Abstract
Here we report a novel role for myeloid cell leukemia 1 (Mcl-1), a Bcl-2 family member, in regulating phosphorylation and activation of DNA damage checkpoint kinase, Chk1. Increased expression of nuclear Mcl-1 and/or a previously reported short nuclear form of Mcl-1, snMcl-1, was observed in response to treatment with low concentrations of etoposide or low doses of UV irradiation. We showed that after etoposide treatment, Mcl-1 could coimmunoprecipitate with the regulatory kinase, Chk1. Chk1 is a known regulator of DNA damage response, and its phosphorylation is associated with activation of the kinase. Transient transfection with Mcl-1 resulted in an increase in the expression of phospho-Ser345 Chk1, in the absence of any evidence of DNA damage, and accumulation of cells in G2. Importantly, knockdown of Mcl-1 expression abolished Chk1 phosphorylation in response to DNA damage. Mcl-1 could induce Chk1 phosphorylation in ATM-negative (ataxia telangectasia mutated) cells, but this response was lost in ATR (AT mutated and Rad3 related)-defective cells. Low levels of UV treatment also caused transient increases in Mcl-1 levels and an ATR-dependent phosphorylation of Chk1. Together, our results strongly support an essential regulatory role for Mcl-1, perhaps acting as an adaptor protein, in controlling the ATR-mediated regulation of Chk1 phosphorylation.
INTRODUCTION
Myeloid cell leukemia 1 (Mcl-1) was first identified as a gene induced during myeloid cell differentiation (Kozopas et al., 1993) and is a prosurvival member of the Bcl-2 family. The activity of Bcl-2 family proteins involves several Bcl-2 homology (BH) domains (Danial and Korsmeyer, 2004) that control protein–protein interactions. These proteins play an important role in cancer by regulating cell death and survival. Compared with other prosurvival members such as Bcl-2 or Bcl-xL, Mcl-1 has been shown to have more transient survival effects (Zhou et al., 1997). Unlike other members of the family, the half-life of Mcl-1 is very short and its expression is highly regulated by growth factors and a variety of other agents. Moreover, Mcl-1 has an extended N-terminus that is not conserved in any other Bcl-2 family protein. The N-terminus contains two PEST domains, which are found in proteins that are rapidly turned over, but deletion of the PEST domains does not extend the short half-life of Mcl-1 (Akgul et al., 2000; Clohessy et al., 2004). When the Mcl-1 gene was knocked out in a mouse model, it resulted in peri-implantation lethality (Rinkenberger et al., 2000), but Mcl-1−/− embryos showed no alterations in the extent of apoptosis, indicating that Mcl-1 plays a role early in development distinct from its prosurvival functions. We and others reported that Mcl-1 expression can lead to suppression of cell proliferation and may have effects on cell cycle machinery (Fujise et al., 2000; Jamil et al., 2005). We showed that a short nuclear form of Mcl-1 (snMcl-1) was associated primarily with the inactive form of Cdk-1 in the nucleus, most prominently during the G2 phase of the cell cycle, which suggested a potential means by which Mcl-1 mediates its anti-proliferative effects (Jamil et al., 2005).
It is now well established that genome surveillance mechanisms function to cause a delay, or arrest, in progression of the cell cycle at the boundary of G1/S or G2/M in response to DNA damage (Zhou and Elledge, 2000; Abraham, 2001; Kastan, 2001; Nyberg et al., 2002; Sancar et al., 2004). These surveillance mechanisms are known to involve a network of interacting proteins that recognize damage and elicit responses including cell cycle delay, DNA repair, and apoptosis (Elledge, 1996; Zhou and Elledge, 2000). Cdk's, which play a central role in cell cycle progression, are key targets of these checkpoints (Iliakis et al., 2003). Yeast model systems have revealed much about the mechanisms of the DNA damage response, as well as the identity of the corresponding genes in higher eukaryotes. A role for a gene such as Mcl-1 would not have been found from studies of more primitive organisms, because Mcl-1 does not appear to have any orthologues other than in vertebrates (e.g., search in OrthoMCL: http://orthomcl.cbil.upenn.edu/cgi-bin/OrthoMclWeb.cgi?rm=index).
Because we had previously shown the interaction of Mcl-1 with Cdk-1 and we and others showed that Mcl-1 expression causes a suppression of cell proliferation, we investigated the potential change in Mcl-1 expression and whether Mcl-1 may possibly play a role when cells are treated with agents that cause DNA damage. We found that Mcl-1 expression increases, particularly in the nucleus, as a result of mild DNA damage. More extensive damage had the expected effect of decreasing expression of Mcl-1, as has been reported previously (Nijhawan et al., 2003). After mild DNA damage, the nuclear Mcl-1 was shown to be associated with active Checkpoint kinase 1 (Chk1). Increased phosphorylation of Chk1 at an activating site was observed after transient expression of Mcl-1, whereas no other evidence of a DNA damage response was evident. Furthermore, knockdown of Mcl-1 expression eliminated the Chk1 phosphorylation that occurs after DNA damage. By comparison of ATM (ataxia telangectasia mutated)-negative and ATR (AT mutated and Rad3 related)-deficient cells, we could conclude that Mcl-1 plays a role in ATR-dependent activation of Chk1, revealing a completely novel function for Mcl-1.
MATERIALS AND METHODS
Cell Lines
HeLa, HL-60, and FDC-P1 cells were obtained from ATCC (Manassas, VA). ATM-deficient HT-144 cells were a kind gift from Dr. P. Olive (BC Cancer Agency). ATR-defective fibroblasts, F02–98, were a kind gift from Dr. P. Jeggo. Hs68, which are primary human foreskin fibroblasts (Wheaton and Riabowol, 2004), maintained within 30–60 mean population doublings, were kindly provided by Dr. K. Riabowol (University of Calgary). HL-60 cells were cultured in RPMI1640 supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM l-glutamine. FDC-P1 cells were grown in RPMI supplemented with 10% fetal calf serum (FCS) and 2.5% WEHI-3-conditioned medium as a source of IL-3. HeLa, Hs68, and F02–98 cells were grown in DMEM and HT 144 cells were cultured in MEM, each with 10% FCS.
