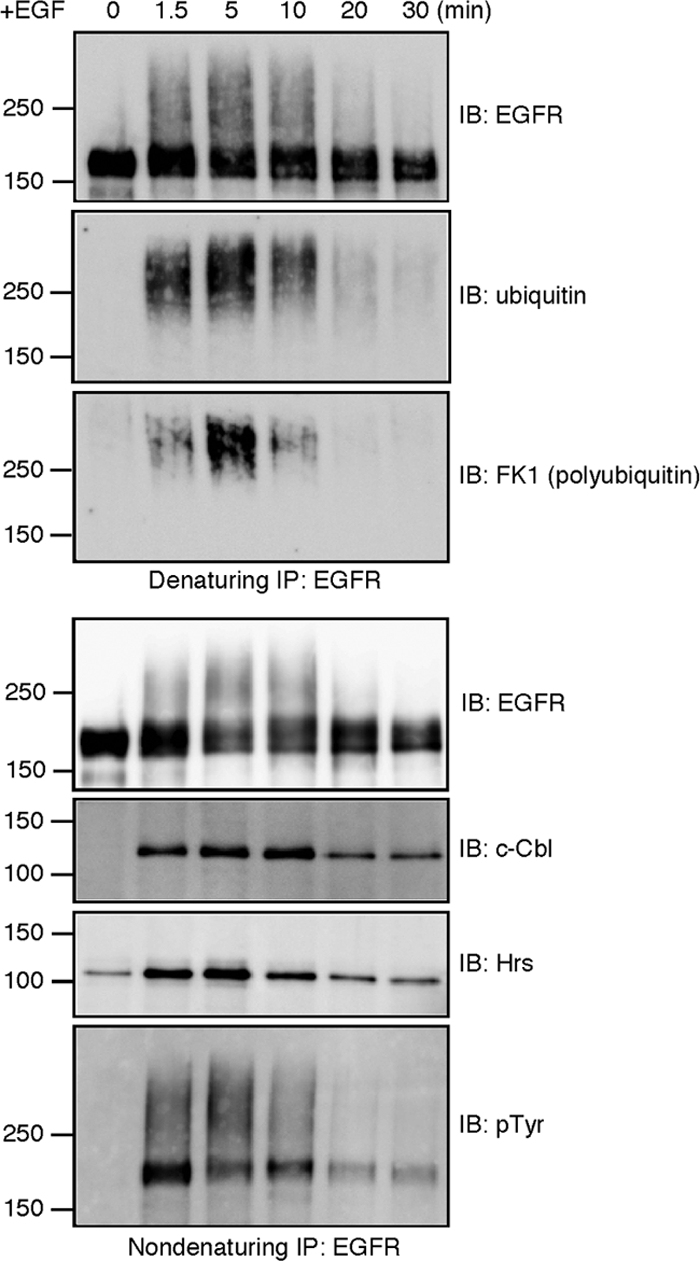
A time-course analysis of EGFR ubiquitination, tyrosine phosphorylation and interaction with c-Cbl and Hrs. HeLa cells were stimulated with EGF for the indicated time periods. The cell lysates were subject to two rounds of EGFR immunoprecipitation. c-Cbl, Hrs, and tyrosine-phosphorylated EGFR were detected from the first, nondenaturing immunoprecipitates. Ubiquitin-conjugated EGFR was detected from the second, denaturing ones.
Among Ubc4/5 proteins, UbcH5A was not depleted in this study. Nonetheless, the knockdown experiment successfully retarded the ubiquitination and degradation of EGFR. This is probably because UbcH5A is expressed at a lower level (Saville et al., 2004). Alternatively, UbcH5A may not be inherently involved in the c-Cbl–mediated EGFR ubiquitination. In the case of IκBα ubiquitination, UbcH5B and -C are capable of the conjugation but UbcH5A not (Gonen et al., 1999).
