Abstract
The ability of gliomas to invade the brain limits the efficacy of standard therapies. In this study, we have examined glioma migration in living brain tissue by using two novel in vivo model systems. Within the brain, glioma cells migrate like nontransformed, neural progenitor cells—extending a prominent leading cytoplasmic process followed by a burst of forward movement by the cell body that requires myosin II. In contrast, on a two-dimensional surface, glioma cells migrate more like fibroblasts, and they do not require myosin II to move. To explain this phenomenon, we studied glioma migration through a series of synthetic membranes with defined pore sizes. Our results demonstrate that the A and B isoforms of myosin II are specifically required when a glioma cell has to squeeze through pores smaller than its nuclear diameter. They support a model in which the neural progenitor-like mode of glioma invasion and the requirement for myosin II represent an adaptation needed to move within the brain, which has a submicrometer effective pore size. Furthermore, the absolute requirement for myosin II in brain invasion underscores the importance of this molecular motor as a potential target for new anti-invasive therapies to treat malignant brain tumors.
INTRODUCTION
Malignant gliomas are a group of primary brain tumors that have remained resistant to therapy and that have a dismal prognosis (Buckner et al., 2007; Stupp et al., 2007). Part of the reason for this state of affairs is that although gliomas rarely metastasize outside the CNS, they are capable of spreading long distances within the brain (Sherer, 1940; Burger and Kleihues, 1989; Hoelzinger et al., 2007). This invasive behavior limits the effectiveness of local therapies and contributes to the high mortality rate seen in these tumors (Lim et al., 2007). Preventing glioma invasion therefore has the potential to convert this highly malignant tumor into a focal disease, which could then be effectively treated with focal therapies, such as radiation and surgery. Realizing this outcome, however, will require a detailed understanding how gliomas invade normal brain.
Gliomas typically invade the brain by migrating long distances through white matter tracts and by infiltrating cortex and subcortical gray matter structures. Brain parenchyma (neuropil) is composed of tightly packed neuronal and glial processes, and it is characterized by extracellular spaces that are in the submicrometer range (Thorne and Nicholson, 2006). This environment therefore represents a particular mechanical challenge to motile cells, such as gliomas, that need to insinuate themselves through a highly constraining brain matrix to migrate. Unfortunately, there is very little information about how glioma cells accomplish this process in vivo. Some insight is provided by studies of embryonic and early postnatal brain, where neural progenitor cells migrate along radial glial cells, and to some extent through the white matter and cortex before stopping and differentiating into mature neurons and glia. Time-lapse microscopy has shown that neural progenitors move in a unique process that has not been described in non-CNS–derived cells and that consists of two spatially separated components. Although the leading portion of the cell undergoes continuous extension of a long finger of cytoplasm, the cell body moves in intermittent, saltatory bursts that are separated by periods of little or no movement (Kakita and Goldman, 1999; Suzuki and Goldman, 2003; Bellion et al., 2005; Schaar and McConnell, 2005; Tsai et al., 2007). However, during early postnatal development, the brain undergoes changes that could have important effects on cell migration: 1) the radial glial scaffolding, which serves as a major substrate for migration, disappears; and 2) neuronal and glial processes become more abundant and tightly packed together, and the dimensions of the extracellular spaces becomes significantly smaller. The majority of migration occurs before these changes are complete. Although there is some evidence that neural progenitors can migrate through adult brain parenchyma, the mechanisms that they use have not been characterized. It thus remains unclear whether invasion of brain by human glioma cells occurs with the amoeboid motility used by other, non-CNS–transformed cells, such as breast carcinomas; the unique two-step mechanism demonstrated by neural progenitor cells in embryonic brain; or something else altogether.
Cell motility requires the formation of cytoplasmic contractile force. Hence, an additional way of examining how glioma cells invade brain is to examine how these cells use myosin II—the major source of cytoplasmic contractile force—and to compare this to other motile cells. For example, fibroblasts and carcinoma cells use myosin II to drive contraction of the cell posterior, to disconnect the motile cell from its extracellular matrix attachments, to expand the leading lamellipodium, and to generate and maintain cell polarity (Ridley et al., 2003; Betapudi et al., 2006). By contrast, myosin II has a more narrowly defined role in neural progenitor cells, where it seems to be specifically required for translocation of the nucleus (Bellion et al., 2005; Schaar and McConnell, 2005; Tsai et al., 2007). It remains unclear whether the differences in the migration mechanisms between fibroblasts and carcinoma cells, on the one hand, and neural progenitor cells, on the other hand, reflects intrinsic differences between CNS-derived and non-CNS–derived motile cells, or whether they are in some way shaped by the mechanics of the different environments through which these cells have to migrate. Although previous in vitro studies have implicated myosin II in glioma migration (Gillespie et al., 1999), they provide no information about what roles this motor plays in driving motility in the physiologically relevant milieu of the brain.
In this study, we have examined how glioma cells invade brain tissue and how they use myosin II to accomplish this process through the use of a novel rodent model of gliomagenesis that reproduces all the key histologic features of human gliomas. We have compared our results in brain to those in a series of in vitro environments, and in so doing, have developed a model that explains how glioma motility, and the role of myosin II in driving this motility, can be shaped by the mechanics of the extracellular microenvironment.
MATERIALS AND METHODS
Reagents and Antibodies
Anti-β-actin monoclonal antibodies, platelet-derived growth factor-AB (human recombinant), poly-l-lysine (0.01% solution), (−)-blebbistatin, and Y27632 were obtained from Sigma-Aldrich (St. Louis, MO). Anti-nonmuscle myosin II heavy (MHC)-A and anti-nonmuscle MHC-B rabbit polyclonal antibodies were purchased from Covance (Berkeley, CA). Antisera against nonmuscle MHC-C were a gift from Dr. R. S. Adelstein (National Heart, Lung, and Blood Institute, National Institutes of Health). Antisera against mts1 was a gift from Dr. Anne Bresnick (Albert Einstein College of Medicine).