Antibodies and Reagents
Anti-Mcl-1 (Sc-19), Chk1 (FL-476), p85α (Z-8), and Oct I (C-21) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-Chk1 (Ser345) was from Cell Signaling (Beverly, MA). Monoclonal anti-human vinculin was from Sigma (St. Louis, MO). Etoposide was purchased from Calbiochem (La Jolla, CA). Isogranulatimide was a kind gift from Dr. M. Roberge (University of British Columbia). [γ-32P]ATP was purchased from ICN Biomedicals (Costa Mesa, CA). RPMI 1640, fetal bovine serum (FBS), Protein G agarose beads were purchased from Invitrogen (Carlsbad, CA).
Cell Treatments
Optimal concentrations of etoposide were determined for each cell line that caused G2 arrest with the least amount of apoptosis. For HL-60 and FDC-P1 cells, 1.5 μM etoposide was used and for HeLa cells, 15 μM was required. For UV treatments, cells were irradiated using a UVB source (5 J m−2 s−1) for 20 s for a total of 100 J/m2, after which cells were washed twice with phosphate-buffered serum (PBS) and then allowed to incubate under normal conditions.
Subcellular Fractionation
Subcellular fractionation was carried out as described (Shiio et al., 2003). Briefly, cells were resuspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, and 10% glycerol), containing protease inhibitors and 1 mM dithiothreitol (DTT). To each extract, 0.1% Triton X-100 was added, and samples were incubated on ice for 7 min. The samples were centrifuged for 5 min at 1300 × g. Supernatant containing cytosolic proteins was centrifuged further for 10 min at 20,000 × g. The pellets containing the nuclei were washed with buffer A, resuspended in buffer B (0.2 mM EGTA, pH 8.0, and 3 mM EDTA, pH 8.0), and incubated on ice for 30 min. The extracts were centrifuged at 1700 × g for 5 min. The pellets were washed and centrifuged for 5 min at 1700 × g. The pellets were resuspended in sample buffer and sonicated for 15 s. In experiments where nuclear proteins were extracted together with chromatin, 5 μg/ml each of DNase I and RNase A were added to the buffer B and the nuclear preparations were sonicated for 10 s.
Indirect Immunofluorescence Staining
Immunofluorescence staining was performed using the protocol as described previously (Liu et al., 1999). Briefly, HeLa cells were seeded on glass coverslips, treated or not with either 15 μM etoposide, and fixed with fresh 4% paraformaldehyde in PBS. Cells were blocked with 10% normal goat serum and probed with 1:250 rabbit anti-Mcl-1 antibody or 1:150 mouse anti-Chk1. Bound antibody was detected by goat anti-rabbit antibody conjugated to Alexa fluor 488 or goat anti-mouse antibody conjugated to Alexa fluor 594. The cell nuclei were stained with 1:10,000 dilution of Hoechst 33342. Stained cells were analyzed using a Zeiss Axioplan 2 imaging microscope (Thornwood, NY).
Immunoprecipitation
Cells were washed with PBS before total cell lysates were obtained by lysing cells in ice-cold solubilization buffer (20 mM Tris HCl, pH 8.0, 1% NP40, 10% glycerol, 137 mM NaCl, and 10 mM NaF) with protease inhibitor cocktail, 200 μM sodium vanadate, and 5 μg/ml each of DNase I and RNase A. Cells were sonicated for 15 s and centrifuged at 32,000 × g for 10 min. Protein concentrations were determined by BCA protein assay. For immunoprecipitation, cytosolic, nuclear or chromatin extracts were precleared with 20 μl of protein G agarose beads for 30 min. Anti-Mcl-1 antibody at 1 μg/ml was added, and after a 2-h incubation, the immunoprecipitates were collected by adding 50 μl of protein G agarose beads. Beads were washed four times with solubilization buffer.
Kinase Assays
For determination of Mcl-1–associated Chk1 activity, total cell lysates were precleared by incubation with agarose G beads for 30 min. The samples were then incubated with anti-Mcl-1 antibody for 2 h followed by addition of agarose G beads for 1 h. After extensive washing, beads were resuspended in assay dilution buffer (25 mM β-glycerophosphate, 20 mM MOPS, 5 mM EGTA, 2 mM EDTA, 20 mM MgCl2, 250 μM DTT, 5 μM β-methyl aspartic acid, pH 7.2). CHKtide at 5 μg/ml (Furnari et al., 1997), [γ-32P]ATP, and cold ATP (25 μM) were added. The reaction was terminated after 20 min by spotting 15 μl on p81 chromatography filter paper (Whatman, Clifton, NJ). The filter papers were washed in 1% O-phosphoric acid, and the activity of each sample was measured in a scintillation counter.
Flow Cytometry
For staining of cells to detect sub-G1 DNA levels and analysis of cell cycle status, cells were fixed with 70% ethanol and subsequently stained with PBS containing 50 μg/ml propidium iodide, 100 μg/ml RNase A, and 0.1% glucose. Stained cells were analyzed using Epics XL flow cytometer (Coulter, Hialeah, FL).
Mcl-1 Small Interfering RNA Interference
For in vitro gene silencing cells were transfected with either Mcl-1 siRNA sequence described by Zhang et al. (2002) or control siRNA (sense UUCUCCGAACGUGUCACGUdTdT, antisense ACGUGACACGUUCGGAGAAdTdT). The purified desalted and double-stranded Mcl-1 siRNA was ordered from Dharmacon Research (Boulder, CO). Control siRNA was purchased from Qiagen (Chatsworth, CA). HeLa cells were plated the day before being transfected with either 20 nM Mcl-1 or control siRNA using SILENTfect (Bio-Rad, Richmond, CA) according to the manufacturer's recommendations. After 24 h the medium was replaced with fresh siRNA for another 24 h.
