Abstract
Despite extensive work on ADP-ribosylation factor (Arf) 1 at the Golgi complex, the functions of Arf2–5 in the secretory pathway, or for that of any Arf at the ER-Golgi intermediate compartment (ERGIC) remain uncharacterized. Here, we examined the recruitment of fluorescently tagged Arf1, -3, -4, and -5 onto peripheral ERGIC. Live cell imaging detected Arfs on peripheral puncta that also contained Golgi-specific brefeldin A (BFA) resistance factor (GBF) 1 and the ERGIC marker p58. Unexpectedly, BFA did not promote corecruitment of Arfs with GBF1 either at the Golgi complex or the ERGIC, but it uncovered striking differences between Arf1,3 and Arf4,5. Although Arf1,3 quickly dissociated from all endomembranes after BFA addition, Arf4,5 persisted on ERGIC structures, even after redistribution of GBF1 to separate compartments. The GDP-arrested Arf4(T31N) mutant localized to the ERGIC, even with BFA and Exo1 present. In addtion, loss of Arf · GTP after treatment with Exo1 caused rapid release of all Arfs from the Golgi complex and led to GBF1 accumulation on both Golgi and ERGIC membranes. Our results demonstrate that GDP-bound Arf4,5 associate with ERGIC membranes through binding sites distinct from those responsible for GBF1 recruitment. Furthermore, they provide the first evidence that GBF1 accumulation on membranes may be caused by loss of Arf · GTP, rather than the formation of an Arf · GDP · BFA · GBF1 complex.
INTRODUCTION
ADP-ribosylation factors (Arfs) play critical roles in membrane traffic within eukaryotic cells by initiating the recruitment of various coat proteins and by modulating the activity of several lipid-modifying enzymes (Donaldson and Jackson, 2000). Mammals express six Arf isoforms, Arf1-6 (Arf2 has been lost in humans), which are grouped into three classes based on primary sequence and gene organization (Kahn et al., 2006). Class I consists of Arf1-3, which share 96% sequence identity and are the most abundantly expressed in cells and tissues examined (Cavenagh et al., 1996). The class II Arfs, Arf4 and Arf5, are 90% identical to each other, occur at much lower abundance and remain poorly characterized. Arf6, the only class III Arf, shares the least degree of sequence identity to the other Arfs.
The best characterized Arfs, Arf1 and Arf6, localize to distinct compartments where they likely perform different functions (Peters et al., 1995). Although Arf1 localizes primarily on the Golgi complex to regulate the assembly of several types of coat complexes, Arf6 has been shown to regulate endosomal membrane traffic and structural organization at the plasma membrane (D'Souza-Schorey and Chavrier, 2006). Although little is known about Arf4, it has been implicated in the sorting of rhodopsin into post-Golgi carriers (Deretic et al., 2005). Despite apparent unique roles for each Arf, recent reports suggest that they may not function independently from each other. Arf knockdown (KD) studies by Kahn and colleagues revealed that distinct pairs of Arfs may be required at each step of protein traffic (Volpicelli-Daley et al., 2005). For example, disruption of ER-to-Golgi traffic resulted only from double KD of Arf1 and Arf4. Requirement for specific Arf pairs suggests that even though there may be some functional redundancy between individual Arfs, each Arf must play some distinct and critical roles. Consistent with this notion, a recent study by Donaldson and colleagues revealed a mechanism in which activated Arf6 promotes recruitment of ARNO for subsequent activation of Arf1 on endosomes (Cohen et al., 2007).
Recruitment of Arf · GDP from cytosol to a membrane allows activation by a guanine nucleotide exchange factor (GEF). Subsequent inactivation of Arf · GTP by a GTPase-activating protein (GAP) completes the cycle and releases Arf · GDP to cytosol (Donaldson and Jackson, 2000). The relative abundance of any given Arf on a particular organelle should depend on the abundance of receptors to recruit Arf · GDP, the specificity of the GEFs present, and the relative rate of inactivation by GAPs. Inactive, GDP-bound Arfs can associate weakly with membranes via a myristoylated amphipathic N-terminal helix (Antonny et al., 1997). However, several studies suggest that membrane proteins, including the p23/24 proteins (Gommel et al., 2001; Majoul et al., 2001) and several ER-Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, such as membrin (Rein et al., 2002; Honda et al., 2005), can function as receptors for Arf1 · GDP and potentially increase Arf1 · GDP levels at the membrane. Receptors for class II and III Arfs have not yet been reported. After recruitment onto membranes of the Golgi complex or the ER-Golgi intermediate compartment (ERGIC), Arf · GDP may be activated by members of either of two Arf-GEF families, the Golgi-specific brefeldin A (BFA) resistance factor 1 (GBF1) and the BFA-inhibited GEFs (BIGs). Even though these two GEF families localize to different subcompartments (Zhao et al., 2002), their ability to activate both class I and II Arfs (Claude et al., 1999; Kawamoto et al., 2002) renders them unlikely to enrich any given Arf on a specific compartment. The specificity of Arf-GAPs localized to early secretory compartments remains uncharacterized (Inoue and Randazzo, 2007).
Several experimental approaches have been developed to characterize Arf-GEFs both in vitro and in vivo. These include drugs, viral proteins, as well as mutations in Arfs and GEFs that interfere with the exchange of GDP for GTP and can stabilize an abortive Arf · GEF complex (Beraud-Dufour et al., 1998; Goldberg, 1998; Mossessova et al., 1998, 2003; Peyroche et al., 1999; Niu et al., 2005; Szul et al., 2005; Wessels et al., 2006). The best-characterized tool, the fungal metabolite BFA, targets the catalytic domain, known as the Sec7 domain (Sec7d) (Mansour et al., 1999; Peyroche et al., 1999; Robineau et al., 2000; Mossessova et al., 2003). BFA inserts at the interface between Arf and the Sec7d, prevents GDP displacement and causes formation of an abortive Arf · GDP · BFA · Sec7d complex. A charge reversal mutation of the catalytic “glutamate finger” in the Sec7d similarly interferes with GDP exchange and causes accumulation of the abortive complex at very low magnesium concentrations (Beraud-Dufour et al., 1998; Renault et al., 2003). Arf1(T31N), a dominant-negative mutant of Arf1 that has a low affinity for GTP, has been proposed to also form such complex and prevent activation of endogenous Arfs. Interestingly, treatment with BFA, as well as expression of GBF1(E794K) or Arf1(T31N), greatly reduce the dynamic exchange between free and membrane-bound GBF1 (Niu et al., 2005; Szul et al., 2005; Zhao et al., 2006). These observations suggest that trapping an Arf · GBF1 complex prevents GBF1 release from membranes and led to the conclusion that nucleotide exchange is linked to release of GBF1 from its membrane receptor (Cherfils and Melançon, 2005; Niu et al., 2005; Szul et al., 2005; Zhao et al., 2006). This conclusion seems consistent with the demonstration that the enterovirus 3A protein blocks protein traffic to the Golgi complex and can form a complex with the GDP bound form of Arf1 [Arf1(T31N)] and GBF1 (Wessels et al., 2006).
In contrast to the detailed information available on the Arf-GEFs that regulate the secretory pathway, much less is known about the function of class II Arfs. The lack of antibodies to detect class II Arfs and the extensive biochemical overlap of different Arf isoforms hindered progress on characterization of Arfs. In this study, we exploit live cell imaging methods in combination with a variety of pharmacological agents that provide evidence for distinct localization and behaviors of class II Arfs at the ERGIC.
MATERIALS AND METHODS
Reagents and Antibodies
BFA and nocodazole (NOZ) were purchased from Sigma-Aldrich (St. Louis, MO) and stored in dimethyl sulfoxide (DMSO) as 10- and 5-mg/ml stock solutions, respectively. Exo1 was purchased from Calbiochem (Gibbstown, NJ) and stored in DMSO at 25 mg/ml (91.5 mM). A serum (9D6) was raised in rabbits against recombinant Arf5 purified as described previously (Berger et al., 1988). Arf5-specific antibodies were affinity purified by passage of 9D6 over an Arf5-conjugated Affigel-10 column (Bio-Rad, Mississauga, ON, Canada) as described previously (Harlow, 1988). Affinity-purified 9D6 was used at 1:25 dilution for immunofluorescence (IF). Additional polyclonal antibodies used for IF were anti-Arf1 (2048; B. Helms, University of Utrecht, Utrecht, The Netherlands) at 1:400 and anti-p58 (Molly6; Saraste et al., 1987; J. Saraste, University of Bergen, Bergen, Norway) at 1:400. Polyclonal antibodies used for immunoblotting were anti-GBF1 (9D4 final bleed; Manolea et al., 2008) at 1:2500 and anti-green fluorescent protein (GFP) (G. Eitzen, University of Alberta, Edmonton, AB, Canada) at 1:1000. The monoclonal antibodies used for IF were anti-GBF1 (clone 25; BD Biosciences, Mississauga, ON, Canada), anti-β-coatomer protein I (COPI) (clone M3A5; Allan and Kreis, 1986; Sigma-Aldrich) at 1:400, and anti-mannosidase II (Man II) (clone 53FC3; Burke et al., 1982) at 1:100. Secondary antibodies used were Alexa 488/594-conjugated goat anti-rabbit and Alexa 488/594-conjugated goat anti-mouse antibodies (Invitrogen, Carlsbad, CA) at 1:600.