Cell Lines and Cell Culture
C6-green fluorescent protein (GFP) rat glioma (Farin et al., 2006) and U251 human glioma cell lines were cultured in 1:1 Dulbecco's modified Eagle's medium:F-12 nutrient mixture (Ham's) supplemented with GlutaMAX and 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA). Cells were maintained at 37°C with 5% CO2.
RNA Interference (RNAi) Knockdown of Myosin II Isoforms
Myosin II siRNA oligonucleotides (oligos) as well as control oligos were chemically synthesized by Dharmacon RNA Technologies (Chicago, IL). Duplex oligos corresponding to the individual human myosin isoforms were as follows: myosin IIA (human MYH9, reference sequence accession no. NM002473) sense strand (5′ GAACAUGGCCCUCAAGAAGUU-3′) and myosin IIB (human MYH10, reference sequence accession no. NM005964) sense strand (5′-UCAGAAACCUCGACAAUUAUU-3′). U251 human glioma cells were transfected with 200 nM myosin II A, myosin II B, or nontargeting control siRNA oligos by using Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer's instructions. The cells were then incubated for 72 h before experimentation.
Transwell Migration Assay
Fluoroblok transfilters (BD Biosciences, San Jose, CA) were coated with either 10% rat tail collagen, type I (BD Biosciences, San Jose, CA) or 3 μg/ml purified human vitronectin (Invitrogen) in sterile deionized water at 37°C for 1 h. C6-GFP cells (5 × 105) in a total volume of 200 μl were seeded on top of the collagen-coated transfilter in serum-free medium, whereas U251 cells were seeded at the same concentration on the vitronectin-coated transfilter. A volume of 500 μl of medium containing chemoattractant (lysophosphatidic acid, platelet-derived growth factor [PDGF], or 10% fetal bovine serum [FBS]) was added to the bottom well a 24-well Transwell dish. The insert with cells seeded on top was incubated of the chemoattractant-containing lower well for 6 h at 37°C. When testing drug inhibition of migration, blebbistatin or Y27632 was added to both the upper and lower wells. After washing both sides of the insert with phosphate-buffered saline (PBS), the cells were fixed in 4% paraformaldehyde on ice for 10 min, and stained with 4,6-diamidino-2-phenylindole (DAPI). The number of fluorescent nuclei on the bottom of the filter were counted in six low power fields (20×) and averaged. All experiments were done in duplicate.
Scrape Migration Assay
C6 and U251 cells were plated on poly-l-lysine–coated or uncoated glass coverslips respectively, and grown to confluence. Cell monolayers were scratched to make a wound and incubated overnight (12–15 h) with blebbistatin (10 μM) or Y27632 (50 μM). Time-lapse images were captured every 2 min after wounding, for up to 15 h, by using a CoolSNAP HQ camera (Roper Scientific, Trenton, NJ) piloted by MetaMorph (Molecular Devices, Sunnyvale, CA). The migrating distance was calculated using MetaMorph by subtracting the width of the wound at the beginning and the end of the experiment and dividing by 2. For each wound the distance was measured on three different positions. Means correspond to the average velocity of three different experiments using two different dishes each time for each condition. Error bars are SEM.
Human Tissue Specimens
Fresh human glioblastoma multiforme and temporal lobe specimens were obtained according to Columbia University Institutional IRB Guidelines (IRB protocol AAAA4666). Tissue samples were obtained and immediately frozen in liquid nitrogen. Samples were stored at −80°C until used for protein.
PDGF-GFP Retrovirus Construction and Production
A 0.8-kb fragment encoding PDGF-B-hemagglutinin (HA) (Shih et al., 2004) cloned into an RCAS vector was obtained from Dr. Eric Holland (Memorial Sloan-Kettering Cancer Center, New York, NY). The PDGF-B-HA was excised from the RCAS vector by cutting with NotI, and it was ligated into the MCS1 region of the retroviral vectors pQ-MCS1-IRES-eGFP and pQ-MCS1-IRES-DsRed. Replication-deficient virus was generated by cotransfecting gp293 cells with a vsv-G protein plasmid and the control vector pQ-MCS1-IRES-eGFP (pQ-GFP), pQ-PDGFHA-IRES-eGFP (PDGF-IRES-GFP), or pQ-PDGFHA-IRES-DsRed (PDGF-IRES-DsRed) (transfection kit; Invitrogen). Conditioned media containing virus particles was collected 24 h after transfection and centrifuged at 1500 rpm, passed through a 0.45-mm filter, and centrifuged at 35,000 rpm for 1 h at 4°C to concentrate the virus. The pellet was resuspended in Opti-MEM (Invitrogen), aliquoted, and stored at −80°C. pNIT-GFP retrovirus was made from transfection of stably pNIT-GFP infected gp293 cells with vsv-G (Kakita and Goldman, 1999). Virus collection and concentration were the same as described above.
Animal Retroviral Injections
P3 neonatal Sprague-Dawley rats were submerged in ice water for 8 min. Animal's heads were placed in a stereotactic apparatus (Stoelting, Avondale, IL). Bregma was identified and a burr hole was made 2 mm lateral and 1 mm rostral. A 33-gauge Hamilton microsyringe (Reno, NV) was inserted to a depth of 2 mm, and 1 μl of virus was injected at a rate of 0.2 μl/min. Animals were anesthetized with ketamine-xylazine 10 d after injection, and they underwent cardiac perfusion with 15 ml of PBS and 15 ml of 4% paraformaldehyde. Brains were postfixed for 24 h, and then they were transferred to PBS until used. Tissue used for immunofluorescence was processed as described below. Tissue used for Western blot analysis was lysed in ice cold SDS lysis buffer (Invitrogen) stored at −20°C until used. All animal experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee, Columbia University.