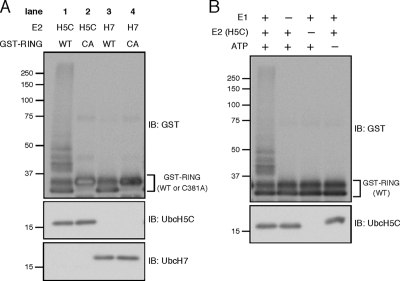
The colocalization and knockdown data strongly suggest that c-Cbl ubiquitinates EGFR in cooperation with Ubc4/5 but not UbcH7. For further confirmation, we examined E2 dependence in an in vitro autoubiquitination assay using GST-fused c-Cbl RING finger domain and purified components. When UbcH5C was used as the E2, ubiquitin-conjugated GST-RING was detected as the high-molecular-weight smear bands (Figure 4A, lane 1). The ubiquitination was dependent on E1, UbcH5C, and ATP (Figure 4B). It has been known that Cys-381 in the c-Cbl RING finger domain is critical for the E3 activity (Joazeiro et al., 1999; Waterman et al., 1999). In this assay, the C381A mutation abrogated the UbcH5C-dependent autoubiquitination (Figure 4A, lane 2). Notably, when UbcH7 was paired with GST-RING, the autoubiquitination activity was not detected (Figure 4A, lane 3). Thus, compared with UbcH7, UbcH5C efficiently catalyzes the in vitro ubiquitination together with the c-Cbl RING.
Figure 4.
UbcH5C cooperates with the c-Cbl RING in vitro. (A) Wild-type or C381A RING finger domain of c-Cbl was purified as GST-fusion, and its autoubiquitination activity was assayed by incubating with ubiquitin, E1, E2 (UbcH5C or UbcH7), and ATP. After the reaction, the samples were subject to immunoblotting. (B) In A, the E1-, E2 (UbcH5C)-, or ATP-dependence was examined.
Taken together, we concluded that the Ubc4/5 family is the primary E2 for the c-Cbl–mediated EGFR ubiquitination. The localization of Ubc4/5 reinforces the notion that the receptor ubiquitination proceeds after internalization.
Polyubiquitination of EGFR Is Required for Efficient Sorting to Lysosomes
To examine whether the ubiquitin moieties of EGFR are extended after internalization, a time-course analysis was done using EGF-stimulated HeLa cells. EGFR was first immunoprecipitated under a nondenaturing condition to detect its association with other proteins. To preserve protein–protein interactions on the cytoplasmic side of the receptor, the anti-EGFR antibody that recognizes the extracellular domain was used for the immunoprecipitation. The aliquots were denatured and again subject to EGFR immunoprecipitation to exclusively detect ubiquitinated forms of the receptor, not of any other associated proteins. All of the proteins were detected at their endogeneous expression levels.
The anti-ubiquitin immunoblotting indicated that the overall ubiquitination of EGFR reaches its peak at 5 min of EGF stimulation (Figure 5). The immunoprecipitated EGFR could also be detected by the polyubiquitin-specific antibody FK1 (Fujimuro et al., 1994), indicating that the receptor is polyubiquitinated (Figure 5). The peaks of the overall ubiquitination and polyubiquitination coincided at 5 min. Considering the localization of EGFR and c-Cbl to Hrs-positive endosomes at this time point (Figure 1C), it is likely that ongoing receptor ubiquitination enhances the receptor polyubiquitination after internalization. In the quantitative Western blot of the immunoprecipitated EGFR at 5 min after the stimulation, the anti-ubiquitin and anti-EGFR antibodies showed mostly linear responses, whereas FK1 did not (Supplementary Figure 4A). The nonlinear reactivity of FK1 obscures the exact quantification of the polyubiquitination; however, the increase in the FK1-positive signal between 1.5 and 5 min after EGF stimulation does indicate that receptor polyubiquitination is promoted after internalization.
Coimmunoprecipitation of c-Cbl and Hrs with EGFR was also examined (Figure 5). The amount of c-Cbl was increased for up to 10 min and then decreased rapidly. Concurrently, the ubiquitinated forms of EGFR were barely detectable after 20 min. The coimmunoprecipitation of Hrs was increased by EGF stimulation and declined after 5 min. Therefore, during the 5–10-min period, EGFR begins to be dissociated from Hrs but remains associated with c-Cbl. The receptor may be interacting with another sorting component, and we have been trying to detect binding between EGFR and the ESCRT-I component Tsg101. However, probably due to the limited sensitivities of the antibodies tested, detection of coimmunoprecipitated endogeneous Tsg101 has been unsuccessful.