RESULTS
Mcl-1 Translocates to the Nucleus in Response to Etoposide
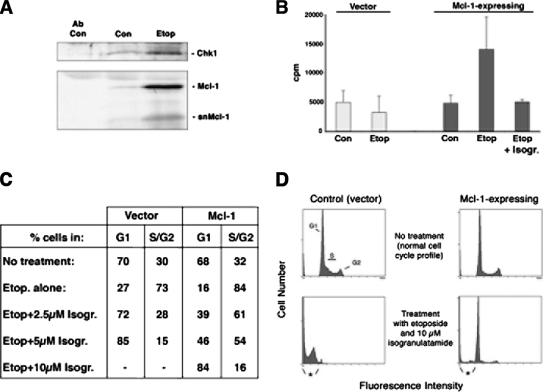
To induce DNA damage, cells were treated with etoposide, which is a topoisomerase II inhibitor that results in double-stranded breaks and single-stranded gaps to trigger cell cycle checkpoint activation (Cliby et al., 2002). To determine whether etoposide treatment affects the cellular location of Mcl-1, we performed subcellular fractionation followed by immunoblotting with anti-Mcl-1 antibody. An increase in Mcl-1 protein levels was observed after 3 h of etoposide treatment in HL-60 cells; a similar increase in Mcl-1 expression was observed in HeLa cells (Figure 1). As we showed previously in HL-60, some Mcl-1 is present in the nucleus even in healthy asynchronous cells (Jamil et al., 2005). However, an increase in nuclear accumulation of Mcl-1 was observed in both HL-60 and HeLa cells, upon etoposide treatment. In both cell types, there is an increase in cytosolic Mcl-1 (where the majority of Mcl-1 is normally present), but there is a much greater increase in the amount of total Mcl-1 in the nucleus. In the nuclear extracts from HL-60 cells, the predominant form was the previously reported short form, snMcl-1 (Jamil et al., 2005), whereas in HeLa cells the predominant form was full-length Mcl-1. The significance of the variable amounts of snMcl-1 in different cell types is not clear at present. It is noteworthy that the concentrations of etoposide used were optimized for each cell line to cause G2 arrest, with the least amount of apoptosis during the course of the experiment. The three cell lines used in this study were shown to arrest at G2 with either 1.5 μM (HL-60 and FDC-P1) or 15 μM (HeLa) etoposide treatment overnight.
Figure 1.

Mcl-1 expression is increased, particularly in the nucleus, in response to etoposide treatment. (A) HL-60 cells were either left untreated (C) or treated with etoposide (Et) for 3 h. Cells were fractionated into cytosolic and nuclear extracts. The blot was probed with anti-Mcl-1 antibody, anti-vinculin antibody to indicate cytosol purity, and anti-Oct1 antibody to indicate purity of nucleus fraction. (B) HeLa cells were untreated (C) or treated with 15 μM of etoposide (Et) for 3 h and prepared as in A. snMcl-1 refers to the protein corresponding to the shorter nuclear form of Mcl-1 that we previously characterized. The samples from cytosol and nuclear extracts represent an equivalent number of cells, resolved on the same gel and blotted together and are thus a direct representation of the relative amounts of Mcl-1. Results are representative of at least five independent experiments.
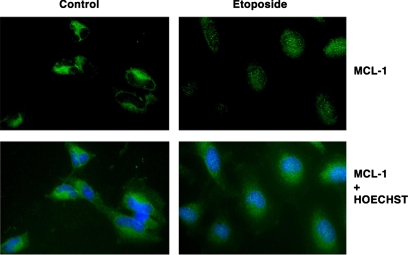
We wanted to demonstrate, by an independent set of analyses, the change in intracellular localization of Mcl-1 after DNA damage. We used immunocytochemistry to stain endogenous Mcl-1 in HeLa cells. Our staining results showed that Mcl-1 is present primarily in the cytosolic compartment in normally proliferating cells, as expected. However, after etoposide treatment, a substantial increase in the nuclear accumulation of Mcl-1 was observed (Figure 2), with the staining appearing as a punctate pattern.
Figure 2.
Mcl-1 translocation to nucleus after etoposide treatment. For intracellular localization of Mcl-1, HeLa cells were left untreated (Control) or treated with 15 μM of etoposide for 2 h (Etoposide). Top, cells were stained with polyclonal anti-Mcl-1 antibody and detected by goat anti rabbit antibody conjugated to Alexa fluor 488. Bottom, cell nuclei were stained with Hoechst 33342 and the combined staining. Stained cells were visualized using a 40× objective.
Mcl-1 Associates with the Checkpoint Regulator Chk1
Because the increased amount of Mcl-1 protein and its accumulation in the nucleus were observed in response to a DNA damaging agent, we sought to determine the potential association of Mcl-1 with components of the DNA damage response machinery. Chk1 plays an essential role in response to genotoxic stress by delaying cell cycle progression through the S and G2 phases (Liu et al., 2000; Takai et al., 2000). Potential physical association of Mcl-1 with Chk1 was investigated in several ways.
Endogenous Chk1 was immunoprecipitated from the nuclear extracts of HeLa cells treated with or without etoposide and immunoblotted for Mcl-1. Our results showed that some Mcl-1 was coimmunoprecipitated with Chk1 from the nuclear extracts of untreated cells, and this association increased substantially in cells treated with etoposide (Figure 3A). It is interesting to note that in etoposide-treated cells, the total level of nuclear Chk1 protein was also increased after etoposide treatment, but increased association with both full-length and short forms of Mcl-1 could be observed in the nucleus. Chk1 has been previously shown to be present in the nucleus in all phases of the cell cycle (Jiang et al., 2003). We next sought to verify that reciprocal coimmunoprecipitation could be observed and at the same time investigate the association of Mcl-1 with Chk1 kinase activity. Total lysates from the murine progenitor cell line, FDC-P1, stably expressing human Mcl-1 or empty vector, were immunoprecipitated with anti-human Mcl-1 (which does not detect murine Mcl-1). These Mcl-1–expressing cells were previously characterized and were shown to have a slower rate of proliferation compared with parental cells (Jamil et al., 2005). As shown in Figure 3B, Mcl-1 immunoprecipitates from etoposide-treated cells contained kinase activity that efficiently phosphorylated the Chk1 peptide substrate, CHKtide (Furnari et al., 1997). In untreated cells, there was no activity detected above background levels. The elevated kinase activity was completely abolished by treatment with the Chk1 inhibitor, isogranulatimide (Jiang et al., 2004). However, this inhibitor has also been shown to inhibit GSK-3β kinase activity, and thus we tested for the potential involvement of this kinase. We found that levels of GSK-3β activity, assayed using its specific substrate, were in fact decreased after etoposide treatment (data not shown). Furthermore, the CHKtide used as a substrate in the Chk1 kinase assay is unlikely to be phosphorylated by GSK-3 because it is not a phosphorylated peptide, which is a prerequisite for GSK-3 phosphorylation. Finally, we also confirmed that immunoprecipitation of endogenous murine Mcl-1 from untransfected FDC-P1 cells also had increased Chk1 activity after etoposide treatment, which was sensitive to 1 μM isogranulatimide (data not shown).
