Abstract
Brahma (BRM) and Brahma-related gene 1 (BRG1) are the ATP-dependent catalytic subunits of the SWI/SNF family of chromatin-remodeling complexes. These complexes are involved in essential processes such as cell cycle, growth, differentiation, and cancer. Using imaging approaches in a cell line that harbors tandem repeats of stably integrated copies of the steroid responsive MMTV-LTR (mouse mammary tumor virus–long terminal repeat), we show that BRG1 and BRM are recruited to the MMTV promoter in a hormone-dependent manner. The recruitment of BRG1 and BRM resulted in chromatin remodeling and decondensation of the MMTV repeat as demonstrated by an increase in the restriction enzyme accessibility and in the size of DNA fluorescence in situ hybridization (FISH) signals. This chromatin remodeling event was concomitant with an increased occupancy of RNA polymerase II and transcriptional activation at the MMTV promoter. The expression of ATPase-deficient forms of BRG1 (BRG1-K-R) or BRM (BRM-K-R) inhibited the remodeling of local and higher order MMTV chromatin structure and resulted in the attenuation of transcription. In vivo photobleaching experiments provided direct evidence that BRG1, BRG1-K-R, and BRM chromatin-remodeling complexes have distinct kinetic properties on the MMTV array, and they dynamically associate with and dissociate from MMTV chromatin in a manner dependent on hormone and a functional ATPase domain. Our data provide a kinetic and mechanistic basis for the BRG1 and BRM chromatin-remodeling complexes in regulating gene expression at a steroid hormone inducible promoter.
INTRODUCTION
The eukaryotic genome is organized into higher order chromatin structures in the nucleus. The regulation of eukaryotic gene expression in the context of chromatin is a complex event and is essential for numerous cellular processes (Lemon and Tjian, 2000;Orphanides and Reinberg, 2002; Labrador and Corces, 2002; Maniatis and Reed, 2002; Felsenfeld and Groudine, 2003; Roberts and Orkin, 2004). Maintenance, establishment, and modification of global chromatin organization and local chromatin structure are modulated by a large number of chromatin-binding proteins that generate transcriptionally permissive or repressed chromatin domains in response to environmental stimuli (Workman and Kingston, 1998; Peterson and Workman, 2000; Jones and Kadonaga, 2000; Wu and Grunstein, 2000; Wolffe and Hansen, 2001; Becker et al., 2002). The association of linker histones and nonhistone and heterochromatin-specific proteins such as high mobility group proteins and HP1 play key roles in the generation of higher order chromatin structures (Arents et al., 1991; Luger et al., 1997; Bustin, 1999; Eissenberg and Elgin, 2000; Hill, 2001; Thomas and Travers, 2001; Woodcock and Dimitrov, 2001; Grewal and Elgin, 2002; Bianchi and Agresti, 2005; Bustin et al., 2005; Verschure et al., 2005). The stearically restricted environment presented by chromatin is in part overcome by multisubunit protein complexes that enzymatically regulate chromatin structure. These complexes can either chemically modify histone tails (acetylation, phosphorylation, ubiquitylation, methylation) or disrupt histone–DNA interactions by using the energy generated through ATP hydrolysis (Strahl and Allis, 2000; Berger, 2002; Narlikar et al., 2002; Neely and Workman, 2002; Shogren-Knaak et al., 2006). The combination of chromatin modification and chromatin-remodeling complexes acting in a concerted manner modulate the accessibility of individual genes to sequence-specific transcription factors, general transcription factors, and components of the RNA pol II transcription machinery.
SWI/SNF is one of three subclasses of ATP-dependent chromatin remodeling enzymes that have been identified in mammalian cells (Peterson, 2002). The SWI/SNF complex exists in one of two forms, containing either one of two highly homologous ATPases, BRG1 or BRM and several shared subunits collectively called BAFs (BRG1- or BRM-associated factors). SWI/SNF is an evolutionarily conserved, ∼2 MDa multisubunit complex interacting with a wide variety of proteins and is functionally implicated in cell cycle, differentiation, and cancer (Muchardt and Yaniv, 2001; Klochendler-Yeivin et al., 2002; Roberts and Orkin, 2004; Cho et al., 2004; Gregory and Shiekhattar, 2004; Imbalzano and Jones, 2005). Several subunits of the SWI/SNF chromatin-remodeling complex possess tumor suppressor activity and play key roles in the functional activity of other tumor suppressor genes including Rb, BRCA1, and c-MYC (Muchardt and Yaniv, 2001; Roberts and Orkin, 2004). For example, a core subunit of SWI/SNF, Snf5 (Ini1) is inactivated in highly aggressive malignant rhabdoid tumors (Versteege et al., 1998; Biegel et al., 1999; Sevenet et al., 1999).
Upon hormone binding, nuclear hormone receptors such as the glucocorticoid receptor (GR) bind hormone response elements and regulate transcription at their target genes through the recruitment of a variety of coactivators, corepressors, chromatin remodeling activities, and components of the basal transcription machinery (Fragoso et al., 1998; Giangrande et al., 2000; Dilworth and Chambon, 2001; McKenna and O'Malley, 2002; Schaaf and Cidlowski, 2003; Belandia and Parker, 2003; Metivier et al., 2003; Hager et al., 2006; Carroll and Brown, 2006; Lee et al., 2006). To further understand the process by which chromatin-remodeling complexes are recruited and regulate target genes to modulate transcription, we have directly visualized the sequence of gene expression events involving the SWI/SNF complex in mouse mammary adenocarcinoma cells that contain a tandem repeat of stably integrated copies of the MMTV-LTR (mouse mammary tumor virus–long terminal repeat). This array which contains 800-1200 binding sites for GR can be visualized by using green fluorescent protein (GFP)-tagged versions of steroid receptors and associated cofactors (Kramer et al., 1999; McNally et al., 2000; Rayasam et al., 2005). We have investigated the molecular basis by which SWI/SNF regulates transcription as well as its influence on the chromatin structure of a steroid hormone–responsive promoter array. Our study provides an integrated view of gene activation events demonstrating the hormone-dependent recruitment of SWI/SNF chromatin-remodeling complexes to the MMTV array and the associated chromatin remodeling, decondensation, and transcriptional events associated with SWI/SNF function. Furthermore, we demonstrate the dynamic interaction of BRG1 and BRM with the MMTV array and determine for the first time, by using in vivo photobleaching microscopy, that BRG1 and BRM chromatin-remodeling complexes have distinct kinetic properties on the MMTV array and dynamically associate with and dissociate from MMTV chromatin in a manner dependent on hormone and a functional ATPase domain. These results further our understanding of SWI/SNF action in chromatin remodeling and gene expression in vivo.
MATERIALS AND METHODS
Expression Vectors
Flag-tagged forms of BRG1, BRM, BRG1-K-R, and BRM-K-R were provided by Anthony Imbalzano (University of Massachusetts Medical School, Worcester, MA). Yellow fluorescent protein (YFP)-BRG1 and YFP-BRG1-K-R were generated by cloning of wild-type BRG1 and mutant BRG1-K-R into pEYFP-C2 and pEGFP-C2 vectors (Clontech, Palo Alto, CA). GFP-BRM and GFP BRG1 have been previously described (Reyes et al., 1997; de la Serna et al., 2001) and was provided by Christian Muchardt, (Institute Pasteur, Paris, France). The full-length MMTV-LTR driving transcription of the luciferase gene, pRSV-GR (glucocorticoid receptor) and pCMV β-Galactosidase (internal control) has been described previously (Lefebvre et al., 1991; Fragoso et al., 1998). All cloned vectors were confirmed by sequencing.
Cell Culture and Stable Cell Lines
The murine mammary adenocarcinoma cell line (3134) contains a large tandem array of a mouse mammary tumor virus/Harvey ras reporter (Kramer et al., 1999). In 3134 cells, 200 copies of the MMTV-LTR with 800-1200 GR-binding sites are stably integrated in a head-to-tail orientation into the centromeric region of chromosome 4 (McNally et al., 2000). The 3617 cell line expressing GFP-GR under control of a tetracycline-repressible (Tet-Off) system is generated by stable transfection of 3134 cells (Walker et al., 1999). The 3617 cell line was stably transfected with a Flag-tagged BRG1-K-R to generate the 5555 cell line. The 1365.1 cell line is derived from NIH 3T3 mouse fibroblast cells by stable transformation with a multicopy episome and contains multiple tandem copies of a stably integrated MMTV-LTR array (Cordingley et al., 1987; Bresnick et al., 1990). Human adrenal carcinoma cells (SW13) are deficient in BRG1, BRM, and GR expression and were obtained from the American Type Culture Collection (ATCC, Manassas, VA). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini, Woodland, CA), 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 5 mg/ml penicillin-streptomycin, and 1 mg/ml G418 (Invitrogen) and kept at 37°C incubator with 5% CO2. Cells were transferred to 10% charcoal-dextran–treated, heat-inactivated fetal bovine serum for 24 h before hormone treatment. 3617 and 5555 cells were supplemented with 10 μg/ml tetracycline (FisherBiotech, Fair Lawn, NJ) to suppress GFP-GR and/or Flag-tagged mutant BRG1-K-R expression. In preparation for biochemical and imaging experiments, cell culture medium was replaced with the same medium without tetracycline and phenol red to induce the expression of GFP-GR or Flag-tagged BRG1-K-R. The cells were grown for an additional 18–24 h and GFR-GR or endogenous GR was activated using dexamethasone at 100 nM for 30 min.