Time-Lapse Microscopy and Quantitative Analysis of Brain Slice Cell Migration
PDGF-IRES-DsRed and pNIT-GFP coinjected rat pups were killed at 10 d after injection by decapitation. Brains were isolated, and 300-μm coronal sections were made using a McIlwain tissue chopper. The sections were transferred onto a 0.4-μm culture plate insert (Millipore, Billerica, MA) and placed on a Video dish (MatTek, Ashland, MA). Data were analyzed using DIAS (Soltech, Oakdale, IA).
Human Xenograft and Immunohistochemical Analysis
Primary cultures of glioma cells were generated from a surgical resection of a human glioblastoma, and cells were infected with GFP-expressing lentivirus (FUW-GFP) and transplanted into the subcortical white matter of adult nude rats (105 cells). Animals were killed at first signs of tumor morbidity (between 51 and 102 d after injection). Brains were fixed by perfusion with 4% paraformaldehyde (PFA) and postfixed overnight in 4% PFA. Then, brains were cryoprotected in 30% sucrose and cryosectioned into 10-μm-thick sections in the coronal plane. Sections were stained with antibodies against GFP, myosin IIA, and IIB, and with DAPI to stain the nuclei.
RESULTS
Glioma Invasion Resembles Neural Migration
To examine the process of glioma invasion in brain, we have used a novel model of glioma in the rat (Assanah et al., 2006). This model uses a PDGF-encoding retrovirus to induce constitutive expression of PDGF in a small population of glial progenitor cells. These PDGF-driven tumors have all of the histological hallmarks of human glioblastoma, including nuclear pleomorphism, marked vascular proliferation, palisading necrosis, and patterns of brain infiltration that closely resemble those seen in human gliomas (Assanah et al., 2006). The retroviruses also express fluorescent reporter genes (GFP or DsRed) and using time-lapse microscopy of brain slices, we can directly monitor the behavior of infected cells as they infiltrate living brain tissue.
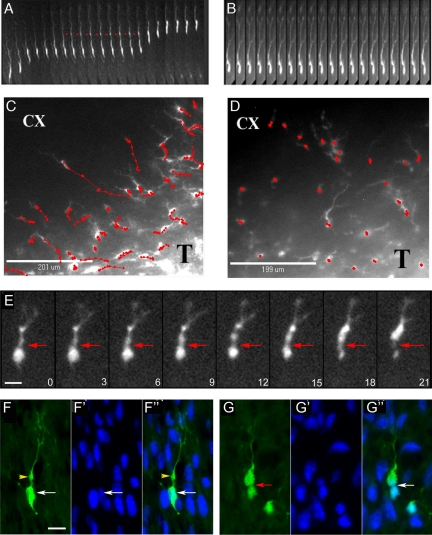
Supplemental Video 1A depicts the process of invasion by a DsRed-labeled glioma cell through the brain. The whole cell kymograph in Figure 1A illustrates several features that occur during this process. A leading cytoplasmic process extends forward and develops a proximal dilatation (red arrows)—similar to what has been described with neural progenitor cell migration in embryonic and early postnatal brain (Kakita and Goldman, 1999; Bellion et al., 2005; Schaar and McConnell, 2005; Tsai et al., 2007). This cytoplasmic process is highly dynamic, retracting, extending, and branching as the cell defines its migration pathway (Supplemental Video 1B). This is followed by a burst of forward movement of the cell body, which undergoes an “hourglass” deformation (Figure 1E). These intermittent bursts of motility are also evident in the time-dependent migration pathways of individual cell bodies, which illustrate periods of rapid movement interspersed with periods of little movement (Figure 1C). Thin sections of fixed tissue (Figure 1, F–F″ and 1G–G″) demonstrate that the glioma cell nucleus undergoes this hourglass deformation at the invasive margin—a region where the tumor cell is surrounded by untransformed brain cells and their densely packed cytoplasmic processes.
Figure 1.
Brain migration by PDGF-transformed, GFP-expressing rat glioma cells. Time-lapse microscopy was performed on 300-μm-thick slice cultures of PDGF-driven tumors at 10 d after injection. (A) Kymograph constructed from images taken every 15 min over the course of 5 h, illustrating the saltatory movement of a glioma cell as it infiltrates toward the cortex; the red arrow head points to the cytoplasmic swelling that forms in the leading process before nuclear translocation. (B) Kymograph (images taken every 15 min) of a cell in the cortex after the slice has been treated with 50 μM blebbistatin. Note that the cell body remains stationary, whereas the leading cytoplasmic process continues to extend forward. (C and D) Migratory paths of cells (red lines) without (C) and with (D) the addition of 50 μM blebbistatin. The migratory paths (red lines) of individual cells at the infiltrative edge of the tumor (T) and overlying cortex (CX) were tracked by marking the position of the cell body at consecutive time points (red dots correspond to position of cell body at 30-min intervals). (E) Time series (micrographs taken at 3-min intervals) showing the deformation of the cell body during a phase of cell body translocation. The red arrowhead points to a focal narrowing that occurs between the cell body and the swelling in the leading process through which the cell body constricts as it moves forward. (F and G) Micrographs of fixed sections stained for GFP (green) and DAPI (blue) showing GFP-expressing cells at different stages of the migratory sequence. F–F″ show a cell with the nucleus (white arrow) separated from a prominent dilatation in the leading process (yellow arrowhead). G–G″ show a cells with a focal deformation of the cell body and nucleus (red arrow). Bars, 10 μm.
Blebbistatin is a small-molecule allosteric inhibitor of myosin II (Limouze et al., 2004), and we used it to examine the role of myosin II in glioma migration in the slice culture. We used DsRed-labeled glioma cells in these studies instead of GFP to avoid the photoinactivation of blebbistatin that occurs when illuminating with blue light (Sakamoto et al., 2005). Adding blebbistatin to 50 μM completely inhibited the forward movement of the cell body, but it did not prevent leading process formation or extension (Supplemental Video 2). This is illustrated in the whole cell kymograph in Figure 1B, in the tracking of the time-dependent migration pathways of individual cell bodies (Figure 1D), and in the migration velocity histogram (Supplemental Figure 1). The effects of Y27632, an inhibitor of rho kinase (Uehita et al., 1997) were essentially the same (Supplemental Figure 1).