Figure 3.
Mcl-1 coimmunoprecipitates with active Chk1 after DNA damage. (A) HeLa cells were untreated (Con) or treated with etoposide (Etop) for 16 h. Immunoprecipitation of endogenous Chk1 was performed from nuclear extracts (or antibody alone, with no extract, was used as a control). Immunoprecipitates were probed for Chk1 and Mcl-1. (B) FDC-P1 cells expressing Mcl-1 or vector alone were untreated (Con) or treated with etoposide (Etop) for 16 h. Anti-Mcl-1 immunoprecipitates were assayed for Chk1 activity. A parallel etoposide-treated sample was incubated in vitro with 2.5 μM of isogranulatimide (Isogr.) for 30 min at 37°C before kinase assay (Etop + Isogr.). Results are the mean ± SD from three independent experiments. (C) FDC-P1 cells with empty vector or overexpressing Mcl-1 were treated with etoposide (Etop.) for 16 h as indicated. Pretreatment with either 2.5, 5, or 10 μM isogranulatimide (Isogr.) was for 10 min before the addition of etoposide. Cells were analyzed by flow cytometry to determine the percentage of viable cells in G1 compared with S and G2. Numbers are averaged from two independent experiments. (D) Representative flow cytometry profiles of empty vector control cells, compared with cells overexpressing Mcl-1, after treatment with etoposide and 10 μM isogranulatimide. Apoptosis is indicated by cells having sub-G1 DNA content (indicated by asterisk).
After etoposide treatment, cells arrest at G2/M, which is dependent on Chk1 activity (Jin et al., 2005), and the Chk1 inhibitor isogranulatimide can reverse the G2/M arrest that is observed after DNA damage (Jiang et al., 2004). Thus, we tested whether the overexpression of Mcl-1 altered the sensitivity of cells to the inhibitory effects of isogranulatimide. Mcl-1–overexpressing and control FDC-P1 cells were used for these experiments. Data in Figure 3C shows that both cell populations have similar cell cycle profiles, as we reported previously (Jamil et al., 2005), despite the fact that the Mcl-1–expressing cells have a reduced proliferation rate. These cells were exposed to etoposide, which caused G2 arrest (Figure 3C). In control cells, 2.5 μM isogranulatimide was able to completely reverse this G2 arrest. However, in cells overexpressing Mcl-1, fourfold higher concentrations of isogranulatimide were required to have a similar effect, indicating that the presence of increased Mcl-1 protein resulted in greater resistance to the Chk1 inhibitor. It should be noted that, compared with control cells, the cells overexpressing Mcl-1 were much more resistant to the toxic effects of the combined treatment with 10 μM isogranulatimide and etoposide (Figure 3D).
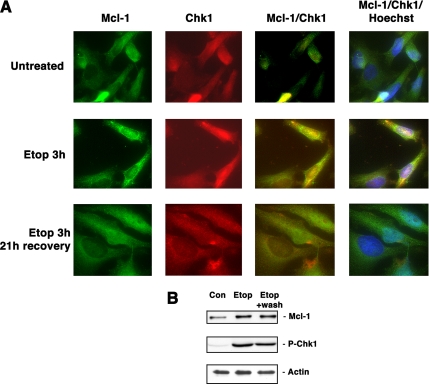
Immunofluorescence staining was also used to monitor potential colocalization of Mcl-1 and Chk1 after etoposide treatment. As shown in Figure 4A, there was an increase in both total and nuclear staining of Mcl-1 and Chk1 after etoposide treatment, and these two proteins were colocalized in the nucleus. Interestingly, after cells were allowed to recover for 21 h after washing out etoposide, there were still elevated levels of both Mcl-1 as well as Chk1 phosphorylated at the activation site Ser345, as shown in Figure 4B. However, the immunofluorescence staining showed that in the majority of cells, little colocalization of the two proteins could be detected. Together, all of these results indicated that Mcl-1 can associate with active Chk1 in the nucleus after DNA damage, although we have not yet determined whether there is a direct association between the two proteins.
Figure 4.
Mcl-1 and Chk1 colocalize after DNA damage. (A) HeLa cells were untreated, treated with 15 μM etoposide for 3 h, or treated with etoposide for 3 h, followed by washing and allowing cells to recover for 21 h. Cells were stained using anti-Mcl-1 or anti-Chk1, or the figures were merged to show colocalization. Additional staining with Hoechst 33342 was used to visualize nuclei. (B) Cell extracts from the same cells used in A were used to immunoblot for the presence of Mcl-1, phospho-Chk1, or actin as a loading control.