Transfections, Immunoblot Analysis, and RNA Interference
In the SW13 transactivation assays, cells were transfected with GR, BRG1, BRM, BRG1-K-R, BRM-K-R, MMTV-LTR-Luc, and CMV β-Gal (as an internal control) using Lipofectamine 2000 according to manufacturer's instructions (Invitrogen). SW13 cells were treated with 100 nM dexamethasone for 4 h, and whole cell extracts prepared. Luciferase and β-galactosidase assays were performed by using the Dual Reporter Assay kit according to the manufacturer's instructions (Tropix, Bedford, MA). All transfections were done in triplicates and all experiments were repeated three times. For Western blots, 5555 cells were grown as described previously in the presence or absence of tetracycline to regulate the expression of Flag-tagged BRG1-K-R. Whole cell extracts were prepared and equal amounts of total cell extracts were fractioned on a 7.5% SDS-PAGE gels and electrotransferred to Immobilon-P (Millipore, Billerica, MA). Mutant BRG1-K-R was detected using a polyclonal anti-Flag antibody (kindly provided by Anthony Imbalzano). Small interfering RNAs (siRNAs) to BRG1 (smart pool) and scrambled siRNAs were purchased from Dharmacon (Chicago, IL). siRNAs were transfected into 3134 cells using Lipofectamine 2000 at a final concentration of 100 nM. After 3 d, cells were treated with 100 nM dexamethasone for 30 min, fixed, and processed for indirect immunofluorescence microscopy combined with RNA fluorescence in situ hybridization (RNA FISH).
Restriction Endonuclease Accessibility Assay
Restriction endonuclease cleavage of MMTV chromatin was conducted as previously described (Mulholland et al., 2003). Nuclei were isolated and digested with SacI restriction enzyme (New England Biolabs, Ipswich, MA). DNA from nuclei was purified and digested to completion with DpnII (New England Biolabs). The digestion products were amplified linearly by primer extension using Taq polymerase and a radiolabeled primer specific to the MMTV-LTR promoter region. The extension products were run on an 8% denaturing sequencing gel and quantified on a phosphorimager using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Nuclease hypersensitivity and chromatin remodeling were expressed as % fractional cleavage, which was determined by dividing the intensity of the SacI digestion product by the sum of the intensities of the SacI and DpnII digestion products.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (Mulholland et al., 2003) with some modifications. Briefly, 3617 and 5555 cells were treated with dexamethasone and tetracycline as described in Figure 4. Cells were fixed with formaldehyde and sonicated on ice with a Branson sonicator (Branson Ultrasonics, Danbury, CT) at a power setting of 20–30 W. After centrifugation, the soluble material was immunoprecipitated overnight with an anti-RNA polymerase II (pol II) antibody (provided by Kevin Gardner, National Cancer Institute, Bethesda, MD). Antibody bound chromatin complexes were immunoprecipitated with protein A-agarose beads, and the bound material was eluted. Formaldehyde cross-links were reversed at 65°C overnight and DNA was purified. DNA from each sample was subjected to PCR (25 cycles) using primer sets specific for the MMTV-LTR nuc-B region. PCR products were run on 6% PAGE gels and stained with sybr green.
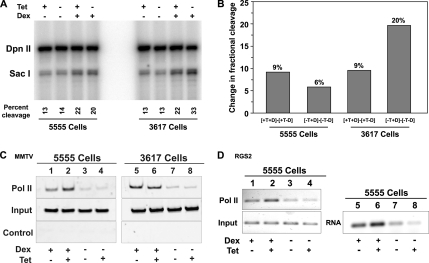
Figure 4.
BRG1 is required for the hormone-dependent remodeling of MMTV chromatin and associated loading of RNA pol II at the MMTV promoter. 5555 and 3617 cells were untreated or treated with 100 nM dexamethasone for 30 min and tetracycline as indicated. Nuclei were isolated and digested with SacI and DpnII restriction enzymes. Digestion products were detected by linear amplification using a radiolabeled primer specific to the MMTV promoter region. Percent cleavage and the fractional change in the accessibility of MMTV promoter to restriction enzymes are indicators of nuclease hypersensitivity and chromatin remodeling of MMTV promoter region. (A) Intensity of SacI digestion product is divided by the sum of the intensities of SacI and DpnII digestion products and presented as percent cleavage at the bottom of each lane. (B) Bar graph shows dexamethasone- or dexamethasone-, GR-, and BRG1-K-R– induced change in percent cleavage in SacI hypersensitivity between 5555 and 3617 cells from A. (C) Expression of BRG1-K-R reduced the loading of RNA pol II to MMTV promoter. 5555 and 3617 cells were untreated or treated with 100 nM dexamethasone for 30 min and tetracycline as indicated. Chromatin was immunoprecipitated using an antibody specific for RNA pol II or no antibody (control). Immunoprecipitated and input DNA were amplified using primers specific to the MMTV promoter. (D) Expression of BRG1-K-R reduced the loading of RNA pol II to the dexamethasone-induced Rgs2 locus. 5555 cells were untreated or treated with 100 nM dexamethasone for 30 min and tetracycline as indicated. cDNA was prepared from RNAs isolated from the indicated conditions. Chromatin was immunoprecipitated using an antibody specific for RNA pol II or no antibody (control). Immunoprecipitated and input DNA or cDNA were amplified using primers specific to the Rgs2 coding region.
Immunofluorescence Microscopy
3134 and 5555 mouse mammary adenocarcinoma cells were grown on 22-mm2 glass coverslips in six-well plates. The cells were fixed in 4% paraformaldehyde and processed for indirect immunofluorescence microscopy as previously described (Parada et al., 2003). The primary antibodies used in this study included anti-BRG1 at 1:100 (provided by Weidong Wang, National Institutes of Health, Bethesda, MD, and by Anthony Imbalzano); anti-ISWI (Snf2h) at 1:200 (provided by Ramin Shiekhattar, Wistar Institute, Philadelphia, PA); and polyclonal anti-BRM and anti-Flag at 1:100 (provided by Anthony Imbalzano). We used species-specific secondary antibodies designed for simultaneous multiple labeling (Jackson ImmunoResearch Laboratories, West Grove, PA). Secondary antibodies were conjugated to FITC or Texas Red. Images were acquired with narrow-band-pass emission filters (Chroma Technology, Rockingham, VT). DNA was stained with DAPI (Invitrogen), and the cells were mounted using Prolong Gold mounting solution (Invitrogen). Cells were imaged on an Olympus IE80 inverted microscope equipped with a 100× 1.35 NA oil immersion objective (Melville, NY) and a Photometrics CCD camera configured at 0.07-μm pixels (Tucson, AZ). Images were analyzed by using Metamorph software (Universal Imaging, Sunnyvale, CA). Colocalization of two distributions were verified by linescan analysis as previously described (Elbi et al., 2002). A line was drawn through a colocalized region and fluorescence intensity peaks from two distributions were measured and then plotted using Metamorph software. Colocalization of the signals were confirmed by examining consecutive optical sections above and below the midplane optical sections covering the entire depth of the cell nuclei.
RNA FISH
3134 and 5555 cell lines were grown on 22-mm2 glass coverslips in six-well plates. Cells were fixed in 4% paraformaldehyde and processed for indirect immunofluorescence microscopy as described above. This was followed by a RNA FISH procedure to detect MMTV transcripts as described previously (Parada et al., 2003; Rayasam et al., 2005). All images were acquired with the same exposure times in order to compare across different treatment conditions. The RNA FISH signals were quantified using MetaMorph software. Thirty five cells from each treatment or control group were randomly selected. Background nuclear fluorescence intensity was subtracted from the RNA FISH fluorescence intensity in each cell. The regions defined by the RNA FISH fluorescence signals were identified by thresholding and the pixel intensities in the regions were averaged to compare across the different conditions. The average integrated intensities were plotted as a bar histogram, with error bars representing SE. We performed one way ANOVA (SPSS software) on all data sets. Where warranted by ANOVA results (p < 0.05), Student-Newman-Keuls post hoc tests (SPSS software) were applied to detect differences (p < 0.05) between experimental conditions.
DNA FISH
3134 cells were grown on 22-mm2 glass coverslips in six-well plates. Cells were fixed in 4% paraformaldehyde and processed for DNA FISH analysis to detect MMTV DNA using a probe specific for the MMTV-LTR array as previously described (Mueller et al., 2001). This was followed by indirect immunofluorescence microscopy. All images were acquired with the same exposure times in order to compare between different treatment conditions. The DNA FISH signals were quantified using MetaMorph software. Thirty-five cells from transfected or untransfected (control) group were randomly selected. Background nuclear fluorescence intensity was subtracted from DNA FISH fluorescence intensity in each cell. The regions defined by the DNA FISH fluorescence signals were identified by thresholding and the areas of MMTV arrays were measured. The areas from each experimental condition were averaged and plotted as a bar histogram, with error bars representing SE. We performed a one-way ANOVA (SPSS software) on all data sets. Where warranted by ANOVA results (p < 0.05), Student-Newman-Keuls post hoc tests (SPSS software) were applied to detect differences (p < 0.05) between experimental conditions.