Myosin II Isoform Expression in Gliomas
Vertebrate myosin II is expressed as three isoforms referred to as IIA, IIB, and IIC (Conti and Adelstein, 2008). Myosins IIA and IIB both contribute to the process of cell migration (Kolega, 2006; Ma et al., 2006; Vincente-Manzanares et al., 2007), and in breast carcinoma cells, they have distinct roles (Betapudi et al., 2006). The enzymatic activity of each of the myosin II isoforms is controlled by phosphorylation of the regulatory light chain (RLC), a process largely catalyzed by two regulatory kinases—myosin light chain kinase and rho kinase (Tan et al., 1992; Amano et al., 1996). Myosin IIA activity is further regulated by metastasin 1 (mts1), which is a member of the S100 family of calcium-binding proteins (Li and Bresnick, 2006). Mts1 is significantly up-regulated in metastatic tumors and its presence in tumor biopsy specimens correlates with a poor prognosis (Zou et al., 2004).
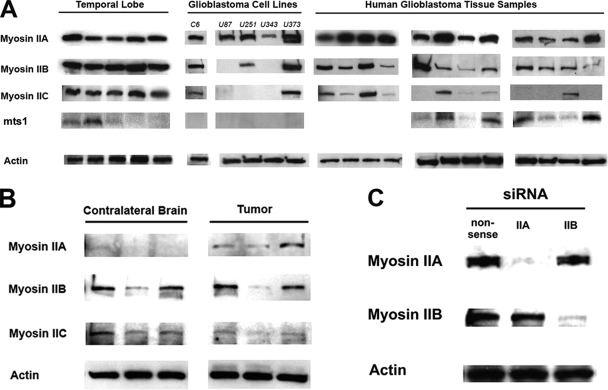
Figure 2A illustrates immunoblots for myosin IIA, IIB, IIC, and mts1 from five human brain specimens, 12 human glioblastoma tissue specimens, and five established glioblastoma cell lines. Whereas brain tissue specimens consistently express all three isoforms of myosin II, both tumor tissue and tumor cell lines only express myosin IIA uniformly, with variability seen in myosin IIB and IIC expression. mts 1 is not expressed in any of the five glioblastoma cell lines examined in this study, although it is expressed to a variable degree in temporal lobe and in tumor tissue specimens. It should be noted, however, that tumor tissue is a mixture of tumor, proliferative vessels, and entrapped gliotic brain, and we cannot rule out that the nontransformed cellular elements are responsible for mts1 immunoreactivity in these specimens.
Figure 2.
Myosin II isoform expression in gliomas. (A) Immunoblot myosin II isoforms from glioma cell and human tissue lysates. β-Actin was used as a loading control. (B) Comparison of myosin II isoform expression in lysates from the rat glioma model generated by intracerebral injection of a PDGF-IRES-GFP–encoding retrovirus, compared with the uninjected contralateral brain. Immunoblot analysis was performed on excisions from three separate rat brains. (C). Immunoblot analysis of myosin IIA and myosin IIB siRNA-transfected U251 cells showing >95% reduction of each isoform. Nonsense siRNA oligos were used as a control.
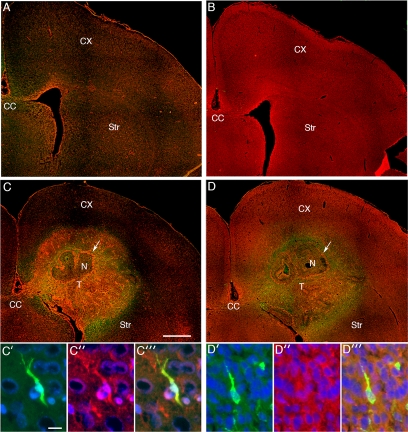
We also examined myosin II isoform distribution in our PDGF-driven rat model of glioma, because this allows us to compare expression levels between tumor and the contralateral, normal brain within the same animal. As Figure 2B illustrates, myosin IIA is minimally expressed in normal brain, and it is significantly up-regulated in tumor, whereas expression of the IIB and IIC isoforms are similar between tumor and normal brain. These findings are also consistent with the results depicted in Figure 3. Immunofluorescence localization of myosin IIA and IIB in our PDGF-driven model was performed on both the contralateral uninvolved cerebral hemisphere (Figure 3A for myosin IIA; Figure 3B for myosin IIB) as well as in the retrovirally induced glioma and surrounding brain (Figure 3C for myosin IIA; Figure 3D for myosin IIB). There is relatively little expression of myosin IIA in normal brain, whereas both normal cortex and white matter express myosin IIB. In comparison with normal brain, elevated levels of myosin IIA are seen throughout the tumor, and particularly in proliferative blood vessels (Figure 3C, white arrow), and this is also evident in GFP-expressing glioma cells that have infiltrated the surrounding brain (Figure 3, C′–C‴). By contrast, immunoreactivity for myosin IIB is similar between tumor and normal brain (Figure 3D), consistent with our immunoblot data (Figure 2B). Furthermore, as with myosin IIA, the highest levels of staining are seen in proliferating vasculature (Figure 3D, white arrow). These results imply that PDGF stimulation directly enhances expression of myosin IIA and that this myosin II isoform is particularly important in the migratory phenotype of malignant gliomas.
Figure 3.