To explore the significance of the receptor polyubiquitination, a ubiquitin mutant in which all seven lysines are mutated to arginines, designated as Ub(no Lys), was expressed. It has been suggested that overexpression of Ub(no Lys) affects lysosomal sorting through the ubiquitin configuration of cargo, without exerting nonspecific effects (Barriere et al., 2007). To monitor the expression, wild-type or the mutant ubiquitin was fused to the N-terminus of GFP (Figure 6A). As reported previously (Tsirigotis et al., 2001), the fusion protein is cleaved at the junction and the resultant free GFP serves as an indicator of the ubiquitin expression. We confirmed the detection of free GFP in cells expressing Ub(WT)-GFP (Figure 6B). The processing was dependent on the C-terminal two glycines of ubiquitin: when a mutant ubiquitin lacking these residues, Ub(ΔGG), was fused to GFP, no cleavage was detected (Figure 6B). To examine the effects on ubiquitination and degradation of EGFR, HEK293T cells, which express a low amount of endogeneous EGFR, were transiently transfected with plasmids encoding EGFR, c-Cbl, and either Ub(WT)-GFP or Ub(no Lys)-GFP. Expression of Ub(no Lys) significantly decreased the FK1 reactivity of the immunoprecipitated EGFR (Figure 6C), indicating that the receptor polyubiquitination is impaired. The FK1 signals were not linear also in this experiment, and the quantitative Western blot suggested that the extent of the polyubiquitination was decreased to <25% by the expression of Ub(no Lys) (Supplementary Figure 4B). Despite the weak FK1 signal, the anti-ubiquitin immunoblotting showed that ubiquitin conjugation of EGFR is not compromised in cells expressing Ub(no Lys) (Figure 6C). We confirmed that this anti-ubiquitin antibody recognizes purified Ub(no Lys) as efficiently as Ub(WT) (Figure 6D). Thus, as a result of Ub(no Lys) conjugation, EGFR is likely to undergo multiple monoubiquitination but not efficient polyubiquitination. Notably, the Ub(no Lys) expression resulted in the lower coimmunoprecipitation efficiency between EGFR and Hrs (Figure 6C). Moreover, Ub(no Lys) retarded the degradation rate of EGFR (Figure 6, E and F): the mean ± SD values from three independent experiments showed that 35 ± 8.1 and 65 ± 4.2% of EGFR were remaining at 45 min in cells expressing Ub(WT) and Ub(no Lys), respectively. These results strongly suggest that the EGFR polyubiquitination facilitates interaction with the sorting machinery and subsequent sorting to lysosomes.
Sustained Tyrosine Phosphorylation Protects EGFR from Deubiquitination and Ensures Lysosomal Sorting
Tyrosine phosphorylation of EGFR underlies its ubiquitination by providing the binding sites for c-Cbl. The time-course analysis in Figure 5 supported this notion. First, the duration of the receptor tyrosine phosphorylation, and the overall ubiquitination coincided: when the immunoprecipitated EGFR was probed with the anti-phosphotyrosine antibody, the phosphorylated receptor was clearly detected during the 1.5–10-min period. The upper smear bands most likely represent ubiquitin-conjugates of the phosphorylated receptor. The tyrosine phosphorylation was evidently decreased after 10 min. Second, concomitant with this decrease, reduced amounts of c-Cbl were coimmunoprecipitated with EGFR. The decrease in the tyrosine phosphorylation is unlikely to reflect receptor degradation, because the anti-EGFR immunoblotting did not show that the receptor was significantly degraded during the 10- and 20-min period. Thus, as previously reported (Burke et al., 2001), dephosphorylation of EGFR seems to precede its degradation in lysosomes.