Fluorescence Recovery after Photobleaching and Image Analysis
1361.5 cells were grown in Lab-Tek one-well chamber slides (Nalge Nunc International, Naperville, IL) for live cell fluorescence recovery after photobleaching (FRAP) experiments. Cells were transfected with YFP-BRG1, YFP-BRG1-K-R, and GFP-BRM and treated with 100 nM dexamethasone for 30 min. FRAP analysis was carried out on a Zeiss 510 laser-scanning confocal microscope (Thornwood, NY). The stage temperature was maintained at 37°C, and images were acquired with a 100× 1.3 NA oil immersion objective and 40 mW argon laser. Five single prebleach images were acquired followed by a brief bleach pulse of 160 ms using 458/488/514-nm laser lines at 100% laser power (laser output, 50%) without attenuation. Single optical sections were acquired at 490-ms intervals by using a 488-nm laser line with laser power attenuated to 0.1%. In all FRAP experiments, signal loss during the recovery period was <5% of the initial fluorescence intensity. The bleach extent and depth were confirmed by analyses of three-dimensional image stacks along the Z-plane of the image axis of fixed cells. Fluorescence intensities in the regions of interest were analyzed, and quantitative FRAP recovery curves were generated using LSM software and Microsoft Excel (Redmond, WA) as previously described (Elbi et al., 2004a). Pseudocolor images of the MMTV array and the area of the bleached region were generated using MetaMorph software. All FRAP recovery curves were generated from background subtracted images, and all quantitative data for FRAP recovery kinetics represent means ± SE from at least 25 cells imaged in three independent experiments.
RESULTS
SWI/SNF Chromatin Remodelers Potentiate the Hormone-dependent Activation of the MMTV-LTR
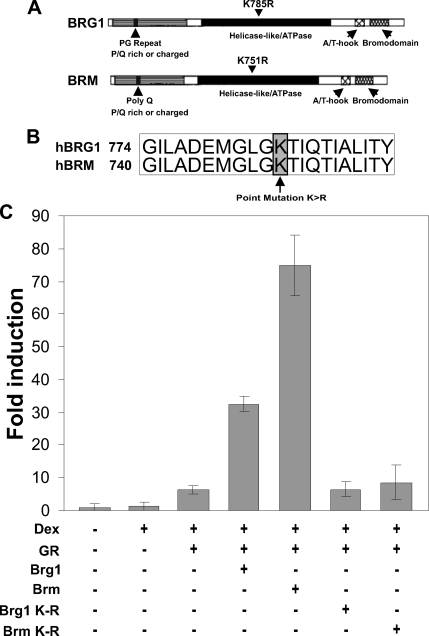
The SWI/SNF chromatin-remodeling complex contributes to the regulation of gene expression by either transcriptional activation or transcriptional repression depending on genetic context and cellular environment (Muchardt and Yaniv, 2001; Klochendler-Yeivin et al., 2002). Although the SWI/SNF complex consists of multiple subunits, the catalytic subunits, BRG1 and BRM, play a key role in chromatin remodeling by utilizing ATP hydrolysis (Peterson, 2002; Narlikar et al., 2002; Roberts and Orkin, 2004). To establish the role of the SWI/SNF complex in the transcriptional regulation of the MMTV-LTR, we carried out transcription reporter assays in the human adrenal carcinoma cell line (SW13). The SW13 cell line expresses all the BAFs and the SWI/SNF complex can be reconstituted by transfecting BRG1 or BRM into this cell line (Muchardt and Yaniv, 2001; Hsiao et al., 2003). This model system has allowed us to assess the contribution of each chromatin remodeling protein to the hormone-dependent regulation of MMTV transcription. Transfection of SW13 cells with a MMTV-Luciferase reporter plasmid and a GR expression plasmid resulted in an eightfold increase of MMTV transcription in the presence of hormone (Figure 1C). However, the transfection of GR along with BRG1 or BRM expression plasmids potentiated the hormone response to 33- and 77-fold, respectively (Figure 1C). ATPase-deficient forms of BRG1 or BRM (BRG1-K-R or BRM-K-R) contain a lysine-to-arginine mutation in the ATP binding pocket (Figure 1, A and B) that abrogates ATP hydrolysis but retains the ability to efficiently incorporate into multisubunit SWI/SNF-like complexes (Rayasam et al., 2005). Consequently, when overexpressed, these mutants function as dominant negatives (de la Serna et al., 2000). Transfection of SW13 cells with ATPase-deficient forms of BRG1 or BRM compromised transcription dramatically (to eightfold), suggesting that optimal MMTV transcription in SW13 cells required a chromatin remodeling competent BRG1 or BRM (Figure 1C). These results suggest that the SWI/SNF family of chromatin remodelers are important regulators in the hormone-dependent activation of the MMTV-LTR.
Figure 1.
SWI/SNF chromatin remodelers potentiate the hormone-dependent activation of the MMTV-LTR. (A) Schematic representation of BRG1 and BRM with conserved domains. (B) Location of lysine to arginine point mutations in the highly conserved ATPase domain of BRG1 and BRM. This mutation abolishes the ability of BRG1 and BRM to hydrolyze ATP and remodel chromatin. (C) BRG1 and BRM potentiate the transcriptional activity of MMTV in BRG1-, BRM-, and GR-deficient human adrenal carcinoma cells (SW13). SW13 cells were transfected with MMTV-LTR-Luciferase and pCMV β-galactosidase (internal control) along with pGR, pBRG1, pBRM, pBRG1-K-R, or pBRM-K-R expression vectors. The cells were treated with 100 nM dexamethasone or vehicle control for 4 h. Luciferase reporter gene activity was assayed and normalized to β-galactosidase reporter gene activity. The data shown is from two independent experiments.
BRG1 and BRM Chromatin-remodeling Complexes Are Selectively Recruited to the MMTV Array in a Hormone-dependent Manner
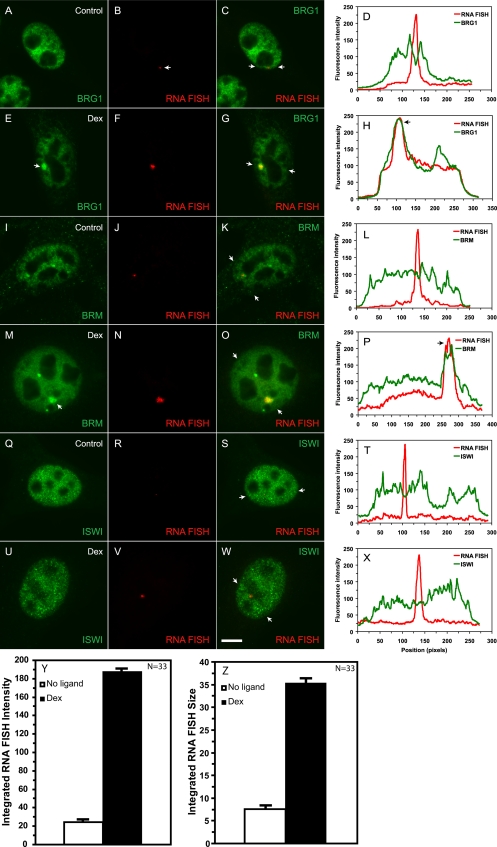
To probe the mechanisms of transcriptional activation by the SWI/SNF complex, we first determined whether BRG1 or BRM can be directly recruited to the MMTV array in vivo by using indirect immunofluorescence microscopy combined with RNA FISH. In 3134 (murine mammary adenocarcinoma) cells, 200 copies of the MMTV-LTR with 800–1200 GR-binding sites are stably integrated in a head-to-tail orientation near the centromere of chromosome 4. Previous studies have indicated that the hormone response of the MMTV-LTR array as a unit is indistinguishable from that of a single integrated copy of the viral LTR (Fragoso et al., 1998; Kramer et al., 1999; McNally et al., 2000). The array, therefore, provides a good model system to examine the in vivo recruitment of chromatin-remodeling complexes to their target promoter. RNA FISH analysis in 3134 cells detects a low level of basal transcription from the MMTV array (Figure 2, B, J, and R); however, the intensity and size of the RNA FISH signal increases dramatically in dexamethasone-treated cells, indicative of GR-mediated activation of the promoter (Figure 2, F, N, V, Y, and Z). In the absence of hormone, endogenous BRG1 or BRM was diffusely distributed in the nucleoplasm and no enrichment of the remodeling proteins was observed at the MMTV array (Figure 2, A–C and I–K). In cells treated with dexamethasone, a single bright and large BRG1 or BRM immunofluorescence signal was detected within the nucleoplasm in addition to a diffuse nuclear distribution (Figure 2, E–G and M–O). The strong fluorescence focal signal was completely coincident with the MMTV RNA FISH signal suggesting that BRG1 and BRM are recruited at the MMTV array in a hormone-dependent manner (Figure 2, G and O). This colocalization was confirmed by a linescan analyses in which BRG1 or BRM fluorescence intensity peaks and nascent MMTV transcripts were shown to be coincident in hormone-treated cells (Figure 2, H and P). In contrast, the distribution of endogenous Snf2h, the catalytic subunit belonging to the imitation switch family of remodeling proteins (ISWI), remained unchanged in the presence or absence of hormone (Figure 2, Q–S and U–W). A strong but diffuse nucleoplasmic staining pattern was noted for ISWI, but unlike BRG1 or BRM, this remodeling protein was not enriched at the array upon dexamethasone treatment, suggesting that hormone-dependent activation of the MMTV promoter involves the selective and class-specific recruitment of remodeling proteins (Figure 2, T and X). The involvement of multiple members of the SWI/SNF family in transcriptional regulation is similar to what has been previously described for the hsp70 promoter (de la Serna et al., 2000). BRG1 and BRM may have separate and distinct roles in the transcriptional process; this is yet to be determined. Alternatively, the recruitment of BRG1 or BRM by GR may be mediated by proteins shared between the BRG1 and BRM complexes. This may enable GR to bring either BRG1- or BRM-containing SWI/SNF complexes to a target promoter in a functionally redundant manner. Recent work has shown that GR makes direct contacts not necessarily with BRG1 but with the BRG1- or BRM-associated factors, BAF 57 and BAF60a (Hsiao et al., 2003).