Construction of Plasmids for Tagged GBF1 and Arfs
A plasmid encoding GBF1 tagged at the N terminus with monomeric Cherry (mCherry) was constructed in the backbone of pcDNA5/TO (Invitrogen). pcDNA5/TO was first modified to include an Nhe1 site by insertion of a synthetic duplex between the HindIII and Kpn1 sites. An inducible plasmid encoding the GFP-GBF1 chimera was then constructed by transferring the Nhe1–Not1 fragment from pIND-GFP-GBF1 (Zhao et al., 2006) into pcDNA5/TO-Nhe1. Construction of pcDNA5/TO-mCherry-GBF1 entailed two sequential steps. A fragment encoding mCherry amplified by polymerase chain reaction (PCR) from pRSETB-mCherry (Dr. R. Tsien, University of California-San Diego, San Diego, CA) was first inserted between the Nhe1 and Xho1 sites of pcDNA5/TO-Nhe1; an Xho1 fragment encoding GBF1 was then transferred from pcDNA5/TO-GFP-GBF1 into the Xho1 sites of pcDNA5/TO-Nhe1-mCherry. This approach yielded an mCherry-GBF1 with linker identical in length and sequence to that of the previously characterized GFP-GBF1 (Zhao et al., 2006).
The plasmids used for Arf-GFP expression in this study were constructed by inserting fragments encoding human Arf1, Arf3, Arf4, and Arf5 between the Xho1 and Kpn1 sites of pEGFP-N1 (Clontech, Mountain View, CA). Fragments were obtained by PCR amplification from vectors harboring human Arf sequences (Berger et al., 1988) by using forward primers that introduced an Xho1 site upstream of the ATG and reverse primers that changed the TGA stop codon to CGC and introduced a Kpn1 site immediately downstream that allowed in frame translation of GFP after a 12-residue linker (AVPRARDPPVAT). The Arf4(T31N) mutant was created by site-directed mutagenesis by using the QuikChange kit (Stratagene, La Jolla, CA), as per manufacturer's instructions. The Arf1(T31N) fragment was amplified using a plasmid encoding hemagglutinin (HA)-tagged bovine Arf1(T31N) (Peters et al., 1995) obtained from V. Hsu (Harvard Medical School, Boston, MA).
The plasmid for Arf1-mCherry was derived from one encoding Arf1-red fluorescent protein (RFP) constructed by Dr. J. Donaldson (National Institutes of Health, Bethesda, MD) by inserting the complete bovine Arf1 cDNA between the BglII and EcoR1 sites of RFP-N1 (Cohen et al., 2007). To generate Arf1-mCherry, we exchanged the BamH1–Not1 fragment encoding RFP with a similar one encoding mCherry. This procedure introduced a 13-residue linker (GILQSTVPRARDP) between the terminal Lys of Arf1 and the initiating Met of mCherry.
To generate mCherry-tagged forms of Arf3, Arf4, and Arf5, we substituted a BamH1–Not1 fragment encoding mCherry into pEGFP-N1 into which Arf cDNAs amplified by PCR from vectors encoding Arf3-HA, Arf4-HA, and Arf5-HA (Peters et al., 1995) had been inserted between the Xho1 and Kpn1 sites. This procedure yielded chimeras containing an eight-residue linker (AVPRARDP) between the terminal Arf residue and the initiating Met of mCherry.
Cell Culture and Normal Rat Kidney (NRK) Cell Line Expressing GFP-tagged GBF1
The cell lines used for this study include HeLa (ECACC, 93021013; Sigma-Aldrich), NRK-52E cells (ATCC CRL-1571, American Type Culture Collection, Manassas, VA), COS1 cells (ATCC CRL-1650; American Type Culture Collection), and T-REx-293 (Invitrogen). The isolation of the stable NRK cell line expressing the N-terminal GFP-tagged GBF1 (NRK-GFP-GBF1) has been described previously (Zhao et al., 2006). Cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C in a 5% CO2 incubator. FuGENE 6 (Roche Diagnostics, Indianapolis, IN) or TransIT-LTI transfection reagent (Mirus, Madison, WI) was used for transient transfections.
Immunofluorescence and Live Cell, Time-Lapse Imaging
For IF experiments, cells were grown and treated with drugs at 37°C on glass coverslips, washed in phosphate-buffered saline (PBS), and fixed with 3% paraformaldehyde in PBS at 37°C for 20 min. Cells double labeled with mouse and rabbit antibodies were processed as described previously (Zhao et al., 2002). Epifluorescence images shown in Figures 1, 6A, 7B, and 8B and in Supplemental Figures 3 and 5 were obtained using an Axioskop II microscope (Carl Zeiss, Thornwood, NY) equipped with a 63× objective (plan-Apocromat, numerical aperture [NA]= 1.4).
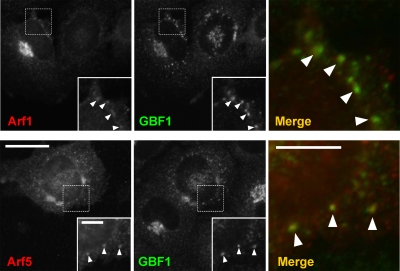
Figure 1.
Arf1 and Arf5 localize to both the Golgi complex and peripheral puncta positive for GBF1. NRK cells were transfected with plasmids encoding untagged Arf1 or Arf5, fixed, and then processed for indirect immunofluorescence with antibodies directed against GBF1 (green channel) and either Arf1 or Arf5 (red channel) as described in Materials and Methods. Bar, 20 μm.
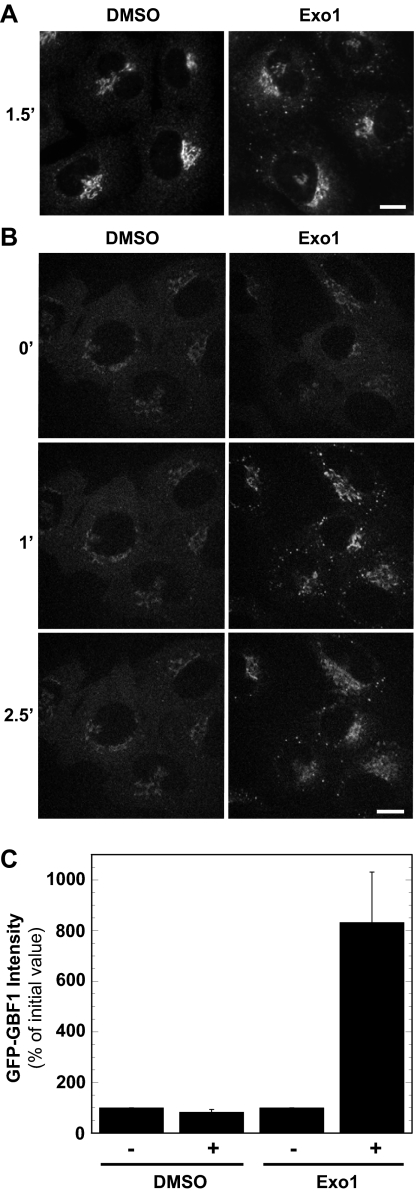
Figure 6.
Exo1 causes GBF1 to concentrate on the Golgi complex and the ERGIC. (A) NRK cells were treated with carrier DMSO or 100 μM Exo1 for 90 s, fixed, and processed for IF by using anti-GBF1 monoclonal antibody as described in Materials and Methods. Images were acquired and processed identically. Bar, 10 μm. (B) NRK-GFP-GBF1 cells were treated with carrier DMSO or 100 μM Exo1 and imaged for 10 min. Images correspond to single frames at the indicated time points. Images representative of at least four experiments in NRK cells. Bar, 10 μm. (C) Quantification of GFP-GBF1 relative signal intensities (percentage of initial value) at the Golgi and peripheral puncta membranes after 1 min of treatment with Exo1. Values correspond to averages with SE obtained from a minimum of 25 cells from at least four different sets of experiments.