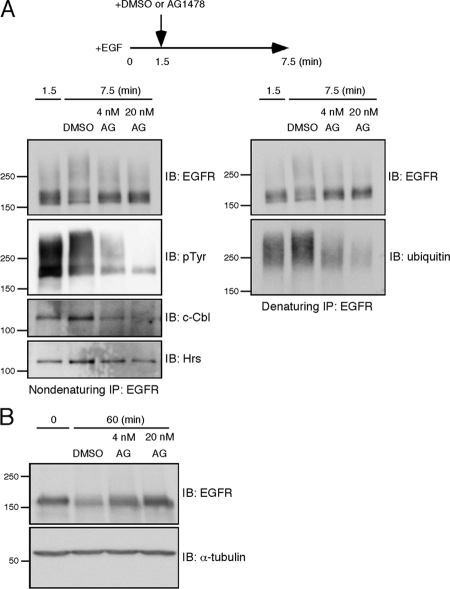
The localization and time-course results support the idea that EGFR is still subject to ubiquitination while it undergoes ubiquitin-dependent sorting in endosomes. The sustained tyrosine phosphorylation could well explain the receptor ubiquitination after internalization. To confirm this hypothesis and to address the significance of the ongoing ubiquitination, we tried to abruptly stop the c-Cbl–mediated ubiquitination by adding the EGFR kinase inhibitor AG1478. After HeLa cells were stimulated with EGF for 1.5 min, DMSO or AG1478 was added to the medium, and the cells were further incubated for 6 min. EGFR was subject to nondenaturing and denaturing immunoprecipitation, as described above. During the AG1478 administration, tyrosine phosphorylation of EGFR was rapidly attenuated, and c-Cbl was concomitantly dissociated from the receptor (Figure 7A). Therefore, after 1.5 min, further ubiquitin molecules should not be conjugated to the receptor. Mere inhibition of the receptor ubiquitination predicts that the ubiquitinated status just before the AG1478 addition is maintained. However, the dissociation of c-Cbl by AG1478 resulted in the dramatic loss of ubiquitin molecules that were conjugated to EGFR during the first 1.5 min (Figure 7A). Similarly, Hrs was dissociated from the receptor during the AG1478 treatment (Figure 7A). The AG1478 administration retarded the degradation of EGFR in a dose-dependent manner (Figure 7B), indicating that the receptor deubiquitination and dissociation from Hrs act against lysosomal sorting. The addition of AG1478 after 1.5 min of EGF stimulation might be too early; however, it should be noted that the duration of EGFR ubiquitination is short in HeLa cells: the receptor was significantly ubiquitinated at 1.5 min and the ubiquitination declined after 5 min (Figure 5). For this reason, we added AG1478 during the initial 5 min, while the receptor ubiquitination was increased. We also tried to do this experiment using HEK293T cells transiently expressing EGFR. When AG1478 was added at 10 min after EGF stimulation, the same effects were observed regarding dephosphorylation and deubiquitination of EGFR and dissociation of c-Cbl and Hrs from the receptor (Supplementary Figure 5). Thus, EGFR is susceptible to deubiquitination and dissociation from Hrs upon the abrupt inhibition of its tyrosine kinase activity. These results argue that the sustained tyrosine phosphorylation maintains the c-Cbl–mediated EGFR ubiquitination after internalization, thereby ensuring lysosomal sorting of the receptor.
Figure 7.
Inhibition of tyrosine phosphorylation leads to rapid deubiquitination of EGFR. (A) HeLa cells were stimulated with EGF for 1.5 min, and then AG1478 was added to the medium at a final concentration of 4 or 20 nM. As a control, an equal volume of DMSO was added. The cells were further incubated for 6 min. EGFR was subject to two rounds of immunoprecipitation. Tyrosine-phosphorylated EGFR, c-Cbl, and Hrs were detected from the first, nondenaturing immunoprecipitates. Ubiquitin-conjugated EGFR was detected from the second, denaturing ones. (B) HeLa cells were treated as described in A. After the addition of DMSO or AG1478, the cells were further incubated for 58.5 min to monitor EGFR degradation. As a control, cells were lysed before EGF stimulation. EGFR was detected by the immunoblotting of the whole cell lysates.
DISCUSSION
In this study, we have shown that Ubc4/5 is the partner of c-Cbl and this E2-E3 pair is colocalized in the plasma membrane as well as endosomes. This finding has advanced the concept of ongoing EGFR ubiquitination after internalization, highlighting the importance of determining the localization of E2s. The obtained results strongly suggest that the ongoing ubiquitination enhances polyubiquitination of EGFR as the receptor is transported to endosomes, and that the polyubiquitin chains are required for efficient lysosomal sorting. It may be taken for granted that cargo proteins remain ubiquitinated until a late step in the MVB sorting, but our results argue that the ubiquitinated status of EGFR is in fact maintained owing to the continuous ubiquitination. The AG1478 treatment acutely dissociated c-Cbl from EGFR, leading to the rapid deubiquitination and impaired degradation of the receptor. This suggests that ubiquitinating and deubiquitinating activities compete along the endocytic pathway. A recent report has indicated that both ubiquitinating and deubiquitinating activities are associated with the proteasome probably to remodel ubiquitin chains of substrates (Crosas et al., 2006). Therefore, it is likely that the competition between the opposing activities is common to both the ubiquitin–proteasome and endocytic pathways.