Overexpression of Mcl-1 Causes Ser345-Chk1 Phosphorylation in the Absence of DNA Damage
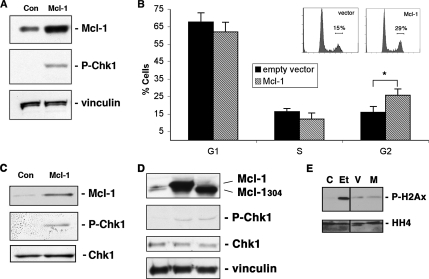
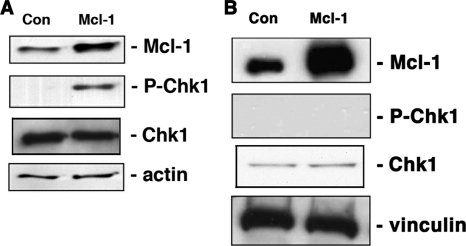
We next investigated the effect of transiently overexpressing Mcl-1 on the events involved in DNA damage response. It is well known that in response to DNA damage, Chk1 is phosphorylated at two sites, Ser345 and Ser317, which are required for kinase activation (Abraham, 2001; Zhao and Piwnica-Worms, 2001; Shiloh, 2003). We found that Mcl-1 transiently overexpressed in HeLa cells resulted in phospho-Ser345 Chk1 in the absence of exogenous DNA damage (Figure 5A). Very little or no phospho-Ser345 Chk1 protein was observed in control cells when empty vector was overexpressed. We next assessed the effect of transient Mcl-1 overexpression on cell cycle status. As shown in Figure 5B, consistent with its enhancement of Chk1 phosphorylation, a significant increase in the percentage of HeLa cells overexpressing Mcl-1 were found to accumulate in G2. This effect of Mcl-1 is observed when high levels are expressed transiently, but it is important to point out that any cells selected to stably express Mcl-1 (such as the FDC-P1 cells used above) never show high levels of Mcl-1 expression, likely due to the inhibitory effect of Mcl-1 on proliferation. We also transiently transfected Mcl-1 in primary human fibroblasts and again, the phosphorylation of Chk1 was observed (Figure 5C). These data suggest that increasing expression of Mcl-1 alone can initiate the events leading to Chk1 phosphorylation.
Figure 5.
Transient overexpression of Mcl-1 increases phosphorylation of Chk1 at Ser345. (A) HeLa cells were transiently transfected with Mcl-1 and whole cell extracts immunoblotted for Mcl-1, phospho-Ser345 Chk1, or vinculin as a loading control. (B) Cells overexpressing Mcl-1, or empty vector, as in A, were analyzed for cell cycle status by flow cytometry. Increased percentage of cells in G2 after Mcl-1 expression showed a significant change (p = 0.05). (C) Hs68 primary fibroblasts were transiently transfected with empty vector (Con) or with Mcl-1 and extracts probed as in A, except for use of anti-Chk1 as a loading control. (D) HeLa cells were transiently transfected with either Mcl-1 or Mcl-1304 as indicated, and extracts were immunoblotted as in A, except for the additional anti-Chk1 blot, which also serves as a loading control. (E) HeLa cells were untreated (C) or treated with etoposide for 3 h (Et) or were transiently transfected with vector alone (V) or Mcl-1–containing vector (M). Chromatin extracts were prepared, separated, and immunoblotted for the presence of phosphorylated histone H2Ax (P-H2Ax) or for histone H4 (HH4) as a loading control. The pairs of samples separated by vertical lines were resolved on a single gel.
To investigate whether Mcl-1-mediated phosphorylation of Chk1 was associated with, or distinct from, the Bcl-2-like prosurvival effects of Mcl-1, HeLa cells were transiently transfected with mutant Mcl-1. Because all three BH domains of Mcl-1 are required for its complete antiapoptotic effects (Clohessy et al., 2006), we generated a mutant Mcl-1 truncated at residue 304 (Mcl-1304), which deletes the BH2 and transmembrane domains. Transient expression of Mcl-1304 in HeLa cells, detected as a band of lower molecular weight than the endogenous Mcl-1, also caused increased phosphorylation of Chk1 (Figure 5D).
In view of these results, we wanted to rule out the possibility that transient transfection with Mcl-1 may result in a generalized stress response leading to Chk1 phosphorylation. We therefore investigated the phosphorylation of H2Ax (γ-H2Ax), which is considered a hallmark of DNA damage. In transiently transfected HeLa cells both empty and Mcl-1 containing vectors showed a slight increase in the γ-H2Ax (Figure 5E). This effect could be attributed to the stress caused to the cells by transfection and was not due to the overexpression of Mcl-1 protein. Compared with the levels of γ-H2Ax observed in transfected cells, a more robust and dramatic increase in γ-H2Ax was observed in cells treated with etoposide. Similar results were observed with Chk2 phosphorylation at Thr68 (an activating site on that kinase), where no significant difference was observed between the empty vector and Mcl-1–overexpressing cells (data not shown). These results rule out the possibility that expression of Mcl-1 itself may cause a generalized DNA damage stress response.
Mcl-1–induced Chk1 Phosphorylation Requires ATR, but not ATM
Chk1 is phosphorylated on Ser345 in response to DNA damage primarily by two members of the phosphoinositide 3-kinase related kinase (PIKK) family of enzymes, ATM or ATR (Abraham, 2001; Shiloh, 2003). We sought to determine the kinase through which the effects of Mcl-1 were being mediated. In HT-144 cells, which have a mutation in the ATM gene (Ramsay et al., 1998), transient transfection of Mcl-1 caused an increase in Chk1 phosphorylation similar to that seen in other cell types (Figure 6A), ruling out an essential role for ATM. Similarly, Mcl-1 overexpression caused similar increases in Chk1 phosphorylation in cells deficient for the related PIKK member, DNA-PK (data not shown). ATR cannot be knocked out because it is an essential gene, and thus we investigated the involvement of ATR by using fibroblasts derived from a Seckel syndrome patient, F02–98 cells, which are known to have reduced ATR activity (O'Driscoll et al., 2003). As shown in Figure 6B, transient transfection of wild-type Mcl-1 in cells harboring the ATR mutation was unable to induce Chk1 phosphorylation, suggesting that ATR is required for Mcl-1–mediated Chk1 phosphorylation.
Figure 6.