Figure 2.
BRG1 and BRM but not ISWI chromatin remodeling activities are recruited to the MMTV array in a hormone-dependent manner. Mouse mammary adenocarcinoma cells (3134) with a single, stably integrated MMTV-LTR array were treated with 100 nM dexamethasone for 30 min. Fixed cells were probed with specific antibodies to detect endogenous BRG1, BRM, or ISWI by indirect immunofluorescence. MMTV RNA was detected by a RNA FISH using a probe specific to the MMTV transcript. The arrows in E and M point to the immunofluorescence detected localization of BRG1 and BRM on the MMTV array. The overlays (yellow) in C, G, K, O, S, and W indicate colocalization of the immunofluorescence and RNA FISH signals. The arrows in C, G, K, O, S, and W point to the end positions of linescans. Linescan analyses in D, H, L, P, T, and X quantitatively show the ligand-dependent recruitment or lack thereof of the BRG1, BRM, or ISWI chromatin-remodeling complexes to the MMTV-LTR array. The linescan in L runs through the RNA FISH signal that is adjacent but not coincident with the BRM fluorescence intensity peak. In H, P, and X, fluorescence intensity peaks for BRG1 and BRM but not for ISWI coincided with MMTV RNA. Bar, 4 um. Treatment of cells with 100nM dexamethasone for 30 minutes increases the intensity (Y) as well as the size (Z) of the MMTV RNA FISH signal.
Transcription from the MMTV-LTR Is Regulated by the ATP-Hydrolysis Activity of BRG1 or BRM
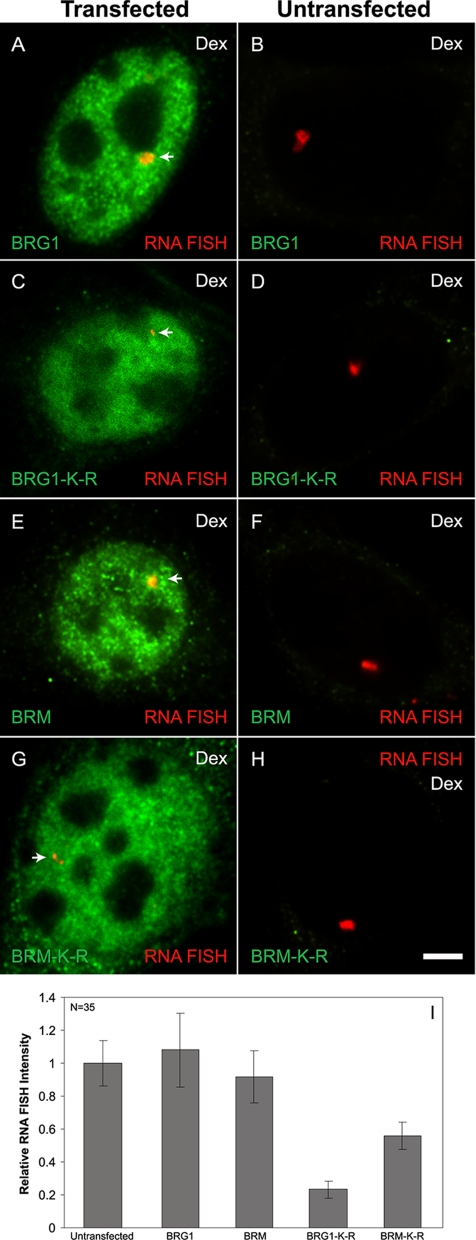
To gain more insight into the functional role of BRG1 or BRM at an integrated GR responsive gene, we evaluated directly the effect of BRG1-K-R or BRM-K-R on transcription at the MMTV array in 3134 cells. 3134 cells, which express endogenous GR, BRG1, and BRM, were transfected with Flag-tagged BRG1, BRM, BRG1-K-R, or BRM-K-R expression vectors. The cells were treated with dexamethasone and analyzed by quantitative RNA FISH combined with anti-Flag indirect immunofluorescence microscopy to distinguish transfected from untransfected cells. In BRG1- or BRM-transfected cells, dexamethasone treatment yielded RNA FISH signals of comparable intensity to untransfected cells (Figure 3, A–B and E–F); hence, overexpression of wild-type BRG1 or BRM did not significantly enhance or inhibit expression from the MMTV array. In BRG1-K-R– or BRM-K-R–transfected cells, the size and the fluorescence intensity of RNA FISH signals were greatly reduced in comparison to neighboring untransfected cells (Figure 3, C-D and G-H). It has been shown that the size and the fluorescence intensity of the MMTV array, as measured by RNA FISH, is correlated with the level of MMTV transcription (Mueller et al., 2001). Quantitation of the RNA FISH signal, from BRG1-K-R– or BRM-K-R–transfected cells, showed an 81 and 45% decrease in MMTV transcription, respectively (Figure 3I). A combined ANOVA analysis and Student-Newman-Keuls post hoc test showed that the BRG1-K-R– and BRM-K-R–transfected cells have statistically significant reduction in RNA FISH signals, whereas the wild-type BRG1-transfected cells do not vary from untransfected populations (p < 0.05). These results demonstrate that BRG1-K-R and BRM-K-R function as dominant negative proteins by interfering with the chromatin-remodeling function of endogenous BRG1 and BRM and as a consequence inhibit hormone-dependent transcription on an integrated MMTV promoter.
Figure 3.
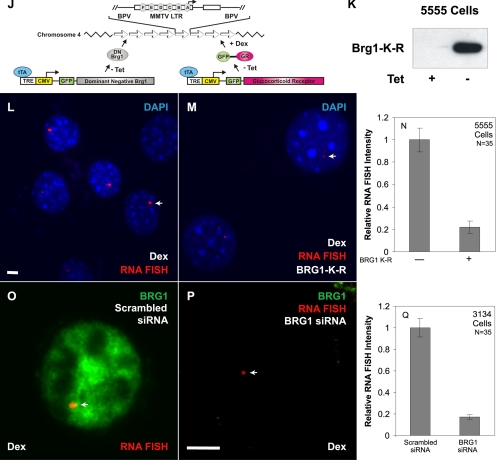
Transcription from the MMTV-LTR is regulated by the ATP-hydrolysis activity of BRG1 or BRM. 3134 mouse mammary adenocarcinoma cells with a stably integrated MMTV-LTR array were transfected with Flag-tagged forms of wild-type BRG1, BRM, and mutant BRG1-K-R, BRM-K-R (A–H). Cells were treated with 100 nM dexamethasone for 30 min and fixed. BRG1, BRG1-K-R, wild-type BRM, and BRM-K-R were detected by indirect immunofluorescence using an anti-Flag antibody and MMTV RNA was detected by RNA FISH using a probe specific to the MMTV transcript. Arrows (A, C, E, and G) point to the localization of MMTV transcript. Average integrated RNA FISH intensities from 35 randomly selected cells in each transfected category (A, C, E, and G) and untransfected category (B, D, F, and H) were measured as described in Materials and Methods and plotted as a bar histogram (I). Error bars, SE. (J–N) A stably expressed dominant negative form of BRG1 (BRG1-K-R) inhibits MMTV transcription. The 5555 mouse mammary adenocarcinoma cells with stably integrated MMTV-LTR array express a Flag-tagged dominant negative form of BRG1 (BRG1-K-R) under the control of a tetracycline-repressible system (J). 5555 cells were grown in the presence or absence of tetracycline to facilitate the expression of BRG1-K-R (K). Western blot analysis was performed using an anti-Flag antibody. 5555 cells were grown in the presence (L) or absence (M) of tetracycline. Cells were treated with 100 nM dexamethasone for 30 min, fixed, and processed for RNA FISH analysis using a probe specific for MMTV transcript. Arrows point to the localization of the MMTV transcript. DNA was stained by DAPI. Average integrated RNA FISH intensity from 35 randomly selected cells grown in the presence (−BRG1 K-R) or absence (+BRG1 K-R) of tet were measured as described in Materials and Methods and plotted as a bar histogram (N). Error bars, SE. siRNA-mediated depletion of BRG1 inhibits MMTV transcription (O–Q). 3134 cells with stably integrated MMTV-LTR array were transfected with a scrambled sequence (O) or with a siRNA pool targeted to BRG1 (P). Cells were treated with dexamethasone for 30 min, fixed, and processed for RNA FISH analysis using a probe specific for MMTV transcripts. Endogenous BRG1 was detected by indirect immunofluorescence using a BRG1-specific antibody. Arrows point to the localization of MMTV transcript. Average integrated RNA FISH intensity of 35 randomly selected cells from cells transfected with siRNAs to BRG1 or cells transfected with scrambled siRNAs were measured and plotted as a bar histogram (Q). Error bars, SE. Bar, 4 um.