Localization of myosin IIA and IIB in normal rat brain and in the PDGF-driven brain tumor model. (A) Low power micrograph showing the distribution of myosin IIA (red) in the contralateral, normal hemisphere of a tumor-bearing rat. The paucity of staining for myosin IIA is consistent with the immunoblot results in Figure 2B. (B) Low power micrograph showing the distribution of myosin IIB (red) in the contralateral, normal hemisphere. Myosin IIB is diffusely distributed in cortex and white matter in the uninvolved hemisphere. (C) Low power micrograph of the tumor-bearing hemisphere, stained with antibody to myosin IIA. There is increased staining for myosin IIA in the tumor (T) compared with the surrounding brain tissue, cortex (CX), striatum (Str) and corpus callosum (CC). Particularly high levels of staining are seen in the tumor vessels (white arrow). Bar, 1 mm. (C′–C‴) High-power micrographs showing immunostaining for myosin IIA (red) in a GFP-expressing cell (green) that is infiltrating the cortex. Nuclei are stained blue with DAPI. Bar, 10 μm. (D) Low-power micrograph of myosin IIB (red) in the tumor-bearing hemisphere. Increased levels of myosin IIB immunoreactivity are seen in the tumor vessels (white arrow), whereas in most of the tumor (T), the levels of myosin IIB immunoreactivity are similar to that of the surrounding brain tissue. (D′–D‴) High-powered micrograph showing myosin IIB immunoreactivity (red) in a GFP-expressing cell (green) infiltrating the cortex. Nuclei are stained blue with DAPI. The white arrow in C and D points to tumor vessels surrounding an area of necrosis (N).
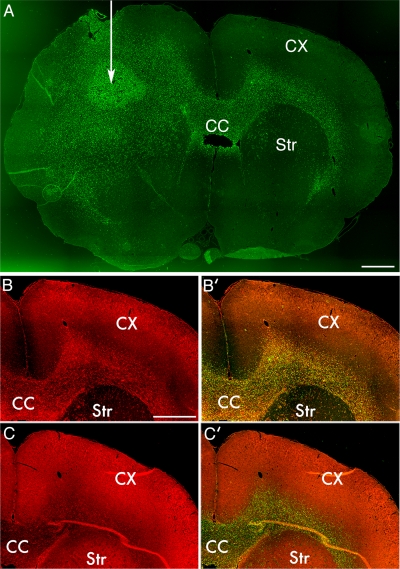
We wanted to confirm that the type of invasive behavior seen in our PDGF-driven model of glioma is representative of what is seen as well in human gliomas. We therefore isolated glioma cells from a surgical specimen of a human glioblastoma and transplanted them into the subcortical white matter of adult nude rats. Histological analysis revealed that the tumors are highly infiltrative with human nuclear antigen-expressing cells widely disseminated from the tumor injection site (Figure 4A, white arrow), primarily along white matter tracts, but also infiltrating the cortex and striatum. These are the same pathways of infiltration that are seen in our PDGF model, as well as in human patients with glioblastoma (Sherer, 1940). As in the PDGF-driven tumors, myosin IIA is markedly up-regulated in tumor compared with brain (Figure 4, B and B′), whereas myosin IIB is expressed at similar levels (Figures 4, C and C′). We also infected the isolated glioblastoma cells with a GFP-expressing lentivirus, and injection of these labeled cells allowed us to track individual tumor cells at the invasive margin of the tumor. Many of the infiltrating GFP-expressing glioma cells showed morphological features that were very similar to those seen in our PDGF-driven tumors (Figure 5). Both myosin IIA and IIB seemed to be diffusely distributed throughout the cytoplasm. The cells had long leading processes and some showed an hourglass-like deformation of the cell body and nucleus, suggesting that they were in the process of squeezing through constrictions in the extracellular space. Immunofluorescence showed that the GFP-expressing cells stained for both myosin IIA and IIB.
Figure 4.
Human glioma xenografts are invasive and demonstrate enhanced myosin IIA expression. (A) Whole brain mount from a nude rat injected with a primary culture derived from a human glioblastoma. White arrow indicates location of the site of tumor inoculation. Tissue sample is stained for human nuclear antigen (green) to demonstrate location of human tumor cells in the rat background. Tumor is seen to have spread across the corpus callosum (CC) to the contralateral white matter, between the cortex (CX) and striatum (Str). Bar, 1 mm. (B) Immunofluorescence localization of myosin IIA demonstrates prominent staining of the infiltrating tumor in the white matter contralateral to the inoculation site. Bar, 1 mm. (B′) Fusion of myosin IIA and human nuclear antigen images demonstrates colocalization. (C) Immunofluorescence localization of myosin IIB demonstrates levels similar to or less than those seen in the surrounding cortex and white matter.
Figure 5.
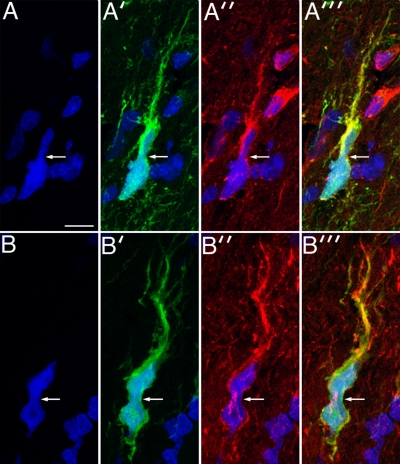
Infiltrating human glioma cells undergo the same cell body and nuclear deformation seen in the PDGF-driven glioma model. Panels A and A′ show a GFP-expressing human glioma cell (green) infiltrating the surrounding, normal brain, with nuclear DAPI staining in A and GFP staining in A′. There is strong immunostaining for myosin IIA (red, A″), and these three images are merged in A‴. Bar, 10 μm. B and B′ correspond to DAPI and GFP staining, respectively, of another infiltrating GFP-expressing human glioma cell. This cell demonstrates strong staining for myosin IIB (red, B″). In both sets of micrographs the white arrows point to focal deformation of the cell bodies.