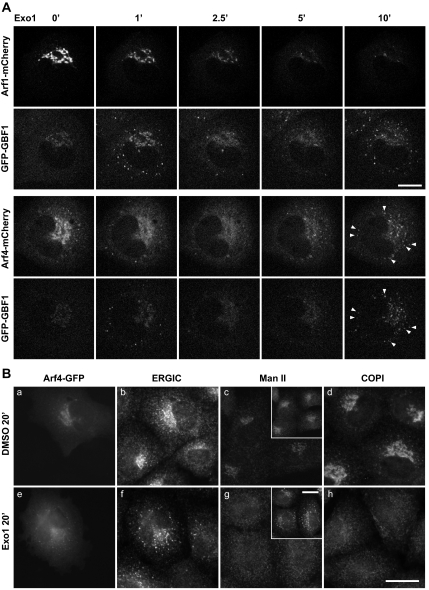
Figure 7.
Exo1 causes rapid loss of Arfs from Golgi membranes but leads to eventual concentration of GFP-GBF1 and class II Arfs at the ERGIC. (A) NRK-GFP-GBF1 cells transfected with plasmids encoding Arfs tagged with mCherry were treated with carrier DMSO or 100 μM Exo1 and imaged for 20 min as described in Materials and Methods. Images correspond to single frames captured at the indicated times. Data obtained with Arf3 and Arf5 are provided as Supplemental Material. Bar, 10 μm. (B) NRK cells were treated with DMSO (a–d) or 100 μM Exo1 (e–h) for 20 min and then processed for double-label IF by using antibodies to the indicated markers. Paired panels a, b and e, f display green, red channels from the same area of cells transfected with plasmids encoding Arf4-GFP. Insets in c and g display images obtained with antibody to the ERGIC marker Molly 6. GBF1 and Arf4-GFP concentrate on ERGIC structures devoid of COPI and the Golgi marker Man II. Images representative of four experiments. Bar, 20 μm.
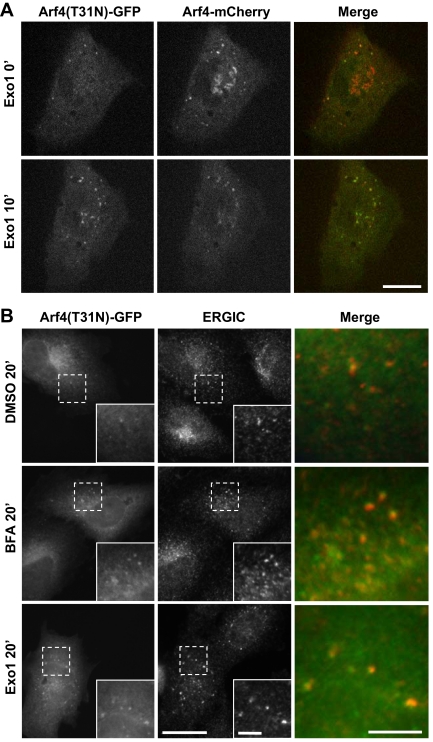
Figure 8.
Inactive Arf4 · GDP localizes to the ERGIC. (A) NRK cells were cotransfected with plasmids encoding wild-type Arf4-mCherry and the GDP-arrested mutant (T31N) of Arf4-GFP. Transfectants were imaged both before and after addition of 100 μM Exo1. Images correspond to single frames obtained immediately prior (t = 0′) or 10 min after Exo1 addition. Wild type (WT), but not the T31N mutant of Arf4-GFP, localizes to the Golgi complex; staining patterns for mutant and WT Arf4 become identical after Exo1 treatment. Bar, 10 μm. (B) NRK cells were transfected with plasmids encoding Arf4(T31N)-GFP. After treatment with carrier DMSO, 5 μg/ml BFA or 100 μM Exo1, cells were fixed and processed for IF as described in text. Insets show enlargement of boxed area. Right panels show merge signal from the enlarged area. Bar, 20 μm.
For live cell imaging, cells were grown in glass-bottomed microwell dishes (Plastek Cultureware; MatTek, Ashland, MA). The medium was switched to CO2-independent DMEM (Invitrogen) supplemented with 10% FBS immediately before imaging on a temperature-controlled (37°C) stage of an Axiovert 200M confocal microscope (Carl Zeiss) equipped with an UltraVIEW ERS 3E spinning disk (PerkinElmer Life and Analytical Sciences, Waltham, MA) and 100× objective lens (plan-Apocromat, NA = 1.4). Live cell imaging was performed in a room maintained either at 23°C (Figures 2, 3, and 5) or 35°C (Figures 4 and 6–8); a thermocouple placed in the glass coverslip registered a few degrees above room temperature even when the heated stage was set at 37°C. Images were captured with a 9100-50 electron multiplier charge-coupled device digital camera (Hamamatsu Photonics, Bridgewater, NJ) and processed with Ultraview Image Suite. Movies with dual labeling were imaged in the same cells by exciting each fluorophore and detecting sequentially (multitrack mode) to avoid channel bleedthrough. Laser intensity and filters were adjusted to give maximum signal but avoid saturation. Experiments involving drug addition were performed by adding 500 μl of medium containing 4 times drug concentration to a dish containing 1.5 ml of medium. For BFA washout experiments (Figure 4), medium was twice aspirated and replaced with medium lacking drug at the indicated time. Focus was adjusted immediately after manipulation as required.
Figure 2.
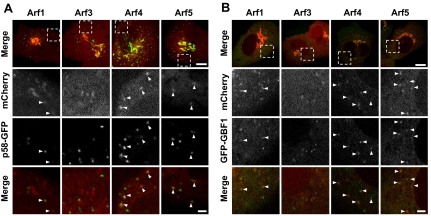
Class II Arf-positive puncta correspond to peripheral ERGIC that also contain GFP-GBF1. (A) NRK cells cotransfected with plasmids encoding p58-GFP and mCherry-tagged forms of either Arf1, Arf3, Arf4, or Arf5 as described in Materials and Methods. Images were acquired from live cells using a spinning disk confocal microscope as described in Materials and Methods. The upper row displays representative merged stills corresponding to a single confocal slice. Bottom rows show enlargement of the boxed area for single and merged images acquired from the mCherry and GFP channels. The majority of cells expressing Arf1 (91%), Arf3 (76%), Arf4 (100%), and Arf5 (100%) contain mCherry-positive puncta. Bar, 10 μm for full images and 2 μm for magnified images. (B) NRK cells stably expressing low levels of GFP-GBF1 (NRK-GFP-GBF1) were transfected with plasmids encoding mCherry-tagged forms of either Arf1, Arf3, Arf4, or Arf5 as described in Materials and Methods, and they were imaged as described for A. Bar, 10 μm for full image and 2 μm for magnified image.
Figure 3.
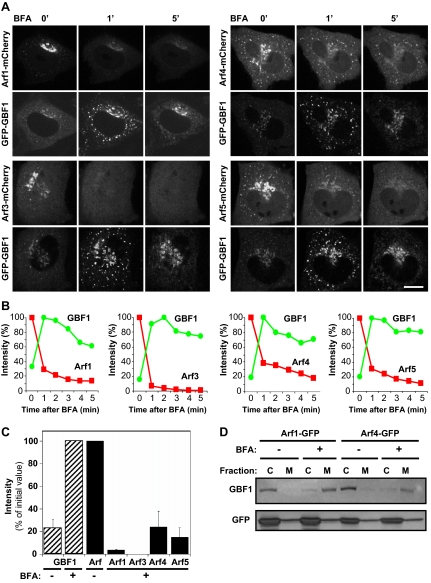
BFA treatment does not cause corecruitment of Arfs with GBF1 on Golgi and ERGIC membranes. (A) NRK-GFP-GBF1 cells were transfected with plasmids encoding the indicated mCherry-tagged Arfs. Images were acquired every 5 s after the addition of 5 μg/ml BFA as described in Materials and Methods. Images shown are from single frames at the indicated time points. Bar, 10 μm. (B) Quantification of the change in Arf-mCherry and GFP-GBF1 signal intensities at the Golgi and peripheral puncta membranes in response to BFA. The signal intensities from stills obtained from each experiment shown in A were measured at the indicated times as described in Materials and Methods and then expressed as a percentage of maximum Arf and GBF1 values. (C) Quantification of GFP-GBF1 and Arf-mCherry relative signal intensities (percentage of maximum) at the Golgi and peripheral puncta membranes after 1 min of treatment with BFA. Values correspond to averages with SE obtained from at least three different sets of experiments. For each cell examined, maxima for Arfs were defined as values measured before BFA addition, whereas maxima for GBF1 were values measured 1 min after BFA addition. (D) T-REx-293 cells were transfected with plasmids encoding Arf1-GFP or Arf4-GFP and treated with carrier DMSO or 10 μg/ml BFA for 2 min at 37°C. Homogenates were prepared and then separated into cytosolic (C) and membrane (M) fractions as described in Materials and Methods. Panels show analysis of fractions following SDS-PAGE and immunoblotting with GBF1 (top) and GFP (bottom) antibodies.