Polyubiquitination of EGFR During Endocytosis
It was thought that EGFR is subject to multiple monoubiquitination (Haglund et al., 2003). Afterward, based on tandem mass spectrometry, Sorkin's group has reported that both mono- and polyubiquitin are conjugated to the receptor (Huang et al., 2006). Our results are consistent with the latter report and underscore the importance of polyubiquitin moieties on EGFR. It should be noted that Haglund et al. could not detect EGFR polyubiquitination using FK1. We have indeed detected the smear bands with the same antibody, but the clear detection was dependent on the time after EGF stimulation. We also found that the chemiluminescent FK1 signals were not in the linear range under our experimental conditions: the signals were drastically reduced by the dilution of the samples. Therefore, it seems that the affinity of FK1 is relatively low and detection of EGFR polyubiquitination using this antibody is particularly influenced by the amounts of the loading materials.
The modest receptor degradation in cells expressing Ub(no Lys) suggests that multiple monoubiquitination can partially sort the receptor to lysosomes. Nonetheless, the obtained results argue that the EGFR polyubiquitination increases the efficiency of Hrs binding and subsequent lysosomal sorting. It has been reported that lysine63-linked polyubiquitin chains facilitate the endocytic rates of some cargo proteins (Galan and Haguenauer-Tsapis, 1997; Geetha et al., 2005; Duncan et al., 2006; Barriere et al., 2007). Notably, a structural study revealed that the ubiquitin-interacting motif of Hrs has double binding sites for ubiquitin, suggesting that one Hrs molecule binds to two ubiquitin moieties within the same polyubiquitin chain (Hirano et al., 2006). Consistent with this, Hrs preferentially binds to lysine63-linked polyubiquitin chains in vitro (Barriere et al., 2007). Considering that lysine63-linkages are most abundant among the polyubiquitin chains on EGFR (Huang et al., 2006), our results strongly suggest that this type of polyubiquitin linkage acts as a lysosomal sorting signal of EGFR, highlighting the emerging concept that the ubiquitin configuration is important in endocytosis.
The Ubc4/5 Family as the Partner of c-Cbl
Despite the previous reports that c-Cbl interacts with UbcH7 (Yokouchi et al., 1999; Zheng et al., 2000), the results presented here did not provide evidence that UbcH7 is crucial for the c-Cbl–mediated EGFR ubiquitination. Rather, we have shown that Ubc4/5 is the partner of c-Cbl. The E3 activity of c-Cbl was originally identified in an in vitro ubiquitination assay using Ubc4 as an E2 (Joazeiro et al., 1999). Furthermore, in vitro reconstitution of EGFR ubiquitination showed that either UbcH5B or UbcH5C can support the reaction (Levkowitz et al., 1999). Our conclusion is consistent with these reports. The amino acid residues in UbcH7 that contact the c-Cbl RING domain, which are positioned in the E2's L1 and L2 loops (Zheng et al., 2000), are conserved in Ubc4, UbcH5A, UbcH5B, and UbcH5C. Indeed, Ubc4 binds to GST-RING(c-Cbl) in a pulldown assay (Joazeiro et al., 1999). The c-Cbl RING can thus bind to both Ubc4/5 and UbcH7. Importantly, evidence is accumulating that binding analyses do not always identify functional E2-E3 pairs. First, as many as 11 E2s can interact with the RING domain of the E3 BRCA1, but not all of them can support BRCA1 autoubiquitination in vitro (Christensen et al., 2007). Second, increasing the affinity between an E2 (Cdc34) and a RING-type E3 (Cdc4) rather impaired the ubiquitinating activity (Deffenbaugh et al., 2003). The authors proposed “Hit and Run” model: rapid release of ubiquitin-charged Cdc34 from Cdc4 is required for efficient ubiquitination. To conjugate ubiquitin molecules to distal lysine residues and/or termini of polyubiquitin chains, flexible positioning of E2 should be advantageous. This idea is very intriguing given that EGFR rapidly undergoes polyubiquitination and many lysine residues serve as ubiquitin acceptor sites (Huang et al., 2006).