Glioma Migration on a Two-Dimensional Surface Resembles How Epithelial Cells Migrate and Does Not Require Myosin II
When plated on a coverslip, human U251MG and rat C6-GFP glioma cells migrate into a cell-free zone with a velocity of 0.13 ± 0.01 and 0.19 ± 0.03 μm/min, respectively. This is depicted for C6-GFP cells in Supplemental Video 3. Unlike movement in the brain, migration on the two-dimensional surface of the coverslip occurs via formation of a broad anterior lamellipodium, much as has been described for epithelial cells (Svitkina et al., 1997), and it occurs continuously and without any deformation of the nucleus. This is illustrated in a kymograph for C6 cells in Figure 6A. Comparable results were observed with U251 cells (data not shown). Likewise, immunofluorescence localization of actin, phosphorylated myosin II RLC, and tubulin in U251 cells subjected to a scrape migration assay (Supplemental Figure 2) demonstrated patterns of staining similar to that described for fibroblasts, endothelial cells, and keratocytes, with colocalization of actin and phosphorylated myosin II along fibrils that were oriented roughly perpendicular to the direction of the scrape (white arrow).
Figure 6.
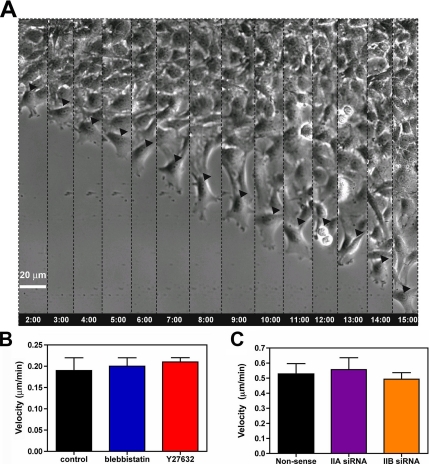
Glioma migration in a scrape assay. (A) Migrating cells at the “wound” edge were imaged for 15 h at a frequency of 1 image every 2 min by phase contrast. The figure shows a leading cell (arrowhead) at different 1-h intervals. Migrating cells form a broad lamellipodium, and the nucleus remains undistorted through the course of migration. At the 12-h time point, the cell divided, and the two daughter cells continued to migrate in different directions. Bar, 20 μm. (B) Migration velocity is not appreciably affected by concentrations of blebbistatin or Y27632 that alter cell morphology or block cytokinesis (see Supplemental Video 4). Bars are the SEM (n = 6). (C) Suppression of either myosin IIA or IIB with RNAi (Figure 1C) has no appreciable effect on migration velocity. Bars are the SEM (n = 6).
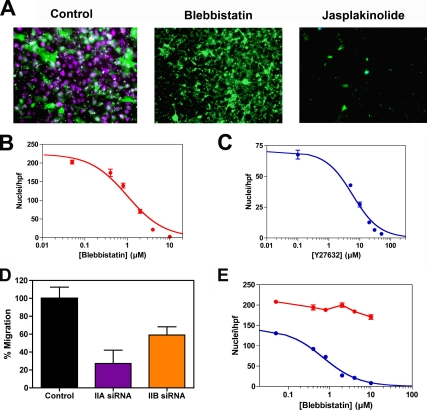
Unlike in brain invasion, migration of glioma cells on a coverslip does not require myosin II. Neither treatment with blebbistatin or Y27632, nor RNAi suppression of myosin IIA or IIB by >95% (Figure 2C) has any appreciable effect on the speed of migration across the cell-free zone (Figure 6, B and C). None of these treatments had any significant effect on cell viability or attachment over 24 h for either C6-GFP or U251 human glioma cell lines (data not shown). To ensure that myosin II was being inhibited we looked for the effect of drug or RNAi on cell morphology and on cytokinesis, because both depend on myosin II function. Both types of treatment produced morphological changes similar to previous descriptions (Supplemental Figure 3), with formation of multiple long, branched cytoplasmic processes (Manning et al., 1998, 2000). Furthermore, both drug and RNAi treatment produced obvious defects in cytokinesis (Supplemental Figure 4). By contrast, treatment with 200 nM jasplakinolide abolished migration across the cell free zone (data not shown). Jasplakinolide maintains actin in a polymerized state and thereby blocks the normal actin dynamics that drive protrusion of leading processes of migratory cells (Bubb et al., 1994). In separate experiments, we determined that this dose of jasplakinolide had no effect on C6-GFP cell viability or attachment to a collagen-coated substrate (data not shown).
Myosin II Is Required for Migration through Pores Smaller than Its Nuclear Diameter
As a next step, we examined glioma migration through a three-dimensional matrix of defined pore size to see whether myosin II is required for migration through this more restricted environment. Figure 7A illustrates that both the cytoplasm (green) and nuclei (magenta) of DAPI-stained, GFP-expressing C6 glioma cells readily migrate through the 3-μm pores of a Transwell membrane after incubation for 6 h. Qualitatively similar results were seen with U251 cells (data not shown). In the presence of 10 μM blebbistatin, however, only the GFP-containing cytoplasmic processes were seen, because the nuclei were unable to protrude through the 3-μm pore (Figure 7A). Identical results were seen with Y27632 (data not shown). Figure 7, B and C, illustrates the dose-response relationship for blebbistatin and Y27632 inhibition of nuclear protrusion for C6 cells, respectively. For both drugs, the data can be fit to hyperbolic isotherms, with IC50 values for blebbistatin of 1.1 ± 0.1 μM and for Y27632 of 6.0 ± 0.6 μM. These compare with IC50 values for blebbistatin of 5.1 μM for myosin IIA and 1.8 μM for myosin IIB, and 5–15 μM for inhibition of rho kinase-mediated contractility of smooth muscle by Y27632 (Uehita et al., 1997; Limouze et al., 2004).
Figure 7.