Dominant negative BRM was consistently a weaker effector of transcription than dominant negative BRG1 (Figure 3I). Purified BRM complexes have been previously shown to be weaker chromatin remodelers than purified BRG1 complexes (Sif et al., 2001). This observation along with our results suggested a more pronounced role for BRG1 in the transcriptional process at the MMTV promoter. Consequently, we focused our efforts on the study of BRG1 by generating a mouse mammary adenocarcinoma cell line that stably expressed BRG1-K-R and GR under the control of a tetracycline-repressible promoter (cell line 5555; Figure 3J). Western blot analysis of total cell lysates prepared from 5555 cells showed a robust induction of BRG1-K-R expression upon removal of tetracycline from culture medium (Figure 3K). We directly assessed the functional role of dominant negative BRG1 on MMTV transcription in vivo by growing the 5555 cell line in the presence (Figure 3L) or absence (Figure 3M) of tetracycline and then challenged these cells with dexamethasone. The expression of BRG1-K-R reduced the robust hormone-dependent RNA FISH signal by 80% (Figure 3, L–N).
ANOVA analysis of the RNA FISH data demonstrated a statistically significant difference between the BRG1-K-R–expressing and nonexpressing conditions (p < 10−8).
To eliminate the possibility that the inhibition of MMTV transcription in the 5555 cell line is merely an outcome of overexpression of an exogenous protein, we used siRNA-mediated gene silencing to knock down endogenous BRG1 in the MMTV array–containing cell line (3134). The absence of nuclear BRG1 immunostaining in cells transfected with a pool of siRNAs designed against BRG1 (Figure 3P) validated the effectiveness of these siRNAs. 3134 cells transfected with siRNAs specific to BRG1 show MMTV RNA FISH signals that were smaller in size and intensity to cells transfected with a scrambled control siRNA (Figure 3, O and P). Quantitation of RNA FISH signal intensities obtained from each transfected group of 3134 cells showed an 84% decrease in the level of MMTV transcription in BRG1 depleted cells (Figure 3Q). ANOVA analysis of the RNA FISH data demonstrated a statistically significant difference between BRG1 siRNA-treated cells and scrambled siRNA -transfected cells (p < 10−8). Interestingly, transcriptional inhibition generated by siRNA-mediated silencing of endogenous BRG1 expression was very similar to the transcriptional inhibition obtained by either the stable or transient expression of dominant negative BRG1 (Figure 3, I, N, and Q). We conclude that interfering with the function of endogenous BRG1 either by the expression of a dominant negative form of BRG1 or by the siRNA-mediated depletion of endogenous BRG1 dramatically compromises transcription from the MMTV promoter.
BRG1 Is Required for the Hormone-dependent Remodeling of MMTV Chromatin and Loading of RNA pol II to the MMTV Promoter
The data presented in Figures 1–3 demonstrate the hormone-dependent recruitment of BRG1 to the MMTV array and the involvement of BRG1 mediated chromatin remodeling in the transcriptional activation of the MMTV promoter. Data from our lab and others have demonstrated that activation of the MMTV promoter by hormone, results in the binding of GR to hormone response elements (GREs) within the nucleosome B-C region of the MMTV promoter (Fragoso et al., 1998; Fryer and Archer, 1998; Fletcher et al., 2002). In the presence of hormone, this region becomes more accessible to a variety of chemical and enzymatic nucleases, the hallmark of a chromatin-remodeling event. We and others have used the restriction endonuclease, SacI, which cuts within the nucleosome B-C region, as a measure of this chromatin transition (Fragoso et al., 1998; Fryer and Archer, 1998; Fletcher et al., 2002). We compared the extent of SacI cleavage in 3617 mouse mammary adenocarcinoma cells (which stably expresses GFP-GR in a tetracycline-repressible system; Walker et al., 1999) with SacI cleavage in the 5555 cell line (generated by stable transfection of 3617 cells with a Flag-tagged BRG1-K-R under the control of the same tetracycline regulator). Both 3617 and 5555 cells express endogenous GR and BRG1. 3617 cells grown in the presence of tetracycline (no expression of exogenous GR) showed an increase in fractional cleavage of 9% at the SacI site, in response to hormone (Figure 4, A and B). Chromatin remodeling in 3617 cells was further increased to 20% in response to hormone when cells were grown in the absence of tetracycline. This increase in cutting most likely reflects the contribution of the additional GR that is expressed in the absence of tetracycline. The additional GR can recruit more BRG1 or BRM to the MMTV array, which in turn can make chromatin even further accessible. The extent of SacI cutting in the presence of tetracycline was similar in both the 3617 and 5555 cell lines (Figure 4, A and B). However, when 5555 cells were grown in the absence of tetracycline, under conditions that induce the expression of BRG1-K-R (and GR), the extent of SacI cutting was diminished to 6%; in contrast to a fractional cleavage of 20% observed in 3617 cells under similar conditions. These results suggest that effective remodeling of MMTV chromatin requires BRG1 with a functional ATPase domain.
To further establish a functional connection between remodeling at the MMTV promoter and transcription from the MMTV promoter, we probed for pol II loading by ChIP under the same conditions used in the chromatin accessibility assay (Figure 4, A and B). 3617 and 5555 cells grown in the presence of tetracycline (no expression of BRG1-K-R) showed a robust increase in the loading of RNA pol II in response to hormone treatment (Figure 4C, lane 2 vs. 4 and lane 6 vs. 8). In contrast, expression of BRG1-K-R in 5555 cells, resulted in a marked decrease in the hormone-dependent loading of RNA pol II (cf. Figure 4C, lanes 1 and 2), whereas tetracycline withdrawal in 3617 cells, in fact, showed a modest increase in pol II loading. These results show that changes in chromatin transitions correlate with changes in the level of RNA pol II loading at the MMTV promoter, thereby providing a functional link between BRG1, chromatin remodeling, and transcription at a hormone-activated promoter. We have extended these studies and examined the contribution of BRG1 to other GR-regulated genes. We find that the robust hormone-dependent induction of transcription from the Rgs2, regulator of G-protein silencing 2, locus (Figure 4D, lane 6 vs. 8) is also accompanied by an increase in pol II occupancy (Figure 4D, lane 2 vs. 4). In contrast, expression of BRG1-K-R in 5555 cells resulted in a significant decrease of hormone-dependent transcription (Figure 4D, lane 5 vs. 6) as well as hormone-dependent pol II occupancy (Figure 4D, lane 1 vs. 2) at the Rgs2 locus. These results further confirm and extend our findings and implicate BRG1 in the regulation of additional GR-regulated genes.