Figure 5.
Stable association of class II Arfs with the ERGIC in the presence of BFA does not require GBF1. (A) Images correspond to the 10-min time points from the Arf4 and Arf5 movies shown in Figure 3A. Bottom panels show single and merged images acquired in the boxed area from the mCherry and GFP channels. Representative of at least three experiments in NRK-GFP-GBF1 cells. Bar, 10 μm for the full images and 2 μm for insets. (B) NRK-GFP-GBF1 cells transfected with either Arf4-mCherry (top row) or Arf5-mCherry (bottom row) were incubated on ice for 2 min followed by treatment with 20 μg/ml NOZ for 15 min on ice. Several minutes (5–10 min) after transfer to a heated stage, cells were treated with BFA and imaged every 5 s for 10 min. Images correspond to frames captured at the indicated times. Bottom panels show single and merged images acquired in the boxed area from the mCherry and GFP channels. Images representative of at least four experiments in NRK-GFP-GBF1 cells. Bar, 10 μm. (C) NRK cells were cotransfected with plasmids encoding p58-GFP, and either Arf4-mCherry (top) or Arf5-mCherry (bottom) and then treated and imaged as described in B. Arf4/5-mCherry–positive structures after treatment with NOZ and BFA contain p58-GFP and correspond to the ERGIC. Images representative of at least two experiments in NRK cells. Bar, 10 μm.
Figure 4.
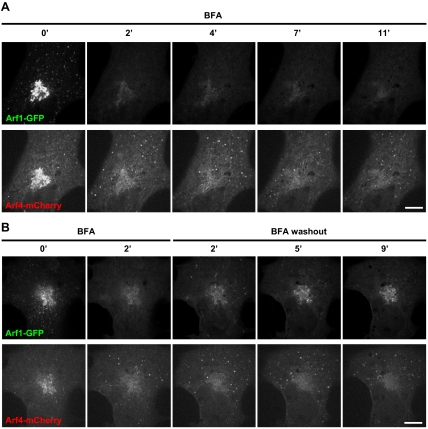
Arf4, but not Arf1, remains associated with the ERGIC after BFA treatment. (A) COS1 cells cotransfected with plasmids encoding Arf1-GFP and Arf4-mCherry were imaged continuously for 11 min after BFA addition. Panels show single channel images at the indicated time points. Representative of at least three experiments. Bar, 10 μm. (B) COS1 cells cotransfected with plasmids encoding Arf1-GFP and Arf4-mCherry were imaged continuously for 2 min after BFA addition and an additional 9 min after BFA washout as described in Materials and Methods. Panels show single channel images at the indicated times. Representative of at least three experiments. Bar, 10 μm.
Cell Fractionation, Preparation of Cell Extracts, and Immunoblots
BFA-mediated membrane recruitment of GBF1 was assayed using highly transfectable T-REx-293 cells. Cells grown on 100-mm plates was transfected with plasmids encoding Arf1-GFP or Arf4-GFP for 17–22 h as indicated. Cells were scraped into 500 μl of buffer (10 mM Tris, pH 8, 150 mM NaCl, protease inhibitor cocktail [Roche Diagnostics], pepstatin A, and O-phenanthroline), and then they were recovered by centrifugation at 1500 × g for 3 min at 4°C. Buffer was removed, and 250 μl of fresh buffer was added to each plate's worth of cells. Cells were treated in suspension with 10 μg/ml BFA or vehicle control (DMSO). After 2-min incubation at 37°C, cells were homogenized by 15 passages through a 23-gauge needle. Low-speed supernatants acquired after centrifugation at 8000 × g for 3 min at 4°C were subsequently centrifuged at 55,000 rpm for 25 min at 4°C. Resultant supernatants (cytosol) were retained and Igepal added to 1%. High-speed pellets (microsomes) were resuspended with equivalent volume of wash buffer containing 1% Igepal. Equivalent amounts of cytosolic and microsomal fractions were separated by Tris-glycine SDS-polyacrylamide gel electrophoresis [PAGE] on 5/15% step gradient gels calibrated with prestained molecular weight standards (Bio-Rad, Hercules, CA). After electrophoresis, proteins were transferred to nitrocellulose membranes, immunoblotted with primary antibodies raised against GBF1 or GFP, and detected using the ECL-Plus system (GE Healthcare, Chalfont St. Giles, United Kingdom).
Image Quantification and Analysis
Quantification of the extent of signal overlap between Arf-mCherry and either GFP-GBF1 or p58-GFP (Figure 2) was performed essentially as described previously (Zhao et al., 2002). Briefly, NIH Image, version 1.62 (http://rsb.info.nih.gov/nih-image) was used to generate separate masks for the green and red signal by using a range of threshold values that retained all discernible peripheral structures. At least five cells were analyzed. Results are expressed as percentage of total spots chosen for analysis in the green mask that were concentric with spots in the red mask.
Signal intensities for GFP-GBF1 and Arf-mCherry were quantified using MetaMorph software, version 6.1 (Molecular Devices, Sunnyvale, CA). The signal intensities exclusively at the Golgi and ERGIC membranes were analyzed by using an inclusive threshold set between 80 and 100 to include total membrane signal without saturation. A median filter of 2 pixels was chosen to eliminate background signal. The integrated intensities exported to Excel (Microsoft, Redmond, WA) were plotted as a function of time.
RESULTS
Arf1 and Class II Arfs Localize to Peripheral ERGIC
GBF1 has been detected at peripheral punctate structures and implicated in the maturation of cargo carriers necessary for assembly and maintenance of the Golgi complex (Garcia-Mata et al., 2003; Szul et al., 2005; Zhao et al., 2006; Manolea et al., 2008). GBF1 likely activates one or more Arf isoforms on these punctate structures to initiate recruitment of COPI, but no Arfs have yet been reported to localize at the ERGIC. The limited availability of selective antibodies to detect endogenous Arfs in fixed cells likely accounts for the lack of information. Antibodies that detect specifically endogenous Arf1 and Arf5 at the Golgi complex in fixed cells are available, but they did not permit further characterization of weak peripheral structures (data not shown). To address this issue, NRK cells were transfected with plasmids encoding Arf1 and Arf5. As shown in Figure 1, overexpressed Arf1 and Arf5 are readily detected at the Golgi complex as well as peripheral puncta.
To examine in more detail the potential function of both class I and class II Arfs at peripheral ERGIC structures, we tagged Arf1, -3, -4, and -5 with both green (GFP) and red (mCherry) fluorescent proteins. We chose not to examine the class III Arf6 because it has been well documented to function in the endo-lysosomal system. A GFP-tagged form of Arf1 has been characterized in detail, and it was shown to exhibit properties identical to those of the untagged protein (Presley et al., 2002). A well-characterized NRK cell line stably expressing low levels of GFP-GBF1 (NRK-GFP-GBF1; Zhao et al., 2006) was transfected with plasmids encoding either Arf1-mCherry, Arf3-mCherry, Arf4-mCherry, or Arf5- mCherry. As shown in Figure 2 (top row), all four Arfs localized to the Golgi complex. The higher magnification images shown in the second row demonstrate that all four Arfs were also readily detected at peripheral punctate structures (arrowheads). Similar results were obtained with Arfs tagged with GFP in live HeLa cells (Supplemental Figure 1), or Arfs tagged with the smaller HA epitope in several other cell types (data not shown). Further examination confirmed that Arf-positive puncta were observed in the majority of live transfected cells; all cells (n > 31) expressing Arf4 or Arf5 displayed Arf-positive puncta, whereas 91% of Arf1 transfectants (n = 34) and 76% of Arf3 transfectants (n = 29) displayed puncta.
To examine in more detail the nature of Arf-positive puncta, NRK cells were cotransfected with plasmids encoding mCherry-tagged Arfs and the ERGIC marker p58 tagged with GFP. As shown in Figure 2A, most Arf4- and Arf5-positive puncta, as well as a significant fraction of Arf1 puncta, also contained p58-GFP. Similar experiments performed in HeLa cells expressing a GFP-tagged form of Sec16L (Connerly et al., 2005) confirmed that, as expected of ERGIC structures, Arf-positive puncta often occurred next to but separate from endoplasmic reticulum exit sites (ERES) (data not shown). Comparison with the distribution of GFP-GBF1 revealed that a significant fraction of the Arf1, -4, and -5–positive structures also contained GBF1 (Figure 2B).