Despite the binding between purified Ubc4 and GST-RING (c-Cbl; Joazeiro et al., 1999), we failed to reliably detect coimmunoprecipitation between full-length c-Cbl and UbcH5C. This might indicate that c-Cbl and Ubc4/5 do not interact in vivo. However, we favor the emerging concept that E2-E3 interaction is transient and weak. It may well be difficult to identify functional E2–E3 pairs on the basis of binding, and here we have provided three lines of evidence (localization, knockdown, and in vitro activity analyses) arguing that Ubc4/5 is the partner of c-Cbl. Therefore, we are negative about the possibility that the failure to detect specific coimmunoprecipitation invalidates our conclusion.
Tyrosine Phosphorylation and Ubiquitin-dependent Sorting of EGFR
Internalized receptor tyrosine kinases remain phosphorylated in endosomes, leading to signal transduction from endosomes (Sorkin and von Zastrow, 2002). As shown here, the sustained tyrosine phosphorylation also underlies the ongoing EGFR ubiquitination. Thus, while tyrosine phosphorylation transduces the signal from EGFR, it facilitates lysosomal sorting of the receptor. It is plausible that tyrosine phosphorylation has dual, opposing functions so that the EGF signaling is not prolonged. It has been known that the sorting machinery such as Hrs and annexin 1 are tyrosine-phosphorylated in response to EGF, to ensure EGFR degradation and to stimulate inward vesiculation in multivesicular endosomes containing EGFR, respectively (White et al., 2005; Stern et al., 2007). The Hrs phosphorylation is dependent, at least in part, on Src family kinases (Bache et al., 2002; Row et al., 2005). Hrs also undergoes ubiquitination (Hoeller et al., 2006), and the possibility that its ubiquitinated status is regulated by the tyrosine phosphorylation remains to be explored.
Notably, when the ongoing EGFR ubiquitination was inhibited by AG1478, the receptor deubiquitination was accelerated. The identity of the responsible deubiquitinating enzyme(s) remains to be revealed. UBPY and AMSH have been shown to deubiquitinate EGFR in vitro and to be implicated in receptor deubiquitination during endocytosis (McCullough et al., 2004; Mizuno et al., 2005; Agromayor and Martin-Serrano, 2006; Kyuuma et al., 2006; Row et al., 2006; Ma et al., 2007). It is possible that they compete with c-Cbl in endosomes. Also, other endosomal and/or cytosolic deubiquitinating enzymes may well act on EGFR especially when c-Cbl is inappropriately dissociated from the receptor.
While the sustained tyrosine phosphorylation underlies the ongoing ubiquitination, dephosphorylation of EGFR may facilitate its deubiquitination by eliminating the binding site for c-Cbl. The dephosphorylation process is not fully understood, but receptor-type protein tyrosine phosphatase-κ has been shown to dephosphorylate activated EGFR (Xu et al., 2005). This phosphatase is a transmembrane protein that orients the catalytic domain in the cytosol, suggesting that EGFR dephosphorylation occurs, at least partially, before MVB sorting. Dissociation of the EGF–EGFR complex, which is triggered by acidification of the endosome lumen, would enhance the receptor dephosphorylation. The timing of the acidification in the context of MVB sorting remains to be elucidated. However, the localization of ESCRT-I, -II, and -III components to late endosomes (Bache et al., 2003, 2006; Slagsvold et al., 2005) raises the possibility that dissociation of EGF from EGFR is commenced while the receptor is interacting with ESCRTs, leading to receptor dephosphorylation and deubiquitination before entry into MVB vesicles.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Minsoo Kim and Tadashi Yamamoto for plasmids and advice during the initial planning of this work, Noriyuki Matsuda and Keiji Tanaka for plasmids and helpful discussions, Hiroshi Ohno for plasmids, and Takeshi Noda for critically reading the manuscript.
Abbreviations used:
- EGFR
epidermal growth factor receptor
- ESCRT
endosomal sorting complex required for transport
- MVB
multivesicular body
- NEM
N-ethylmaleimide.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-0988) on May 28, 2008.