Large-Scale Chromatin Decondensation and Condensation Are Regulated by BRG1 and BRM Chromatin-remodeling Complexes
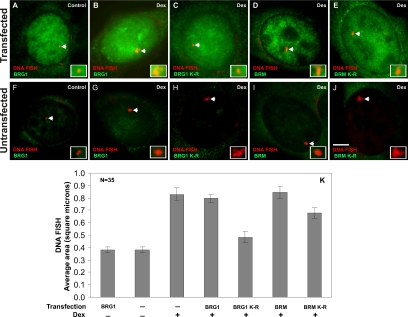
Results obtained from chromatin accessibility experiments (Figure 4) provide limited information on the nature of chromatin remodeling in vivo. We assessed the impact of chromatin-remodeling complexes on the large-scale MMTV chromatin structure and topology in 3134 cells. 3134 cells contain a 2-Mb stably integrated array containing 200 copies of the MMTV-LTR. Combining DNA FISH and indirect immunofluorescence microscopy, Mueller et al. (2001) observed that hormone treatment resulted in an increase of the size of the array, suggesting that the array decondenses concomitant with transcriptional activation. To address the contribution of remodeling proteins in array decondensation, 3134 cells were transfected with BRG1, BRG1-K-R, BRM, or BRM-K-R, treated with dexamethasone, and processed for DNA FISH analysis combined with indirect immunofluorescence microscopy. As previously observed, in BRG1- or BRM-transfected cells, a decondensation of the array was detected in response to hormone (Figure 5, B and D; Mueller et al., 2001). In contrast, in BRG1-K-R–transfected cells we detected a relative decrease in the size of large arrays even in the presence of hormone, suggesting that the inability to remodel chromatin resulted in a less pronounced decondensation event (cf. Figure 5, C to B). A similar decrease in the size of large arrays was also detected in BRM-K-R–transfected cells, although it was far less in magnitude in comparison to BRG1-K-R (cf. Figure 5, E to D). The effect on the size of the MMTV array was specific to cells transfected with the various remodelers, because in the neighboring untransfected cells, robust and large DNA FISH signals were observed (Figure 5, F–K). ANOVA analysis and Student-Newman-Keuls post hoc tests show that the BRG1-K-R– and BRM-K-R–transfected cells have a statistically significant reduction in the size of DNA FISH signals, whereas the wild-type BRG1-transfected cells do not vary from untransfected cells (p < 0.05). The size of the DNA FISH signals in BRG1-K-R–transfected cells also differs significantly from the DNA FISH signals in BRM-K-R–transfected cells. We conclude that the expression of dominant negative BRG1 or dominant negative BRM inhibits the hormone-induced, large-scale decondensation of the 2-Mb MMTV array. The effect of BRG1 on chromatin remodeling appears to be more pronounced than that of BRM. These findings provide a strong in vivo correlation between chromatin remodeling, chromatin decondensation, and transcription.
Figure 5.
Large-scale chromatin decondensation is regulated by BRG1 and BRM chromatin-remodeling complexes. 3134 cells were transfected with Flag-tagged forms of wild-type BRG1 (A and B), mutant BRG1-K-R (C), wild-type BRM (D), and mutant BRM-K-R (E). Cells were treated with 100 nM dexamethasone for 90 min, fixed, and processed for DNA FISH analysis. BRG1, BRG1-K-R, BRM, and BRM-K-R were detected by indirect immunofluorescence using an anti-Flag antibody and MMTV DNA was detected by DNA FISH using a probe specific to the entire MMTV-LTR array. The average DNA FISH signal areas obtained from 35 randomly selected cells in the transfected (A–E) and untransfected (F–J) populations were measured and plotted as a bar histogram (K). Error bars, SE. The inset rectangle shows an enlarged image of the DNA FISH signal. Expression of BRG1-K-R and BRM-K-R prevent hormone-induced decondensation of MMTV chromatin. Bar, 4 um.
Chromatin-remodeling Complexes Have Distinct Kinetic Properties at Their Target Gene and Dynamically Associate with the MMTV Array in a Hormone-dependent Manner
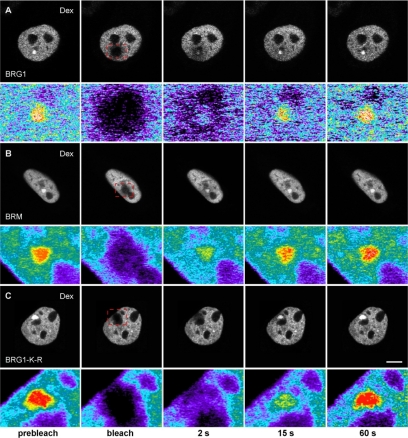
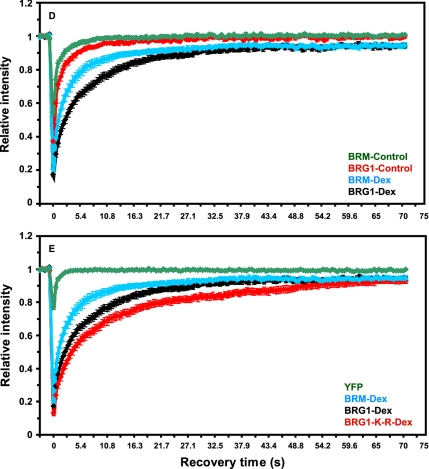
The dynamics of chromatin-remodeling complexes bound to a specific target site on a regulated promoter or the modulation of the kinetic properties of chromatin-remodeling complexes in response to hormone has never been demonstrated in living cells. In fact, this modulation is one of the key mechanisms essential for the functional role of nuclear hormone receptors (Stenoien et al., 2001; Schaaf and Cidlowski, 2003; Stavreva et al., 2004; Elbi et al., 2004b; Farla et al., 2005; Rayasam et al., 2005). In vivo FRAP has been used to study the dynamic properties of chromatin proteins and that the FRAP recovery kinetics of chromatin proteins are directly related to their chromatin-binding properties (Lefebvre et al., 1991; Fragoso et al., 1998; Lever et al., 2000; Kimura and Cook, 2001; Kimura et al., 2002; Maruvada et al., 2003; Phair et al., 2004; Becker et al., 2005; Chen et al., 2005). Because BRG1 and BRM were selectively recruited to the MMTV array in response to hormone (Figures 2 and 3), we used FRAP to study the binding kinetics of chromatin-remodeling complexes at the amplified MMTV target in vivo. Array-containing cells were transfected with YFP- or GFP-tagged BRG1, BRM, BRG1-K-R, or YFP alone and treated with dexamethasone for 30 min. Consistent with the immunofluorescence experiments (Figure 2), a clear enrichment of YFP- or GFP-tagged BRG1, BRM, or BRG1-K-R was observed at the MMTV array when cells were treated with hormone (Figure 6, A–C). BRG1, BRM, or BRG1-K-R bound to the MMTV array was bleached using a brief laser pulse. The recovery of fluorescence signal in the bleached region was monitored using in vivo time-lapse confocal microscopy. We correlated the dynamics of array-bound BRG1 and array-bound BRM with the dynamics of BRG1 and BRM distributed in the nucleoplasm. As a standard we used YFP, which moves freely in the nucleoplasm, and as anticipated the recovery of YFP fluorescence was very rapid, reaching 90% of prebleach levels in <0.4 s (Figure 6E). The recovery kinetics of YFP-BRG1 on the MMTV array (Figure 6D, BRG1-Dex) in response to hormone was significantly slower than YFP-BRG1 in the nucleoplasm (Figure 6D, BRG1-Control) in the absence of hormone with t1/2 of 3.9 ± 0.49 and 0.9 ± 0.25 s, respectively (p < 0.001; Figure 6, A and D). A similar difference in kinetic behavior was observed with array-bound BRM and nucleoplasmic BRM. Nucleoplasmic BRG1 or BRM presumably represent nonspecific DNA-binding events (Karpova et al., 2004). The observed difference in the recovery kinetics of BRG1 and BRM on the MMTV array (Figure 6D, BRG1-Dex vs. BRM-Dex) in response to hormone was significant, with t1/2 of 3.9 ± 0.49 and 1.95 ± 0.46 s, respectively (p < 0.001), suggesting a stronger interaction of BRG1 with MMTV chromatin than BRM. These interaction differences between BRG1 and BRM might account for their differential effects on transcription and remodeling. Interestingly, in the presence of dexamethasone the recovery kinetics of BRG1-K-R on the MMTV array was the slowest, reaching 50% of prebleach levels within 5.5 s (t1/2 of 5.49 ± 0.86; Figure 6, C and E, BRG1-K-R-Dex), suggesting an ATP-dependent functional interaction of BRG1 with MMTV chromatin. Cells expressing different levels of the fluorescently tagged proteins showed similar FRAP recovery curves (Supplementary Figure S1) indicating that differential expression levels cannot account for the kinetic differences observed in FRAP experiments. Our photobleaching experiments suggest that BRG1, BRM, and BRG1-K-R chromatin-remodeling complexes have distinct kinetic properties on the MMTV array and dynamically associate with and dissociate from MMTV chromatin in vivo in a hormone and ATP-dependent manner.
Figure 6.
Chromatin remodeling complexes have distinct kinetic properties and dynamically associate with MMTV-LTR array in a ligand and ATPase-dependent manner. (A–C) Qualitative FRAP analysis of BRG1, mutant BRG1-K-R and BRM in 1365.1 cells. 1365.1 mouse fibroblast cells were transfected with YFP-BRG1, YFP-BRG1-K-R, or GFP-BRM and treated with 100 nM dexamethasone for 30 min. BRG1 (A), BRM (B), or BRG1-K-R (C) bound to the MMTV-LTR array was imaged before and during recovery after photobleaching of the array for 120 ms. Images were acquired at the indicated times after the end of the bleach pulse. The MMTV-LTR array and the area of the bleached region is indicated by a red rectangle and shown as an enlarged pseudocolor image in the bottom panels. (D) Quantitative FRAP analysis of YFP-BRG1 or GFP-BRM in the nucleoplasm (control) or bound to the MMTV-LTR array after treatment with dexamethasone for 30 min (Dex). BRG1 and BRM bound to the MMTV-LTR array showed slower recovery kinetics after ligand treatment. (E) Quantitative FRAP analysis of YFP-BRG1, YFP-BRG1-K-R, or GFP-BRM bound to the MMTV-LTR array after treatment with dexamethasone for 30 min. The recovery kinetics of mutant BRG1-K-R bound to the MMTV array was slower than the wild-type BRG1 or wild-type BRM bound to MMTV-LTR array. All quantitative data values in the FRAP studies represent averages ± SE from at least 25 cells imaged in three independent experiments. Bar, 4 um.