Quantitative analysis of images acquired from live cells (similar to those shown in Figure 2) revealed that although the majority of the Arf4- (84 ± 4%; n = 169) and Arf5 (78 ± 2%; n = 195)-positive structures contained the ERGIC marker p58-GFP, this fraction was reproducibly smaller for Arf1 (44 ± 4%; n = 255) and Arf3 (27 ± 6%; n = 123) puncta. Similar analysis of cells coexpressing Arfs-mCherry and GFP-GBF1 confirmed that most (>80%) class II Arf-positive puncta also contained GBF1. We conclude that class II Arfs-positive peripheral structures correspond to the ERGIC.
Arfs Do Not Accumulate on Membranes with GBF1 after BFA Treatment
The dissociation of GBF1 from membranes has been proposed to be tightly linked to nucleotide exchange and to only occur after the release of GDP and binding of GTP on the Arf substrate. This model is derived from the observation that BFA treatment or overexpression of exchange-deficient mutants of Arf1 or GBF1 greatly reduces the kinetics of GBF1 release from membranes and causes its accumulation on a limited subset of intracellular membranes (Niu et al., 2005; Szul et al., 2005; Zhao et al., 2006). We set out to exploit these observations to identify the preferred Arf substrate of GBF1 at the ERGIC. The model predicted that immediately after BFA addition, GBF1 would accumulate at the membrane with concomitant retention of a corresponding Arf signal. We reasoned that formation of the 1:1 Arf · GBF1 complex would be best detected using proteins modified with similar fluorescent tags to avoid differences in signal strength resulting from the use of antibodies with varying affinities.
Analysis of mCherry-tagged Arfs revealed that the majority of all four Arfs quickly redistributed from the Golgi complex upon treatment with BFA (Figure 3 and accompanying Supplemental Movies). Contrary to expectation, little class I Arf signal remained on either the Golgi complex or the ERGIC, even as GBF1 signal dramatically increased on these structures (Figure 3A). Furthermore, the response of class II Arfs on the ERGIC matched neither our expectation, nor the behavior of class I Arfs. The level of Arf4-mCherry and Arf5-mCherry at puncta remained relatively constant and did not increase to match the signal observed with GFP-GBF1 (Figure 3B and Supplemental Movies). Further testing to validate the outcome by exchange of fluorescent tags (i.e., Arf-GFP and mCherry-GBF1) and expression in other cell lines such as HeLa or COS1 cells yielded identical results (data not shown). Furthermore, even high GBF1 overexpression did not lead to Arf accumulation on membranes upon treatment with inhibitory levels (5 μg/ml) of BFA (data not shown). Interestingly, several class II Arf-positive puncta displayed limited motion and seemed stable (Figure 3A and Supplemental Movies), as reported by Hauri and colleagues (Ben-Tekaya et al., 2005; Appenzeller-Herzog and Hauri, 2006).
Quantification of membrane-associated GBF1 and Arf signal from each experiment shown in Figure 3A confirmed that whereas GBF1 accumulated on membranes and reached a maximum around 1 min, none of the Arfs increased in signal after BFA treatment (Figure 3B). Interestingly, although all Arfs released rapidly from membranes upon drug addition, a significant fraction of class II Arfs dissociated more slowly from membranes, consistent with retention of Arf4 and Arf5 signal at puncta reported in Figure 3A. Quantitative analysis of membrane-bound Arf and GBF1 fluorescence signal after 1-min treatment in several similar experiments confirmed these observations (Figure 3C). Complementary analysis of subcellular fractions further confirmed that BFA does not lead to Arf accumulation in the membrane fraction (Figure 3D). Both GBF1 and Arfs were released to the cytosol (C) fraction upon homogenization, as reported previously (Boman and Kahn, 1995; Claude et al., 1999). However, whereas GBF1 dramatically accumulated on membranes (M) when cells were treated briefly with BFA, Arfs remained largely in the cytosol fraction, with no increase in the membrane fraction. Finally, despite multiple efforts to isolate the putative Arf · GBF1 complex, analysis of GBF1 immunoprecipitates failed to detect enrichment of any of the Arfs (data not shown). These results suggest that contrary to expectation, BFA does not trap Arfs with GBF1 in an abortive complex on membranes in vivo.
The dramatically different behavior of class I and class II Arfs prompted us to examine their response to BFA in more detail. COS1 and NRK cells were cotransfected with plasmids encoding either Arf1-GFP or Arf4-mCherry to directly compare the response of Arf1 and Arf4 to BFA treatment. Both Arfs could be readily detected on peripheral puncta in both COS1 (Figure 4) and NRK cells (data not shown) before drug addition. However, as observed before, Arf1 rapidly dissociated from all membranes under conditions where Arf4-mCherry clearly remained associated on peripheral puncta (Figure 4A). Similar results were observed in cells expressing Arf1-mCherry and Arf4-GFP. Interestingly, unlike what was observed in NRK-GFP-GBF1 cells (Figure 3), Arf4 gradually accumulated on peripheral puncta in both COS1 (Figure 4A) and NRK (data not shown) cells. Recovery of Arf1 and Arf4 membrane association after BFA washout revealed further differences between these Arfs (Figure 4B and Supplemental Movies). Shortly after BFA removal, Arf1-GFP signal recovered onto peripheral structures, several of which were positive for Arf4-mCherry. However, at longer times post-BFA washout, the majority of the Arf1 signal accumulated in the juxtanuclear Golgi, whereas Arf4 remained primarily on peripheral structures. These results confirm that Arf1 and Arf4 behave differently in response to treatment with BFA.
Class II Arfs Can Associate with the ERGIC Independently of GBF1
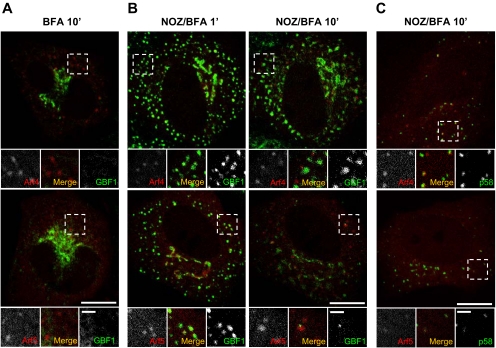
The retention of class II Arfs at peripheral puncta after BFA treatment seem consistent with the predicted formation of membrane-bound Arf · GBF1 complex. However, two additional observations established that retention of class II Arfs on membranes does not involve GBF1. First, many puncta retained Arf4-mCherry and Arf5-mCherry signal even after GFP-GBF1 had redistributed to the ER at longer times (Figure 5A). Our second test took advantage of the fact that nocodazole (NOZ), a microtubule-depolymerizing agent, prevents redistribution of GBF1 to the ER (Zhao et al., 2006). We reasoned that retention of GBF1 at punctate structures in absence of microtubules would better test whether Arfs form a membrane-bound complex with GBF1 after BFA treatment. NRK-GFP-GBF1 cells were treated with NOZ and then imaged after BFA addition (NOZ/BFA; Figure 5B). Although NOZ treatment prolonged the presence of GFP-GBF1 on the Golgi complex and peripheral ERGIC (Figure 5B), class I Arfs quickly dissociated from puncta (data not shown), as observed previously in absence of NOZ (Figure 3). In contrast, the class II Arfs remained associated with peripheral puncta (Figure 5, B and C, and Supplemental Figure 2). However, whereas Arf4/5-positive puncta colocalized with GBF1 shortly after BFA addition, most of the GBF1 redistributed to slightly larger structures that gradually separated from the puncta containing class II Arfs (Figure 5B). We suspect that class II Arfs remained on ERGIC structures, whereas membrane-bound GBF1 redistributed to a distinct membrane structure because BFA treatment causes GBF1 to relocalize to the ER when microtubules are present (Zhao et al., 2006).
To confirm that the class II Arf-positive structures observed after extended treatment with NOZ/BFA correspond to the ERGIC, we repeated this experiment with cells cotransfected with a plasmid encoding p58-GFP. As shown in Figure 5C, most of the structures onto which class II Arfs persisted after 10-min treatment with NOZ/BFA were positive for p58-GFP. Quantitative analysis established that the majority of the Arf4- (91 ± 5%) and Arf5 (89 ± 6%)-positive puncta also contained p58-GFP, confirming that they correspond to the ERGIC. These results demonstrate that class II Arfs can localize to specific membranes independently of GBF1 and suggest the presence of separate receptors for class II Arfs and GBF1.