Glioma migration through Transwell membranes. (A) Micrograph of fixed, DAPI-stained C6-GFP cells treated with 10 μM blebbistatin or 200 nM jasplakinolide after being seeded on the upper surface of a 3-μm Fluoroblok transfilter and incubated for 6 h. Although blebbistatin does not prevent the cells from extruding their cytoplasm (green) through the pores, it does block extrusion of the nuclei, as evidenced by the lack of DAPI staining (blue). Jasplakinolide prevents extrusion of both cytoplasm as well as nuclei. (B) Dose response for inhibition of nuclear translocation across the 3-μm membrane by blebbistatin. Data were fit to a hyperbolic isotherm, with Ki = 1.0 ± 0.1 μM. (C) Corresponding dose–response relationship for Y27632, with Ki = 5.8 ± 0.6 μM. (D) Effect of siRNA suppression of myosin IIA and myosin IIB on U251 cell invasion through 3-μm Transwell membranes. After migration toward 10% FBS for 24 h, the cells were fixed and stained with DAPI. Nuclei were counted in several 20× fields of magnification. Data are expressed as mean ± SD per high-powered field. (E) Effect of varying Transwell pore size on the need for myosin II. Dose–response curves for transmigration of C6-GFP cells treated with various concentrations of blebbistatin or Y27632. Cells were fixed and stained with DAPI after 6 h of incubation with either a 3-μm (blue) or 8-μm (red) Fluoroblok transfilter. Data for the 3-μm pores for this set of experiments fit a hyperbolic dose–response relationship, with Ki = 0.7 ± 0.1 μM, very similar to the value derived from the data in Panel B. Nuclei were counted in several 20× fields of magnification. Data are expressed as mean ± SD per high-powered field.
If the Transwell assay is performed with C6-GFP cells in the presence of 200 nM jasplakinolide, neither cytoplasmic processes nor nuclei are seen (Figure 7A). These results imply that glioma migration through the 3-μm pores of the Transwell membrane occurs in at least two stages that have different cytoskeletal requirements and that correspond to the two major components of motility seen in the brain slice studies (Figure 1A). The first is extension of a leading cytoplasmic process through the pore, which depends on actin dynamics and can occur in the absence of myosin II function. The second, protrusion of the nucleus-containing cell body, requires myosin II as well.
We selectively suppressed myosin IIA and IIB expression in human U251 cells with RNAi and performed Transwell assays by using 3-μm pores. As Figure 7D shows, suppression of myosin IIA reduces the number of nuclei that have squeezed through the 3-μm pores to 27 ± 15% of control, whereas suppression of myosin IIB reduces the number of nuclei to 59 ± 9% of control. These results suggest that both IIA and IIB are needed for migration through a 3-μm pore and that neither can fully compensate for loss of the other.
One explanation of our results is that glioma cells need myosin II when they have to squeeze through an opening smaller than the short axis of their elliptically shaped nucleus. We measured the short and long axes of the nuclei of C6-GFP cells plated on glass (see Materials and Methods), and we found that they measured 7.7 ± 0.7 and 10.5 ± 1.1 micrometers (n = 20). We therefore reexamined the effect of myosin II inhibition of C6-GFP migration by using Transwell membranes with 8-μm pores and compared results to those using 3-μm pore membranes. As Figure 7E shows, blebbistatin does not inhibit the ability of C6-GFP nuclei to maneuver through the 8-μm pores (red), whereas it inhibits migration through a 3-μm pore membrane (blue).
DISCUSSION
Glioma migration is a multistep process that uses the extracellular matrix, proteases, integrins, signaling networks, ion channels, and the cytoskeleton (Mohanam et al., 1994; Gillespie et al., 1999; Goldbrunner et al., 1999; Fillmore et al., 2001; Wick et al., 2001; Ernest et al., 2005; Salhia et al., 2008). Because glioma invasion remains a vexing problem that limits all current brain tumor therapies, many of these components have been investigated as targets for the development of anti-invasive treatments. However, clinical trials with agents that target these invasive elements have not demonstrated broad-scale or durable responses (Steward and Thomas, 2000; Mamelak et al., 2006; Prados et al., 2006; Nabors et al., 2007). There thus remains a pressing need to identify other anti-invasive strategies, and addressing this will require a detailed understanding of how glioma cells migrate within the brain.
We therefore examined how glioma cells move in both physiologically relevant and spatially defined environments to answer three questions. First, do glioma cells migrate like other transformed cells, such as carcinomas, or like other motile cells that share a similar environment, such as neural progenitors? Second, what role does the effective pore size in the extracellular environment play in shaping how gliomas migrate? Finally, how do glioma cells use myosin II to drive this migration, and how does the use of myosin II by these cells differ from non-CNS motile cells?
Our data from Figures 1 and 6 clearly demonstrate that how glioma cells move depends on the environment through which they are moving. In the absence of spatial constraints, gliomas resemble epithelial and mesenchymal cells, extending a broad lamellipodium, moving continuously and without nuclear distortion, and localizing activated myosin II along actin filaments that are roughly perpendicular to the direction of movement. By contrast, when migrating through brain white matter, gliomas move in a manner remarkably similar to that previously described for neural progenitor cells (Bellion et al., 2005, Schaar and McConnell, 2005; Tsai et al., 2007). Given the submicrometer size of the extracellular spaces that are present in the tightly packed neuropil of the brain (Bruehlmeier et al., 2003; Thorne and Nicholson, 2006), motile cells would likely find it impossible to migrate in a fibroblast-like manner, because there would be no room to allow a lamellipodium to form. Thus, we propose that the unique form of motility shared by gliomas and neural progenitors reflects adaptations that these cells have to make to a uniquely challenging environment with small effective pore sizes. One of these adaptations is in how glioma cells use myosin II. Myosin II is absolutely required for migration in brain, where we propose that its major role is to push the bulky nucleus and cell body through the small pores found within the brain matrix. Our results with the Transwell assay support this conclusion, because myosin II is required to extrude the nucleus only when the pore size is smaller than the effective nuclear diameter (Figure 7). By contrast, myosin II is not required to push the cytoplasm through either brain parenchyma or 3-μm Transwell pores (Figures 1 and 7), which suggests that the plasma membrane is sufficiently flexible that it can protrude through small pores through the action of actin dynamics alone. This is consistent with a recent study that found that the nuclear envelope is much stiffer than the plasma membrane (Dahl et al., 2004). It is also consistent with our finding that in a two-dimensional environment, in which all spatial constraints are removed, glioma cells can move with normal velocities in the absence of myosin II activity (Figure 6).