DISCUSSION
It is well-established that the remodeling of chromatin structure is an essential process that has a profound effect on basic cellular functions including transcription, DNA recombination, repair, and replication (Fletcher and Hansen, 1996; Fyodorov and Kadonaga, 2001; Becker et al., 2002; Elgin and Workman, 2002). The mechanisms of gene activation are highly complex and because of this complexity most studies focus on individual events using biochemical or genetic approaches. Here our study provides an integrated kinetic view of gene activation events on a target gene in vivo involving hormone-dependent recruitment of chromatin- remodeling complexes, dynamic interaction of chromatin-remodeling complexes with a target promoter, chromatin remodeling, regulation of higher order chromatin structure, RNA pol II loading, and transcriptional activation. Our results demonstrate that individual gene regulatory events are coordinated in vivo by members of the SWI/SNF chromatin-remodeling complex thereby providing a mechanistic basis for BRG1 and BRM chromatin-remodeling complexes in the transcriptional process.
The 3134 (murine mammary adenocarcinoma) cell line contains 200 copies of the MMTV-LTR array stably integrated in a head-to-tail orientation at a single integration event near the centromere of chromosome 4 (Kramer et al., 1999). The hormone responsiveness of the MMTV array is identical to that of a single copy MMTV promoter, thereby making it a useful model system to directly visualize gene expression events such as the recruitment of chromatin-remodeling complexes and nuclear receptors to a target promoter in real time (Fragoso et al., 1998; Fletcher et al., 2002). Belmont and colleagues (Memedula and Belmont, 2003) have used an amplified gene array based on the lac operator/repressor system to analyze the sequential recruitment of chromatin-remodeling complexes by the acidic activator VP16 to a condensed chromatin locus. Tsukamoto et al. (2000) and Janicki et al. (2004) have used a modified lac operator/repressor artificial array to demonstrate the recruitment of a lac repressor-VP16 chimera that resulted in chromatin decondensation. However in these studies, the contribution of chromatin-remodeling complexes on chromatin decondensation were not directly investigated. Here we extended these studies by observing the in vivo functional link between local chromatin remodeling, higher order chromatin reorganization, and transcriptional activation using various approaches including quantitative in vivo microscopy, chromatin accessibility, and decondensation assays as well as photobleaching approaches. Importantly, we have determined for the first time that BRG1 and BRM chromatin-remodeling complexes have distinct kinetic properties on the MMTV array, and they dynamically associate with and dissociate from MMTV chromatin in a manner dependent on hormone and a functional ATPase domain.
Three subclasses of ATP-dependent chromatin-remodeling complexes have been identified in mammalian cells: SWI/SNF, ISWI, and Mi-2/CHD (Narlikar et al., 2002). We find that the members of the SWI/SNF remodeling complex, BRG1 and BRM, are preferentially recruited to the MMTV promoter in a hormone-dependent manner. Under the same experimental conditions, we failed to detect any enrichment of the ISWI (Snf2h) chromatin-remodeling complex at the MMTV array (Figure 2). Although, we cannot define the molecular basis of this specificity, the subunit composition of individual chromatin-remodeling complexes is likely a contributory factor (Hsiao et al., 2003). We have confirmed the contribution of BRG1 and BRM ATPases in the transcriptional activation of the MMTV promoter by biochemical and imaging approaches in well defined genetic backgrounds. Using SW13 cells that are deficient in BRG1, BRM, and GR expression, we find that both BRG1 and BRM potentiated transcription by GR on a transiently introduced MMTV reporter template (Figure 1). Furthermore, transactivation required a functional BRG1, BRM, and ATP hydrolysis because the ATPase-deficient forms of BRG1 and BRM failed to stimulate transcription under similar conditions. The introduction of ATPase-deficient remodeling complexes can also dramatically compromise transcription from the stably integrated MMTV repeat (Figure 3). We have further confirmed our observations by siRNA-mediated silencing of endogenous BRG1 expression (Figure 3). At this point, we are unable to ascertain if BRG1 and BRM make distinct contributions to MMTV activation. BRG1 and BRM may have unique functions in the transcriptional process; alternatively, GR might be able recruit either BRG1 or BRM via shared BAFs. The use of cell lines lacking BRG1 or BRM might provide some insight into the complex(es) that contributes to MMTV activation.
ATP-dependent chromatin-remodeling complexes and histone-modifying complexes dynamically modulate chromatin structure both at the nucleosome as well as at a higher order level (Vignali et al., 2000; Jenuwein and Allis, 2001). We explored the consequences of ATPase-deficient remodeling proteins on chromatin structure by using a restriction enzyme accessibility assay to assess the disruption of local chromatin structure. Our studies demonstrated the expected hormone-dependent increase in restriction enzyme cutting in control cells (from 9 to 20%) compared with an inhibition of this hormone-dependent increase in endonuclease cutting in cells expressing the dominant negative form of BRG1 (Figure 4). Interestingly, our ChIP analysis showed that this reduction in chromatin remodeling in cells expressing BRG1-K-R was accompanied by a reduction RNA pol II loading and transcription. These experiments provide data that implicate chromatin remodeling by BRG1 as a necessary prerequisite for optimal transcription of the MMTV promoter.
We have also used quantitative DNA FISH analysis in conjunction with indirect immunofluorescence microscopy, to examine higher order chromatin reorganization events in vivo. We observed a large-scale chromatin decondensation, of the MMTV array, in response to hormone when wild-type BRG1 and BRM is expressed, as has been previously described (Mueller et al., 2001). When BRG1-K-R or BRM-K-R was expressed, the hormone-dependent decondensation events were inhibited significantly by BRG1-K-R and less so by BRM-K-R, in keeping with the differential transcriptional effects of these remodeling-deficient proteins (Figure 5). These findings suggest that chromatin remodeling mediated by BRG1 and BRM ATPases can lead to higher-order chromatin unfolding and reorganization and this, in turn, correlates well with increased transcription from the MMTV array.
The dynamics of BRG1 and BRM chromatin-remodeling complexes at a specific promoter and the modulation of their kinetic properties in response to environmental stimuli have never been demonstrated in native chromatin in living cells. In our study, we found that BRG1, BRM, and BRG1-K-R dynamically exchange at the MMTV promoter with distinct kinetic properties in a manner dependent on hormone and a functional ATPase domain. The dynamic exchange of remodeling proteins on the MMTV array are consistent with our in vitro results obtained from rapid UV laser cross-linking where purified SWI/SNF binds to and is displaced from purified MMTV chromatin (Fletcher et al., 2002; Nagaich et al., 2004). Because the FRAP recovery kinetics of chromatin proteins are directly related to their chromatin-binding properties (Lefebvre et al., 1991; Fragoso et al., 1998; Lever et al., 2000; Kimura and Cook, 2001; Hager et al., 2002; Kimura et al., 2002; Maruvada et al., 2003; Phair et al., 2004; Becker et al., 2005; Chen et al., 2005), we conclude that the remodeling proteins with the slowest exchange rate reside longest on the MMTV promoter and associate most strongly with MMTV chromatin. A comparison of the kinetic properties of chromatin-remodeling complexes revealed that BRG1 was more strongly associated with the MMTV array than BRM (Figure 6D). Interestingly, the remodeler with the slowest exchange rate is the dominant negative BRG1 (BRG1-K-R). Molecular chaperones have been demonstrated to regulate the dynamic properties of GR and PR in the nucleus and recently the high mobility group box 1 protein, HMGB1 has been found to influence the residence time of GR in chromatin (Stavreva et al., 2004; Wagner et al., 2004; Elbi et al., 2004a,b; Agresti et al., 2005). Considering that the dominant negative BRG1 (BRG1-K-R) is simply a single amino acid change in the ATPase domain, our study reveals the importance of ATP hydrolysis in the dynamic properties of BRG1 and BRM. Further studies will be necessary for a complete understanding of the regulation of chromatin protein dynamics and its role in gene expression.
ACKNOWLEDGMENTS
We acknowledge Jim McNally for valuable discussions on FRAP. We thank Pamela Badger and Anindya Hendarwanto for technical assistance on FRAP analysis and tissue culture. We also thank Weidong Wang (NIA, Baltimore), Tony Imbalzano (U Mass, Worcester), and Kevin Gardner (NIH, NCI) for providing antibodies to BRG1, BRM, and pol II, respectively. We also thank Christian Muchardt (CARS, France) for providing us with BRM expression constructs. Imaging was carried out in the Fluorescence Imaging Facility, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute (NCI). This research was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0123) on May 28, 2008.