Stimulation of Arf · GTP Hydrolysis by Exo1 Causes GBF1 to Transiently Concentrate on Golgi and ERGIC Membranes
Our inability to detect BFA-dependent accumulation of stable 1:1 stoichiometric Arf · GBF1 complexes prompted us to consider an alternate explanation for the membrane recruitment of GBF1 caused by BFA treatment and expression of the Arf1(T31N) and GBF1(E794K) mutants. We hypothesized that loss of Arf · GTP, rather than formation of an abortive complex, traps GBF1 on membranes. To test this alternate mechanism, we turned to Exo1, a cell-permeable methylanthranilate analog that rapidly releases Arf1 from Golgi membranes. Exo1 does not affect Arf-GEFs and has been suggested to modify Golgi Arf1 activity by increasing the rate of GTP hydrolysis through an Arf-GAP-dependent step (Feng et al., 2003). We reasoned that if GBF1 accumulation on Golgi and ERGIC membranes were linked to loss of Arf · GTP, Exo1 should cause membrane accumulation of GBF1 similar to that observed with BFA.
As shown in Figure 6, treatment with Exo1 led to rapid but transient recruitment of both endogenous and GFP-GBF1 at the Golgi complex and the ERGIC. Examination of NRK cells by standard IF using anti-GBF1 antibodies first established that Exo1 causes membrane accumulation of endogenous untagged GBF1, most conspicuously at peripheral puncta (Figure 6A). This effect was specific to GBF1, because as reported previously for BFA (Zhao et al., 2006), Exo1 did not cause accumulation of BIG1 on peripheral puncta; similar effects were observed in HeLa and COS1 cells (Supplemental Figure 3). As observed with BFA, GFP-GBF1 accumulated rapidly on membranes, reaching a maximum ∼1 min after Exo1 addition and decreasing thereafter (Figure 6B). Quantification of several experiments similar to that shown in Figure 6B revealed an approximately eightfold increase in membrane-associated GFP-GBF1 signal 1 min after Exo1 treatment (Figure 6C). These striking results demonstrate that GBF1 accumulation on Golgi and ERGIC membranes can result from loss of Arf · GTP and does not require formation of a stable, abortive complex. More importantly, they suggest a potentially novel regulatory mechanism for GBF1 recruitment.
Class II Arfs and GBF1 Accumulate at the ERGIC after Extended Treatment with Exo1
The dramatic effects of Exo1 on GBF1 recruitment led us to investigate its impact on the class I and class II Arfs. As reported previously for Arf1 (Feng et al., 2003), Exo1 caused rapid release of all tested Arfs from the Golgi complex, suggesting that Exo1 promotes GTP hydrolysis on all Golgi-localized Arfs (Figure 7A and accompanying Supplemental Movies, and Supplemental Figure 4). Interestingly, as observed with BFA (Figure 2), class II Arfs but not class I Arfs remained on puncta shortly after Exo1 addition. Consistent with results presented in Figure 6, GBF1 accumulated on structures positive for class II Arfs. However, unlike what was observed with BFA (Figure 3), class II Arfs did not persist but rather redistributed with GBF1 2–3 min after Exo1 addition (Figure 7A, and Supplemental Figure 4 and accompanying Supplemental Movies). The most striking difference between the two drugs was observed at later times when class II Arfs and GBF1-positive punctate structures reappeared (Figure 7A, arrowheads). Puncta containing both Arf4 and GBF1 signal started to occur at ∼7 min and grew progressively brighter for up to 20 min. Similar results were obtained in fixed NRK cells expressing Arf4-mCherry with an antibody to detect endogenous GBF1 (data not shown). These observations confirm that class I and class II Arfs behave differently, and they reveal that BFA and Exo1 impact ERGIC dynamics differently.
To determine whether the reappearing punctate structures correspond to the ERGIC or fragmented Golgi mini-stacks, we repeated our analysis with a wider range of markers in fixed cells. We chose to examine marker distribution 20 min after Exo1 addition, because this treatment yielded maximum signal in live cells. Images shown in Figure 7B, e and f, demonstrates that Arf4 accumulated in puncta positive for an ERGIC marker. Further labeling of Arf4-GFP transfectants for COPI or the Golgi marker Man II-confirmed complete dispersal of both markers after treatment with Exo1 (Figure 7B, g and h). These observations demonstrate that Arf4-positive puncta (insets) do not correspond to Golgi fragments or mini-stacks that reformed near peripheral ERES and the ERGIC. The absence of COPI from Exo1-induced puncta further demonstrated effective loss of activated Arfs. The association of class II Arfs with the ERGIC in presence of either BFA or Exo1 suggests the presence of an organelle specific receptor with affinity for the inactive GDP-bound form of these Arfs.
Class II Arfs Concentrate on Peripheral Puncta in Their Inactive, GDP-bound Form
To confirm that the inactive GDP-bound form of Arf4 can associate specifically with ERGIC membranes, we examined the localization of an inactive variant of Arf4-GFP containing the T31N mutation, shown previously in Arf1 to prevent stable GTP binding (Dascher and Balch, 1994) and reported to inactivate Arf4 (Kim et al., 2003). Overexpression of Arf1(T31N) interferes with the recruitment of COPI to Golgi membranes and causes disassembly of the Golgi complex (Zhang et al., 1994) (Dascher and Balch, 1994; Garcia-Mata et al., 2003). Live NRK cells expressing similar levels of both wild type Arf4-mCherry and Arf4(T31N)-GFP were imaged in absence and presence of Exo1. Interestingly, and for reasons we cannot explain, expression of high levels of the Arf4 dominant-negative mutant did not prevent assembly of the Golgi complex when low levels of Arf1(T31N) readily dispersed Golgi markers (Supplemental Figure 5; Dascher and Balch, 1994). Instead, Arf4(T31N)-GFP localized primarily to peripheral puncta and only weakly to the Golgi complex that remained decorated with wild-type Arf4 (Figure 8A). Treatment with Exo1 caused loss of wild-type Arf4 from the Golgi region and further accumulation of Arf4(T31N) on peripheral puncta positive for wild-type Arf4.
To confirm that the Arf4(T31N)-GFP puncta correspond to the ERGIC, we repeated this analysis in fixed cells labeled with an antibody to the ERGIC. As shown in Figure 8B, Arf4(T31N)-GFP associated weakly with puncta in mock-treated cells, and these structures were labeled with the ERGIC antibody. Treatment with either BFA or Exo1 caused further accumulation of Arf4(T31N)-GFP at the ERGIC (Figure 8B). These results clearly demonstrate that Arf4-GFP is bound to the ERGIC membranes in the GDP-bound form and indicates the presence of a binding site for Arf4 that localizes to the ERGIC.
DISCUSSION
GBF1, the only known Arf-GEF localized to the ERGIC and cis-Golgi complex (Claude et al., 1999; Kawamoto et al., 2002; Zhao et al., 2002; Garcia-Mata et al., 2003) plays a critical role in cargo transport from the ER to the Golgi complex (Zhao et al., 2006; Szul et al., 2007; Manolea et al., 2008). However, until now, the preferred Arf substrates for GBF1 at the ERGIC remained unknown. In this study, we discovered that both class I and class II Arfs localize to peripheral structures. Surprisingly, whereas only ∼50% of Arf1-positive puncta colocalize with the ERGIC marker p58, the majority of Arf4- and Arf5-positive punctate structures correspond to the ERGIC. Attempts to use BFA to trap GBF1 in abortive complexes with its preferred Arf substrates failed to detect corecruitment of any Arf with GBF1. However, these experiments revealed that class II Arfs remain on peripheral ERGIC membranes, even after BFA treatment and subsequent redistribution of GBF1 to other compartments. Further experiments confirmed that Arf4 associates selectively with ERGIC membranes in its GDP-bound form. Our observations demonstrate the presence of receptors specific for class II Arfs on the ERGIC and suggest potential regulatory function for these Arfs in ER-to-Golgi traffic.