Mcl-1-induced phosphorylation of Chk1 is dependent on ATR. ATM-deficient HT-144 cells (A) and ATR-defective fibroblasts, F02–98 (B) were transiently transfected with empty vector (Con) or Mcl-1, and extracts probed as indicated.
Increase in Mcl-1 Expression with UV Irradiation Is ATR Dependent
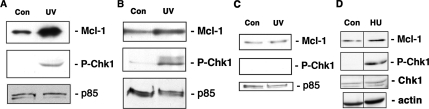
ATR is activated mainly by stalled replication forks and agents that generate bulky lesions like UV irradiation (Guo et al., 2000; Unsal-Kacmaz et al., 2002). We therefore examined the protein levels of Mcl-1 after exposure of cells to UV radiation, which primarily activates ATR (Heffernan et al., 2002). A low dose of UV resulted in the induction of Mcl-1 expression within 2 h in both HeLa cells and in primary human fibroblasts (Figure 7, A and B). In these cells in which Mcl-1 was increased, a corresponding increase in phospho-Chk1 was also observed. We and others have shown that higher doses of UV cause Mcl-1 degradation (Nijhawan et al., 2003; Germain and Duronio, 2007), but we consistently observed increased Mcl-1 expression in response to low-level UV irradiation. In contrast to the increase in Mcl-1 observed in other cell lines, F02–98 cells showed little to no increase in Mcl-1 levels in response to UV irradiation (Figure 7C). As expected, in these cells with low levels of ATR activity, no increase in Chk1 phosphorylation was evident after exposure to UV. However, when these same F02–98 cells were treated with a dose of hydroxyurea known to induce DNA damage response in these cells (Alderton et al., 2004), a significant increase in Chk1 phosphorylation could be observed, with little or no change in the amount of Mcl-1. These results demonstrate that the lack of Chk1 phosphorylation observed in response to low dose of UV was not due to their general inability to phosphorylate Chk1.
Figure 7.
Low-level UV radiation induces Mcl-1 expression and Chk1 phosphorylation. HeLa cells (A), Hs68 cells (B), or F02–98 cells (C) were untreated (Con) or irradiated with UV, and extracts prepared after 2 h. Immunoblots of whole cell lysates were probed with anti-Mcl-1, anti-phospho-Ser345 Chk1 (P-Chk1), or anti-p85 antibodies. (D) F02–98 cells were untreated or treated with 2 mM hydroxyurea (HU) for 3 h. Whole cell lysates were immunoblotted as indicated. For D, all panels were reblots of the same gel, and the pairs of lanes shown were at different portions of the same gel and thus separated by a vertical line.
Mcl-1 Is Required for Ser345 Chk1 Phosphorylation after DNA Damage
The question of whether Mcl-1 plays an essential role in Chk1 activation was tested in cells in which Mcl-1 expression was reduced using siRNA. HeLa cells were transfected with Mcl-1–specific siRNA and cells were largely depleted of Mcl-1 after 48 h (Figure 8A). The amount of total protein in each cell extract was equivalent based on the immunoblots for the p85 subunit of PI 3-kinase. Furthermore, the expression levels of the prosurvival protein Bcl-2 in Mcl-1–depleted cells did not change over the time course of this experiment. In control cells treated with an irrelevant siRNA, treatment with etoposide resulted in phosphorylation of Chk1 at Ser345 as well as increases in Mcl-1 expression. However, in cells where Mcl-1 expression was knocked down, phosphorylation of Chk1 was abolished after etoposide treatment (Figure 8A). Decreased Mcl-1 expression did not have any effect on the endogenous level of Chk1 protein, showing that the lack of Chk1 phosphorylation in the Mcl-1–depleted cells could not be attributed to decreased levels of Chk1. It is important to note that neither down-regulation of Mcl-1 expression, nor treatment with etoposide, over this short time period resulted in any noticeable cell death as determined by trypan blue dye exclusion test (not shown) or by propidium iodide (PI) staining for subdiploid DNA (Figure 8B).
Figure 8.
Mcl-1 is required for Ser345 Chk1 phosphorylation after DNA damage. (A) HeLa cells were transfected with control (left two lanes) or Mcl-1 siRNA (right two lanes); 48 h after initial transfection, cells were left untreated (lanes 1 and 3) or were treated with etoposide for 1 h (lanes 2 and 4). Total cell proteins were extracted and immunoblotted with anti-Mcl-1, anti-phospho Chk1 (Ser 345), anti-Chk1, anti-Bcl-2, and anti-p85 antibodies. (B) HeLa cells were transfected with control siRNA or Mcl-1–specific siRNA for 48 h, followed by treatment without or with etoposide for 1 h. Cells were stained with PI and analyzed by flow cytometry for DNA content.
DISCUSSION
This study has revealed an unexpected function of the Bcl-2 family protein, Mcl-1, in DNA damage responses. There is increasing evidence from some recent studies of involvement of other members of the Bcl-2 family in DNA damage response or repair (Kamer et al., 2005; Youn et al., 2005; Zinkel et al., 2005; Wang et al., 2008), and thus several of these proteins may have such alternate functions. Given that overexpression of Mcl-1 alone is sufficient to direct the upstream kinase(s) to activate the key checkpoint signal transducer Chk1 and that suppression of Mcl-1 expression blocks the ability of cells to respond to DNA damage by activating Chk1, Mcl-1 must play a key role in this pathway. Our results have ruled out an essential role for ATM or DNA-PK in the Mcl-1–regulated phosphorylation of Chk1 and have placed Mcl-1 upstream of Chk1 in the ATR-Chk1 pathway.
Interestingly, the functions of Mcl-1 appear to be very similar to those that have been reported for Claspin. Down-regulation of Claspin has been shown to inhibit Chk1 activation in response to DNA damage (Kumagai and Dunphy, 2000; Chini and Chen, 2003). However, unlike Claspin, Mcl-1's association with Chk1 does not depend on DNA damage because we observed some association in untreated cells (Figure 3B). It is possible that Mcl-1 plays a role in directing the phosphorylating kinase to Chk1 during S and G2 phases of cell cycle in unperturbed cells, and Claspin provides an additional layer of control by further activating Chk1 and consolidating the checkpoint response upon DNA damage.