REFERENCES
- Agresti A., Scaffidi P., Riva A., Caiolfa V. R., Bianchi M. E. GR and HMGB1 interact only within chromatin and influence each other's residence time. Mol. Cell. 2005;18:109–121. doi: 10.1016/j.molcel.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Baumann C. T., John S., Walker D., Vigneron M., McNally J. G., Hager G. L. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Becker A., Miyara F., Han Z., Kihara M., Brown D. T., Hager G. L., Latham K., Adashi E. Y., Misteli T. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol. Biol. Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B., Parker M. G. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell. 2003;114:277–280. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- Berger S. L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Biegel J. A., Zhou J. Y., Rorke L. B., Stenstrom C., Wainwright L. M., Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- Bresnick E. H., John S., Berard D. S., Lefebvre P., Hager G. L. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc. Natl. Acad. Sci. USA. 1990;87:3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Catez F., Lim J. H. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Carroll J. S., Brown M. Estrogen receptor target gene: an evolving concept. Mol. Endocrinol. 2006;20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. S., Elizondo L. I., Boerkoel C. F. Advances in chromatin remodeling and human disease. Curr. Opin. Genet. Dev. 2004;14:308–315. doi: 10.1016/j.gde.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- de la Serna I. L., Carlson K. A., Hill D. A., Guidi C. J., Stephenson R. O., Sif S., Kingston R. E., Imbalzano A. N. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna I. L., Carlson K. A., Imbalzano A. N. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- Dilworth F. J., Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Elgin S. C. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Elbi C., Walker D. A., Lewis M., Romero G., Sullivan W. P., Toft D. O., Hager G. L., DeFranco D. B. A novel in situ assay for the identification and characterization of soluble nuclear mobility factors. Science STKE. 2004a;2004:PL10. doi: 10.1126/stke.2382004pl10. [DOI] [PubMed] [Google Scholar]
- Elbi C., Walker D. A., Romero G., Sullivan W. P., Toft D. O., Hager G. L., DeFranco D. B. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA. 2004b;101:2876–2881. doi: 10.1073/pnas.0400116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbi C., Misteli T., Hager G. L. Recruitment of the Dioxin Receptor to Active Transcription Sites. Mol. Biol. Cell. 2002;13:2001–2015. doi: 10.1091/mboc.13.6.mk0602002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Workman J. L. Chromosome and expression mechanisms: a year dominated by histone modifications, transitory and remembered. Curr. Opin. Genet. Dev. 2002;12:127–129. doi: 10.1016/s0959-437x(02)00276-9. [DOI] [PubMed] [Google Scholar]
- Farla P., Hersmus R., Trapman J., Houtsmuller A. B. Antiandrogens prevent stable DNA-binding of the androgen receptor. J. Cell Sci. 2005;118:4187–4198. doi: 10.1242/jcs.02546. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fletcher T. M., Hansen J. C. The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- Fletcher T. M., Xiao N., Mautino G., Baumann C. T., Wolford R. G., Warren B. S., Hager G. L. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 2002;22:3255–3263. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G., Pennie W. D., John S., Hager G. L. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol. Cell Biol. 1998;18:3633–3644. doi: 10.1128/mcb.18.6.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J., Archer T. K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Fyodorov D. V., Kadonaga J. T. The many faces of chromatin remodeling: SWItching beyond transcription. Cell. 2001;106:523–525. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- Giangrande P. H., Kimbrel E. A., Edwards D. P., McDonnell D. P. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol. Cell. Biol. 2000;20:3102–3115. doi: 10.1128/mcb.20.9.3102-3115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I., Shiekhattar R. Chromatin modifiers and carcinogenesis. Trends Cell Biol. 2004;14:695–702. doi: 10.1016/j.tcb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Elgin S. C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- Hager G. L., Elbi C., Johnson T. A., Voss T. C., Nagaich A. K., Schiltz R. L., Qiu Y., John S. Chromatin dynamics and the evolution of alternate promoter states. Chromosome. Res. 2006;14:107–116. doi: 10.1007/s10577-006-1030-0. [DOI] [PubMed] [Google Scholar]
- Hager G. L., Elbi C. C., Becker M. Protein dynamics in the nuclear compartment. Curr. Opin. Genet. Dev. 2002;12:137–141. doi: 10.1016/s0959-437x(02)00278-2. [DOI] [PubMed] [Google Scholar]
- Hill D. A. Influence of linker histone H1 on chromatin remodeling. Biochem. Cell Biol. 2001;79:317–324. [PubMed] [Google Scholar]
- Hsiao P. W., Fryer C. J., Trotter K. W., Wang W., Archer T. K. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano A. N., Jones S. N. Snf5 tumor suppressor couples chromatin remodeling, checkpoint control, and chromosomal stability. Cancer Cell. 2005;7:294–295. doi: 10.1016/j.ccr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Janicki S. M., et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T. Exploring the transcription-chromatin interface. Genes Dev. 2000;14:1992–1996. [PubMed] [Google Scholar]
- Karpova T. S., Chen T. Y., Sprague B. L., McNally J. G. Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep. 2004;5:1064–1070. doi: 10.1038/sj.embor.7400281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Cook P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Sugaya K., Cook P. R. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A., Muchardt C., Yaniv M. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 2002;12:73–79. doi: 10.1016/s0959-437x(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Kramer P., Fragoso G., Pennie W. D., Htun H., Hager G. L., Sinden R. R. Transcriptional state of the mouse mammary tumor virus promoter can effect topological domain size in vivo. J. Biol. Chem. 1999;274:28590–28597. doi: 10.1074/jbc.274.40.28590. [DOI] [PubMed] [Google Scholar]
- Labrador M., Corces V. G. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Northrop J. P., Kuo M. H., Stallcup M. R. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P., Berard D. S., Cordingley M. G., Hager G. L. Two regions of the mouse mammary tumor virus LTR regulate the activity of its promoter in mammary cell lines. Mol. Cell Biol. 1991;11(5):2529–2537. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B., Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Lever M. A., Th'ng J. P., Sun X., Hendzel M. J. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution [see comments] Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Maruvada P., Baumann C. T., Hager G. L., Yen P. M. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J. Biol. Chem. 2003;278:12425–12432. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- McNally J. G., Mueller W. G., Walker D., Wolford R. G., Hager G. L. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Memedula S., Belmont A. S. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 2003;13:241–246. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- Metivier R., Penot G., Hubner M. R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. When the SWI/SNF complex remodels the cell cycle. Oncogene. 2001;20:3067–3075. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- Mueller W. G., Walker D., Hager G. L., McNally J. G. Large scale chromatin decondensation and recondensation in living cells and the role of transcription. J. Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland N. M., Soeth E., Smith C. L. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene. 2003;22:4807–4818. doi: 10.1038/sj.onc.1206722. [DOI] [PubMed] [Google Scholar]
- Nagaich A. K., Walker D. A., Wolford R. G., Hager G. L. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neely K. E., Workman J. L. Histone acetylation and chromatin remodeling: which comes first? Mol. Genet. Metab. 2002;76:1–5. doi: 10.1016/s1096-7192(02)00014-8. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Parada L. F., Elbi C. C., Dundr M., Misteli T. Gene expression. In: Davey J., Lord M., editors. Cell Function. Vol. 2. Oxford: Oxford University Press; 2003. pp. 47–77. [Google Scholar]
- Peterson C. L. Chromatin remodeling enzymes: taming the machines. Third in review series on chromatin dynamics. EMBO Rep. 2002;3:319–322. doi: 10.1093/embo-reports/kvf075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Workman J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Phair R. D., Scaffidi P., Elbi C., Vecerova J., Dey A., Ozato K., Brown D. T., Hager G. L., Bustin M., Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam G. V., Elbi C., Walker D. A., Wolford R. G., Fletcher T. M., Edwards D. P., Hager G. L. Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell. Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J. C., Muchardt C., Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. W., Orkin S. H. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Schaaf M. J., Cidlowski J. A. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol. Cell. Biol. 2003;23:1922–1934. doi: 10.1128/MCB.23.6.1922-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N., Sheridan E., Amram D., Schneider P., Handgretinger R., Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am. J. Hum. Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Sif S., Saurin A. J., Imbalzano A. N., Kingston R. E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva D. A., Muller W. G., Hager G. L., Smith C. L., McNally J. G. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D. L., Nye A. C., Mancini M. G., Patel K., Dutertre M., O'Malley B. W., Smith C. L., Belmont A. S., Mancini M. A. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol. Cell. Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Travers A. A. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Hashiguchi N., Janicki S. M., Tumbar T., Belmont A. S., Spector D. L. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Verschure P. J., van d. K., I, de L. W., van d., V, Carpenter A. E., Belmont A. S., van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I., Sevenet N., Lange J., Rousseau-Merck M. F., Ambros P., Handgretinger R., Aurias A., Delattre O. Truncating mutations of hSNF5/INI1 in aggressive pediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Vignali M., Hassan A. H., Neely K. E., Workman J. L. ATP-dependent chromatin-remodeling complexes. Mol. Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Chiosea S., Ivshina M., Nickerson J. A. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J. Cell Biol. 2004;164:843–850. doi: 10.1083/jcb.200307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D., Htun H., Hager G. L. Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods. 1999;19:386–393. doi: 10.1006/meth.1999.0874. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Hansen J. C. Nuclear visions: functional flexibility from structural instability. Cell. 2001;104:631–634. doi: 10.1016/s0092-8674(02)01453-8. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Wu J., Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]