Class II Arfs Localize to the Golgi Complex and the ERGIC
We present the first direct evidence for the localization of both class I and class II Arfs at the ERGIC. Despite clear expectation that Arfs participate in COPI recruitment at the ERGIC, no studies had yet reported the presence of either Arf class on these structures. Imaging of fixed cells overexpressing untagged or HA-tagged forms of Arf1 and Arf5 readily detected Arfs at the Golgi complex, but only a fraction of the transfectants displayed clear Arf-positive peripheral puncta. Two serendipitous observations enabled the study reported here. First, we observed more GBF1 and Arfs localization to the ERGIC in cells kept at temperatures below 37°C. In addition, although only a fraction of transfectants showed Arf-positive puncta after fixation, Arfs were observed at puncta in nearly all transfected live cells. Almost all live cells expressing class I and II Arfs tagged with fluorescent proteins displayed Arf-positive peripheral puncta over a wide range of Arf expression. Nearly all of those containing Arf4 and Arf5 corresponded to the ERGIC because they were also positive for the ERGIC marker p58-GFP. Interestingly, a consistently smaller fraction of class I Arf-positive puncta contained p58-GFP. These observations suggest a potentially unique function for class II Arfs at the ERGIC.
GBF1 Concentrates on Golgi and ERGIC Membranes without Forming an Abortive Complex with Arfs
To examine whether class II Arfs were activated preferentially by GBF1 at peripheral ERGIC, we subjected live cells to specific treatments intended to promote the formation of a complex between GBF1 and its Arf substrates. As described in more detail in Results, previous work predicted that treatment with BFA would trap one or more Arfs on membranes commensurate with GBF1 recruitment and formation of a putative abortive Arf · GDP · BFA · GBF1 complex (Cherfils and Melançon, 2005; Niu et al., 2005; Szul et al., 2005; Zhao et al., 2006). Contrary to expectation, our data strongly suggest that BFA treatment does not promote association of class I or class II Arfs with GBF1. Analysis of live imaging experiments in Figure 3 revealed no concomitant increase of Arf-mCherry signal as GFP-GBF1 accumulated on Golgi and ERGIC membranes shortly after BFA treatment. Class II Arfs remained on punctate structures, but several experiments established that class II Arfs associated with membranes independently of GBF1.
Our results remain consistent with the well-characterized mechanism of action of BFA. The demonstration that BFA is an uncompetitive inhibitor that can form ternary complexes with Arf and isolated Sec7 domains is incontrovertible (Mansour et al., 1999; Peyroche et al., 1999; Robineau et al., 2000; Mossessova et al., 2003; Renault et al., 2003). However, our results suggest that such complexes must be transient and do not accumulate in vivo. Rapid inhibition of GBF1 activity may lead instead to modification of either GBF1 or its receptor that traps GBF1 on its receptor and decreases GBF1 activity toward Arfs (Figure 9). Formation of a transient abortive Arf · GDP · BFA · GBF1 complex is likely what allowed Jackson and colleagues to detect BFA-dependent interaction between GBF1 and Arf1 in their bimolecular fluorescence complementation assay (Niu et al., 2005). It may be important to note that reconstitution of the split yellow fluorescent protein is largely irreversible (Kerppola, 2006) and likely traps a transient intermediate rather than reveals a stable Arf · GEF complex. Finally, we cannot exclude the possibility that GBF1 forms undetectable but stable complexes with endogenous Arfs that could exclude tagged-Arfs. This possibility could explain association of Arf1 and Arf4 with GBF1 reported by Szul et al. (2007), but this could not be unambiguously tested because methods to efficiently detect complexes containing endogenous Arfs have not been established. Instead, we demonstrated that accumulation of GBF1 on membranes in response to BFA treatment can result from loss of Arf · GTP from membranes and need not depend on formation of stable abortive complex with BFA (Figure 9). Our “Arf · GTP loss” model was tested directly using Exo1, a drug that causes rapid release of Arf1 from membranes by promoting GTP hydrolysis on Arf and clearly does not block the guanine nucleotide exchange by Arf-GEFs (Feng et al., 2003). As predicted by our model, Exo1 promoted release of Arfs and caused dramatic recruitment of both endogenous and GFP-GBF1 on Golgi and ERGIC membranes (Figures 6 and 7). These results established that GBF1 can accumulate on ERGIC and Golgi membranes independently of a physical interaction with an Arf. This Arf · GTP loss model is consistent with previous studies with GBF1(E794K) and Arf1(T31N) that cause similar loss of Arf · GTP and lead to accumulation of GBF1 at the membrane (Garcia-Mata et al., 2003; Szul et al., 2005).
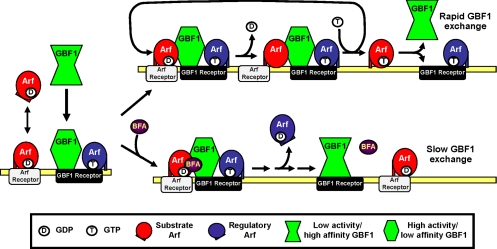
Figure 9.
Arf · GTP loss model for the effect of BFA on GBF1 association with membranes. GBF1 and Arf · GDP are recruited to specific membranes through interaction with distinct receptors. BFA absent (top), in the presence of a regulatory Arf · GTP (blue), membrane-bound GBF1 acquires an active conformation that can interact with membrane-bound substrate Arf · GDP (red) and promote release of GDP. GTP loading causes the release of a new Arf · GTP (red) and frees up active GBF1 for another cycle of nucleotide exchange. GBF1 dissociates rapidly from its receptor under these conditions but can activate multiple Arf substrates per binding cycle. BFA present (bottom), BFA forms an abortive complex Arf · GDP · BFA · GBF1 complex with membrane-bound GBF1 and blocks nucleotide exchange. Arf-GAP activity leads to rapid loss of regulatory Arf · GTP from the membrane, which induces a conformational change in GBF1. In absence of Arf · GTP, membrane-bound GBF1 interacts poorly with Arf · GDP and dissociates slowly from its receptor. Some Arf · GDP remain associated with their receptors.
As proposed in our Arf · GTP loss model, reduction in Arf · GTP levels caused by either BFA or Exo1 leads to a conformational change in GBF1 or its receptor that ultimately slows down dissociation of GBF1 from membranes. Arf · GTP could regulate this change either directly as illustrated in Figure 9 or indirectly through another effector or lipid remodeling. The absence of Arf · GDP · BFA · GBF1 complexes suggests that membrane-trapped GBF1 displays low activity and no longer interacts productively with Arfs. Alternatively, we cannot rule out that membrane-bound GBF1 remains fully active but forms only weak Arf · GDP · BFA · GBF1 complexes. The expectation of a tight abortive complex arose from previous studies performed primarily with the isolated Sec7 domain of ARNO mutants rendered sensitive to BFA (Peyroche et al., 1999; Mossessova et al., 2003; Renault et al., 2003). However, these studies do not allow any prediction as to the stability of complexes formed with full-length GBF1. For example, analysis with the isolated Sec7 domain of Gea2 revealed a dissociation rate of BFA from the Arf · Gea2 complex that was 20-fold faster than observed for the ARNO complex (Robineau et al., 2000). Such variations could explain why full-length GBF1 may form weak Arf · GDP · BFA · GEF complexes that cannot accumulate under in vivo conditions.
BFA Traps GBF1 and Class II Arfs on Separate Receptors
Another unexpected result from our study was the clear separation of GFP-GBF1 from Arf4-mCherry and Arf5-mCherry after treatment with BFA. This clear spatial separation between class II Arfs and GBF1 was best illustrated in cells lacking microtubules; in response to BFA, class II Arfs again remained localized to the ERGIC, but over time GBF1 redistributed to more diffuse, uncharacterized globular structures. The appearance of Arf4- and 5-positive puncta containing p58-GFP but devoid of GFP-GBF1 suggests the presence of specific binding sites for class II Arfs at the ERGIC. We hypothesized that these binding sites act as receptors for class II Arfs in their GDP-bound form, because they are retained on ERGIC membranes after treatment with either BFA or Exo1. This hypothesis was tested by examining the distribution of Arf4(T31N), a mutant expected to accumulate in the GDP form. As predicted, Arf4(T31N) localized to punctate structures that overlapped with p58, even when cells were treated with BFA or Exo1 to eliminate Arf · GTP.
Several lines of evidence support the idea that separate receptors exist for GBF1 and Arf · GDP. Although a membrane receptor for GBF1 has yet to be identified, hGmh1 (Chantalat et al., 2003), p115 (Garcia-Mata and Sztul, 2003), and Rab1b (Monetta et al., 2007) have been identified as interacting partners that may contribute to membrane recruitment of GBF1. In contrast, several candidate receptors for Arf1 · GDP have been reported. Arf1 · GDP can associate with membranes by interacting with p23, a member of the p24 family of transmembrane proteins (Gommel et al., 2001). Donaldson and colleagues provided similar evidence that the SNARE membrin functions to recruit Arf1 · GDP to early Golgi compartments (Honda et al., 2005). Although the nature of the putative class II Arf receptors remains unknown, we expect their properties to be distinct from those reported for the Arf1 · GDP receptors. Indeed, in contrast to the Golgi-localized p23 and membrin, the putative Arf4 · GDP receptor should localize primarily at the ERGIC. Furthermore, the affinity, abundance, or both of class II Arf receptors must be greater at the ERGIC because much of Arf4 remains at the ERGIC after BFA or Exo1 treatment, whereas significantly less Arf1 signal is retained on the Golgi complex.