It is noteworthy that Claspin was recently shown to be in the same complex as proliferating cell nuclear antigen (PCNA; Brondello et al., 2007), which is an important cell cycle regulatory protein, suggesting Claspin may be a common link between DNA damage response and cell cycle machineries. Interestingly, Mcl-1 is the only member of Bcl-2 family that has previously been shown to interact directly with PCNA (Fujise et al., 2000) placing Mcl-1 at the interface of apoptosis and cell cycle regulation. Based on these observations, a more relevant role of Mcl-1 in normal cell cycle regulation may be suggested, because we have found Mcl-1 (or the shorter snMcl-1 form of the protein) in the nucleus in several cell types (Jamil et al., 2005), and Mcl-1 has also been shown to predominantly localize to the nucleus, where it associates with PCNA, in U2OS cells (Fujise et al., 2000).
As we have mentioned earlier, our findings are based on treatments with etoposide or UV radiation that are at lower doses than those used in most studies. We were careful to use doses that caused G2 arrest, but did not cause significant apoptosis during the time course of the experiments. In fact, it is clear that higher doses than we have used here, of either etoposide or UV radiation, cause apoptosis together with loss of Mcl-1 expression (Nijhawan et al., 2003; Germain and Duronio, 2007). We cannot rule out that the increased Mcl-1 expression is also contributing to cell survival in cells treated with lower levels of DNA-damaging agents. Thus, a dual function for Mcl-1 can be suggested. It is possible that when damage is sustained, up-regulated Mcl-1 is involved in mediating the checkpoint response that allows members of the DNA damage response machinery to function. At the same time as the DNA is being repaired, Mcl-1 at the mitochondria may also play a role in providing survival until the repair has been completed.
Perhaps the most intriguing aspect of this study is that the findings may offer an explanation for the essential function of Mcl-1 that was demonstrated in studies of Mcl-1 knockout mice (Rinkenberger et al., 2000), because those studies showed that there was no change in the extent of apoptosis in the Mcl-1−/− embryos. Similar to Mcl-1 knockout mice, knockout of either ATR or Chk1 are also embryonic lethal at a preimplantation stage (de Klein et al., 2000; Liu et al., 2000; Takai et al., 2000). Besides playing a key role in DNA damage response, the essential functions of these genes are also likely to be to maintain normal DNA replication, as has been suggested for Chk1 (Kaneko et al., 1999). As we and others have reported previously, growth of cells overexpressing Mcl-1 is dramatically inhibited, which is also consistent with a proposed role for Mcl-1 in orchestrating a checkpoint response. Indeed, we find that when high levels of Mcl-1 are present after transient overexpression, we detected an accumulation of cells in G2, consistent with the expected effects of increased Chk1 phosphorylation. During response to DNA damage, Chk1 phosphorylation is enhanced, and as we have also shown here, this can be independent of ATM (Kaneko et al., 1999) and thus more dependent upon ATR. The results of this study also show that regulation of Chk1 phosphorylation in response to DNA damage is dependent on the presence of Mcl-1, because siRNA-mediated knockdown of Mcl-1 expression results in loss of DNA damage–induced Chk1 phosphorylation.
To better understand the molecular events that are controlling Mcl-1 functions in the nucleus, future studies should be aimed at finding the direct binding partners of Mcl-1. We have established in this study the presence of Mcl-1 in a complex that includes the checkpoint regulator Chk1, supporting the suggestion that a key function of Mcl-1 may be in the coordination of events required to generate appropriate response to DNA damage and maintenance of chromosome integrity. These newly characterized properties of an antiapoptotic member of the Bcl-2 family that has been extensively studied in recent years may lead to important new discoveries that expand our understanding of the important functions of Mcl-1.
ACKNOWLEDGMENTS
We thank Dr. Michel Roberge for helpful discussions regarding Chk1 and for providing isogranulatimide, Dr. Karl Riabowol for providing primary human fibroblasts, Dr. P. Olive for providing HT-144 cells and Dr. Susan Lees-Miller (University of Calgary) for providing the ATR-defective cells that were obtained from Dr. P. Jeggo. We also thank Dr. Marco Garate for technical advice and Dr. Andrew Sandford for critical review of the manuscript. This work was supported by a grant to V.D. from the Canadian Institutes of Health Research.
Abbreviations used:
- ATM
ataxia telangectasia mutated
- ATR
AT mutated and Rad3 related
- BH
Bcl-2 homology
- Chk1
checkpoint kinase 1
- Mcl-1
myeloid cell leukemia-1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1171) on May 21, 2008.