REFERENCES
- Agromayor M., Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 2006;281:23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Raiborg C., Mehlum A., Madshus I. H., Stenmark H. Phosphorylation of Hrs downstream of the epidermal growth factor receptor. Eur. J. Biochem. 2002;269:3881–3887. doi: 10.1046/j.1432-1033.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G. L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere H., Nemes C., Du K., Lukacs G. L. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell. 2007;18:3952–3965. doi: 10.1091/mbc.E07-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke P., Schooler K., Wiley H. S. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen D. E., Brzovic P. S., Klevit R. E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Crosas B., et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Deffenbaugh A. E., Scaglione K. M., Zhang L., Moore J. M., Buranda T., Sklar L. A., Skowyra D. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell. 2003;114:611–622. doi: 10.1016/s0092-8674(03)00641-x. [DOI] [PubMed] [Google Scholar]
- Duan L., et al. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 2003;278:28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., Lehner P. J. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M., Sawada H., Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Galan J. M., Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T., Jiang J., Wooten M. W. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell. 2005;20:301–312. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Gonen H., Bercovich B., Orian A., Carrano A., Takizawa C., Yamanaka K., Pagano M., Iwai K., Ciechanover A. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J. Biol. Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- Grøvdal L. M., Stang E., Sorkin A., Madshus I. H. Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp. Cell Res. 2004;300:388–395. doi: 10.1016/j.yexcr.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Gruenberg J. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hirano S., Kawasaki M., Ura H., Kato R., Raiborg C., Stenmark H., Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat. Struct. Mol. Biol. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- Hoeller D., et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang F., Marusyk A., Sorkin A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol. Biol. Cell. 2003;14:858–870. doi: 10.1091/mbc.E02-08-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C. A. P., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kyuuma M., et al. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 2006;31:159–172. doi: 10.1247/csf.06023. [DOI] [PubMed] [Google Scholar]
- Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Longva K. E., Blystad F. D., Stang E., Larsen A. M., Johannessen L. E., Madshus I. H. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. M., Boucrot E., Villen J., Affar E. B., Gygi S. P., Gottlinger H. G., Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M. J., Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Myromslien F. D., Grovdal L. M., Raiborg C., Stenmark H., Madshus I. H., Stang E. Both clathrin-positive and -negative coats are involved in endosomal sorting of the EGF receptor. Exp. Cell Res. 2006;312:3036–3048. doi: 10.1016/j.yexcr.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bremnes B., Mehlum A., Gillooly D. J., D'Arrigo A., Stang E., Stenmark H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Ravid T., Heidinger J. M., Gee P., Khan E. M., Goldkorn T. c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J. Biol. Chem. 2004;279:37153–37162. doi: 10.1074/jbc.M403210200. [DOI] [PubMed] [Google Scholar]
- Row P. E., Clague M. J., Urbé S. Growth factors induce differential phosphorylation profiles of the Hrs-STAM complex: a common node in signalling networks with signal-specific properties. Biochem. J. 2005;389:629–636. doi: 10.1042/BJ20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Saville M. K., Sparks A., Xirodimas D. P., Wardrop J., Stevenson L. F., Bourdon J. C., Woods Y. L., Lane D. P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- Schmidt M. H. H., Dikic I. The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Aasland R., Hirano S., Bache K. G., Raiborg C., Trambaiolo D., Wakatsuki S., Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Sorkin A., von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- Stern K. A., Visser Smit G. D., Place T. L., Winistorfer S., Piper R. C., Lill N. L. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol. Cell. Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien C. B. F., Langdon W. Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–305. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- Tsirigotis M., Zhang M., Chiu R. K., Wouters B. G., Gray D. A. Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J. Biol. Chem. 2001;276:46073–46078. doi: 10.1074/jbc.M109023200. [DOI] [PubMed] [Google Scholar]
- Umebayashi K. The roles of ubiquitin and lipids in protein sorting along the endocytic pathway. Cell Struct. Funct. 2003;28:443–453. doi: 10.1247/csf.28.443. [DOI] [PubMed] [Google Scholar]
- Umebayashi K., Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Ismail A., Gao X., Fu G., Li X., O'Malley B. W., Nawaz Z. The ubiquitin-conjugating enzyme UBCH7 acts as a coactivator for steroid hormone receptors. Mol. Cell. Biol. 2004;24:8716–8726. doi: 10.1128/MCB.24.19.8716-8726.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman H., Levkowitz G., Alroy I., Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- White I. J., Bailey L. M., Aghakhani M. R., Moss S. E., Futter C. E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2005;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Tan L. J., Grachtchouk V., Voorhees J. J., Fisher G. J. Receptor-type protein-tyrosine phosphatase-kappa regulates epidermal growth factor receptor function. J. Biol. Chem. 2005;280:42694–42700. doi: 10.1074/jbc.M507722200. [DOI] [PubMed] [Google Scholar]
- Yokouchi M., Kondo T., Houghton A., Bartkiewicz M., Horne W. C., Zhang H., Yoshimura A., Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.