Previous reports suggested that the Arf1 · GDP receptors membrin and p23 function to concentrate Arf1 on membranes to facilitate its activation by membrane-associated GEFs (Rein et al., 2002; Honda et al., 2005). Distinct Arf · GDP receptors for different Arfs may similarly facilitate activation of distinct subsets of Arfs on specific compartments. The presence of separate receptors for the different classes of Arfs agrees with the observed corequirement for both Arf1 and Arf4 in ER to Golgi traffic (Volpicelli-Daley et al., 2005). For example, one possible role for class II Arf · GDP receptors at the ERGIC might be to concentrate Arf4 at the ERGIC. Subsequently, Arf4 · GTP would regulate GBF1 recruitment/activation to facilitate sequential activation of Arf1. Unlike what was reported for ARNO recruitment by Arf6 · GTP for subsequent Arf1 activation (Cohen et al., 2007), Arf4 cannot function as a receptor for GBF1 because they can be spatially separated. Furthermore, whatever role Arf4 plays at the ERGIC seems not to be essential because Arf4(T31N) expression does not prevent Golgi assembly. Proper testing of the interdependent activities of class I and II Arfs and the specific role of Arf4 in ER-to-Golgi traffic will require future identification of receptors for both GBF1 and Arfs.
Supplementary Material
ACKNOWLEDGMENTS
We thank X. Zhao for preliminary data on the effect of BFA on Arf4, M. Schneider for construction and initial characterization of Arf mutants, and Dr. A. Simmonds, Dr. X. Sun, and H. Chan (University of Alberta) for technical help with confocal microscopy. We thank F. Manolea for helpful discussions, and Dr. B. Antonny for helpful discussion and critical reading of the manuscript. This study was supported by a grant to P. M. from the Canadian Institutes of Health Research. J. C. is the recipient of a Province of Alberta Graduate Fellowship.
Abbreviations used:
- Arf
ADP-ribosylation factor
- BFA
brefeldin A
- BIGs
BFA inhibited GEFs
- COP
coatomer protein
- ERES
endoplasmic reticulum exit sites
- ERGIC
endoplasmic reticulum-Golgi intermediate compartment
- GBF1
Golgi-specific brefeldin A resistance factor 1
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- IF
immunofluorescence
- Man II
mannosidase II
- mCherry
monomeric Cherry
- NOZ
nocodazole
- Sec7d
Sec7 domain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0373) on June 4, 2008.
REFERENCES
- Allan V. J., Kreis T. E. A microtubule-binding protein associated with membranes of the Golgi apparatus. J. Cell Biol. 1986;2229:2239. doi: 10.1083/jcb.103.6.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B., Beraud-Dufour S., Chardin P., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Hauri H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Ben-Tekaya H., Miura K., Pepperkok R., Hauri H. P. Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 2005;118:357–367. doi: 10.1242/jcs.01615. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J., Hauber J., Hauber R., Geiger R., Cullen B. R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- Boman A. L., Kahn R. A. Arf proteins: the membrane traffic police? Trends Biochem. Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- Burke B., Griffiths G., Reggio H., Louvard D., Warren G. A monoclonal antibody against a 135-K Golgi membrane protein. EMBO J. 1982;1:1621–1628. doi: 10.1002/j.1460-2075.1982.tb01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh M. M., Whitney J. A., Carroll K., Zhang C., Boman A. L., Rosenwald A. G., Mellman I., Kahn R. A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Chantalat S., Courbeyrette R., Senic-Matuglia F., Jackson C. L., Goud B., Peyroche A. A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol. Biol. Cell. 2003;14:2357–2371. doi: 10.1091/mbc.E02-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Melançon P. On the action of Brefeldin A on Sec7-stimulated membrane-recruitment and GDP/GTP exchange of Arf proteins. Biochem. Soc. Trans. 2005;33:635–638. doi: 10.1042/BST0330635. [DOI] [PubMed] [Google Scholar]
- Claude A., Zhao B. P., Kuziemsky C. E., Dahan S., Berger S. J., Yan J. P., Armold A. D., Sullivan E. M., Melançon P. GBF 1, A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol. Biol. Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerly P. L., Esaki M., Montegna E. A., Strongin D. E., Levi S., Soderholm J., Glick B. S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Dascher C., Balch W. E. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- Deretic D., Williams A. H., Ransom N., Morel V., Hargrave P. A., Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc. Natl. Acad. Sci. USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Jackson C. L. Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Feng Y., Yu S., Lasell T. K., Jadhav A. P., Macia E., Chardin P., Melançon P., Roth M., Mitchison T., Kirchhausen T. Exo 1, a new chemical inhibitor of the exocytic pathway. Proc. Natl. Acad. Sci. USA. 2003;100:6469–6474. doi: 10.1073/pnas.0631766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 2003;4:320–325. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Szul T., Alvarez C., Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell. 2003;14:2250–2261. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Gommel D. U., Memon A. R., Heiss A., Lottspeich F., Pfannstiel J., Lechner J., Reinhard C., Helms J. B., Nickel W., Wieland F. T. Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J. 2001;20:6751–6760. doi: 10.1093/emboj/20.23.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Honda A., Al-Awar O. S., Hay J. C., Donaldson J. G. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J. Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Randazzo P. A. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto K., Yoshida Y., Tamaki H., Torii S., Shinotsuka C., Yamashina S., Nakayama K. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic. 2002;3:483–495. doi: 10.1034/j.1600-0854.2002.30705.x. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Hayashi M., Lo J. F., Yang Y., Yoo J. S., Lee J. D. ADP-ribosylation factor 4 small GTPase mediates epidermal growth factor receptor-dependent phospholipase D2 activation. J. Biol. Chem. 2003;278:2661–2668. doi: 10.1074/jbc.M205819200. [DOI] [PubMed] [Google Scholar]
- Majoul I., Straub M., Hell S. W., Duden R., Soling H.-D. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev. Cell. 2001;1:139–145. doi: 10.1016/s1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Manolea F., Claude A., Chun J., Rosas J., Melançon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol. Biol. Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. J., Skaug J., Zhao X. H., Giordano J., Scherer S. W., Melançon P. p200 ARF-GEP 1, a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc. Natl. Acad. Sci. USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monetta P., Slavin I., Romero N., Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol. Biol. Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E., Corpina R. A., Goldberg J. Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell. 2003;12:1403–1411. doi: 10.1016/s1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Gulbis J. M., Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human arno and analysis of the interaction with ARF GTPase. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. Overexpression of wild-type and mutant ARF1 and ARF 6, distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C. L. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Presley J. F., Ward T. H., Pfeifer A. C., Siggia E. D., Phair R. D., Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Rein U., Andag U., Duden R., Schmitt H. D., Spang A. ARF-GAP-mediated interaction between the ER-Golgi v-SNAREs and the COPI coat. J. Cell Biol. 2002;157:395–404. doi: 10.1083/jcb.200112092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L., Guibert B., Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- Robineau S., Chabre M., Antonny B. Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA. 2000;97:9913–9918. doi: 10.1073/pnas.170290597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J., Palade G. E., Farquhar M. G. Antibodies to rat pancreas Golgi subfractions: identification of a 58-kD cis-Golgi protein. J. Cell Biol. 1987;105:2021–2029. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szul T., Garcia-Mata R., Brandon E., Shestopal S., Alvarez C., Sztul E. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic. 2005;6:374–385. doi: 10.1111/j.1600-0854.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- Szul T., Grabski R., Lyons S., Morohashi Y., Shestopal S., Lowe M., Sztul E. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J. Cell Sci. 2007;120:3929–3940. doi: 10.1242/jcs.010769. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. Isoform-selective effects of the depletion of Arfs1–5 on membrane traffic. Mol. Biol. Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E., et al. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zhang C., Rosenwald A. G., Willingham M. C., Skuntz S., Clark J., Kahn R. A. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Claude A., Chun J., Shields D. J., Presley J. F., Melançon P. GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J. Cell Sci. 2006;119:3743–3753. doi: 10.1242/jcs.03173. [DOI] [PubMed] [Google Scholar]
- Zhao X., Lasell T. K., Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.