REFERENCES
- Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Akgul C., Moulding D. A., White M. R., Edwards S. W. In vivo localisation and stability of human Mcl-1 using green fluorescent protein (GFP) fusion proteins. FEBS Lett. 2000;478:72–76. doi: 10.1016/s0014-5793(00)01809-3. [DOI] [PubMed] [Google Scholar]
- Alderton G. K., Joenje H., Varon R., Borglum A. D., Jeggo P. A., O'Driscoll M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- Brondello J. M., Ducommun B., Fernandez A., Lamb N. J. Linking PCNA-dependent replication and ATR by human Claspin. Biochem. Biophys. Res. Commun. 2007;354:1028–1033. doi: 10.1016/j.bbrc.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Chini C. C., Chen J. Human claspin is required for replication checkpoint control. J. Biol. Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Cliby W. A., Lewis K. A., Lilly K. K., Kaufmann S. H. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 2002;277:1599–1606. doi: 10.1074/jbc.M106287200. [DOI] [PubMed] [Google Scholar]
- Clohessy J. G., Zhuang J., Brady H. J. Characterisation of Mcl-1 cleavage during apoptosis of haematopoietic cells. Br. J. Haematol. 2004;125:655–665. doi: 10.1111/j.1365-2141.2004.04949.x. [DOI] [PubMed] [Google Scholar]
- Clohessy J. G., Zhuang J., de Boer J., Gil-Gomez G., Brady H. J. Mcl-1 interacts with truncated Bid and inhibits its induction of cytochrome c release and its role in receptor-mediated apoptosis. J. Biol. Chem. 2006;281:5750–5759. doi: 10.1074/jbc.M505688200. [DOI] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A. M., Lehmann A. R., Hoeijmakers J. H. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- Elledge S. J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fujise K., Zhang D., Liu J., Yeh E. T. Regulation of apoptosis and cell cycle progression by MCL1. Differential role of proliferating cell nuclear antigen. J. Biol. Chem. 2000;275:39458–39465. doi: 10.1074/jbc.M006626200. [DOI] [PubMed] [Google Scholar]
- Furnari B., Rhind N., Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Germain M., Duronio V. The N Terminus of the Anti-apoptotic BCL-2 Homologue MCL-1 Regulates Its Localization and Function. J. Biol. Chem. 2007;282:32233–32242. doi: 10.1074/jbc.M706408200. [DOI] [PubMed] [Google Scholar]
- Guo Z., Kumagai A., Wang S. X., Dunphy W. G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan T. P., Simpson D. A., Frank A. R., Heinloth A. N., Paules R. S., Cordeiro-Stone M., Kaufmann W. K. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G., Wang Y., Guan J., Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- Jamil S., Sobouti R., Hojabrpour P., Raj M., Kast J., Duronio V. A proteolytic fragment of Mcl-1 exhibits nuclear localization and regulates cell growth by interaction with Cdk1. Biochem. J. 2005;387:659–667. doi: 10.1042/BJ20041596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Pereira E., Maxfield M., Russell B., Goudelock D. M., Sanchez Y. Regulation of Chk1 includes chromatin association and 14–3-3 binding following phosphorylation on Ser-345. J. Biol. Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- Jiang X., Zhao B., Britton R., Lim L. Y., Leong D., Sanghera J. S., Zhou B. B., Piers E., Andersen R. J., Roberge M. Inhibition of Chk1 by the G2 DNA damage checkpoint inhibitor isogranulatimide. Mol. Cancer Ther. 2004;3:1221–1227. [PubMed] [Google Scholar]
- Jin Z. H., Kurosu T., Yamaguchi M., Arai A., Miura O. Hematopoietic cytokines enhance Chk1-dependent G2/M checkpoint activation by etoposide through the Akt/GSK3 pathway to inhibit apoptosis. Oncogene. 2005;24:1973–1981. doi: 10.1038/sj.onc.1208408. [DOI] [PubMed] [Google Scholar]
- Kamer I., Sarig R., Zaltsman Y., Niv H., Oberkovitz G., Regev L., Haimovich G., Lerenthal Y., Marcellus R. C., Gross A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Kaneko Y. S., Watanabe N., Morisaki H., Akita H., Fujimoto A., Tominaga K., Terasawa M., Tachibana A., Ikeda K., Nakanishi M. Cell-cycle-dependent and ATM-independent expression of human Chk1 kinase. Oncogene. 1999;18:3673–3681. doi: 10.1038/sj.onc.1202706. [DOI] [PubMed] [Google Scholar]
- Kastan M. B. Cell cycle. Checking two steps. Nature. 2001;410:766–767. doi: 10.1038/35071218. [DOI] [PubMed] [Google Scholar]
- Kozopas K. M., Yang T., Buchan H. L., Zhou P., Craig R. W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. USA. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Liu J. L., Ye Y., Qian Z., Qian Y., Templeton D. J., Lee L. F., Kung H. J. Functional interactions between herpesvirus oncoprotein MEQ and cell cycle regulator CDK2. J. Virol. 1999;73:4208–4219. doi: 10.1128/jvi.73.5.4208-4219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M., Ruiz-Perez V. L., Woods C. G., Jeggo P. A., Goodship J. A. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Ramsay J., Birrell G., Baumann K., Bodero A., Parsons P., Lavin M. Radiosensitive melanoma cell line with mutation of the gene for ataxia telangiectasia. Br. J. Cancer. 1998;77:11–14. doi: 10.1038/bjc.1998.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger J. L., Horning S., Klocke B., Roth K., Korsmeyer S. J. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 2000;14:23–27. [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Shiio Y., Eisenman R. N., Yi E. C., Donohoe S., Goodlett D. R., Aebersold R. Quantitative proteomic analysis of chromatin-associated factors. J. Am. Soc. Mass Spectrom. 2003;14:696–703. doi: 10.1016/S1044-0305(03)00204-6. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Takai H., Tominaga K., Motoyama N., Minamishima Y. A., Nagahama H., Tsukiyama T., Ikeda K., Nakayama K., Nakanishi M., Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K., Makhov A. M., Griffith J. D., Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. USA. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Gao F., May W. S., Zhang Y., Flagg T., Deng X. Bcl-2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol. Cell. 2008;29:488–498. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton K., Riabowol K. Protein kinase C delta blocks immediate-early gene expression in senescent cells by inactivating serum response factor. Mol. Cell. Biol. 2004;24:7298–7311. doi: 10.1128/MCB.24.16.7298-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn C. K., Cho H. J., Kim S. H., Kim H. B., Kim M. H., Chang I. Y., Lee J. S., Chung M. H., Hahm K. S., You H. J. Bcl-2 expression suppresses mismatch repair activity through inhibition of E2F transcriptional activity. Nat. Cell Biol. 2005;7:137–147. doi: 10.1038/ncb1215. [DOI] [PubMed] [Google Scholar]
- Zhang D., Li F., Weidner D., Mnjoyan Z. H., Fujise K. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and fortilin. The potential role of MCL1 as a fortilin chaperone. J. Biol. Chem. 2002;277:37430–37438. doi: 10.1074/jbc.M207413200. [DOI] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. B., Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zhou P., Qian L. P., Kozopas K. M., Craig R. W. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 1997;89:630–643. [PubMed] [Google Scholar]
- Zinkel S. S., Hurov K. E., Ong C., Abtahi F. M., Gross A., Korsmeyer S. J. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]