It has previously been shown that carcinoma and melanoma cells can switch between two distinct modes of motility when migrating a three-dimensional matrix—a rounded bleb-associated form that is dependent on Rho kinase and an elongated form that is Rho kinase independent and involves Rac-mediated F actin protrusion (Sahai and Marshall, 2003). The progenitor-like migration that we observe glioma cells use when invading brain tissue does not fit into either of these categories. Glioma cells do manifest an elongated morphology, but their leading cytoplasmic process develops a proximal dilatation, which in progenitor cells is associated with centrosome movement (Tsai et al., 2007), and their motility is clearly dependent on Rho kinase. Thus, we propose that progenitor-like migration represents a distinct mode of motility that is specifically adapted for migrating through the densely packed neuropil of the brain. Although we do not know whether other types of cancer cells have the capacity for progenitor-like migration, it is a well-established histopathological finding that metastatic carcinoma cells do not infiltrate brain in the way that glioma cells do, but rather grow as a well circumscribed, expansile mass. This might be expected if carcinoma cell migration requires formation of a broad lamellipodium, which would be sterically blocked by the tightly packed neuropil.
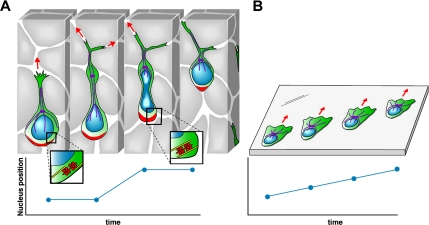
Figure 8A depicts a model that summarizes how we envision myosin II drives glioma invasion in situ, and it may be useful to compare this model to those developed for other non-CNS migratory cells, such as keratocytes, fibroblasts, and endothelial cells (Svitkina et al., 1997; Kolega 2006). In these cells, movement is initiated by extending a broad lamellipodium at the leading edge of the cell that contains a meshwork of elongating actin filaments. Myosin II is recruited to this meshwork and cross-links intersecting actin filaments, causing them to become aligned and producing a force perpendicular to the axis of these filaments. This generates a pulling force that draws the cell body forward. Coupled to this is a myosin II-mediated contraction at the cell rear, which disconnects the cell from its ECM attachments. This mechanism requires that myosin II be rapidly transported through the dense actin network in the lamellipodium—a process facilitated by mts1, which depolymerizes myosin IIA (Li and Bresnick, 2006). By contrast, if myosin II is specifically required to squeeze a stiff nucleus through small pores, and if the brain environment prevents lamellipodium formation, then rapid redistribution of myosin II might not be required, and mts1 might be unnecessary. This may explain why we were unable to detect any mts1 immunoreactivity in five highly invasive glioma cell lines (Figure 2A). If migration of other, non-CNS–derived cells involves myosin II-coordinated pulling from the front and pushing from the rear, is the same true for glioma cell migration in brain? If so, this creates a problem for glioma cells, because given the spatial constraints in the brain, myosin II would not be able to generate a cross-linked, aligned network of actin filaments perpendicular to the axis of movement. Neural progenitor cells seem to have solved this problem by using dynein to generate a pulling force, which complements a pushing from the rear by myosin II (Tsai et al., 2007). We note that glioma cells migrating in brain develop a dilatation in the leading cytoplasmic process (Figure 1A, red arrows), which in neural progenitor cells is associated with a dynein-mediated displacement of the centrosomes that is central to the process of nuclear protrusion. Further work will be required to determine whether glioma migration in brain also requires the action of dynein as well.
Figure 8.
Model of how a glioma cell migrates through brain tissue. (A) Starting with the left-most image, the infiltrating glioma cell is depicted as having a prominent leading process that often branches at its distal end. A dilatation forms in the leading process (black arrow). Evidence form neural progenitors show that the centrosome and associated microtubules (purple) move forward into the dilatation (Tsai et al., 2007). The mechanical constraints generated from the small intercellular spaces impede forward movement of the nucleus and cell body until contraction of actomyosin II at the rear of the cell (red) provide the necessary force to squeeze the nucleus through a narrowing in the extracellular space. The graph of nuclear position over time illustrates the resulting saltatory movement. (B). Glioma cells crawling on a two-dimensional surface move in a different manner that more closely resembles the migration of fibroblasts. Migration is associated with formation of broad lamellipodium, the nucleus remains undistorted, and its forward movement is continuous and unimpeded, as illustrated by the linear plot of nuclear position over time. Although myosin II is involved in maintaining cell polarity and shape, its activity is not required for cell motility in this barrier-free environment.
Myosin II consists of three isoforms, and our studies of isoform expression and RNAi suppression speak to the importance of myosin IIA and IIB in driving glioma invasion. In particular, myosin IIA expression is up-regulated relative to normal brain in invasive human glioma xenografts and the PDGF-driven tumors (Figures 2–4). PDGF also plays an important role in tumor angiogenesis, in part by promoting the recruitment of pericytes and vascular smooth muscle cells to newly formed tumor blood vessels (Guo et al., 2003). Blebbistatin and Y27632 block PDGF-stimulated migration of vascular smooth muscle cells (Nishiguchi et al., 2003; Wang et al., 2006), and this suggests that myosin II is also a downstream effector of PDGF-mediated angiogenesis—a point emphasized in Figure 3 by the relatively high levels of expression of myosins IIA and IIB in tumor vasculature. The importance of myosin IIA in driving in situ migration of gliomas is also underscored by our results with RNAi suppression (Figure 7D), which demonstrates that suppression of myosin IIA produces a significantly greater inhibition of Transwell migration than suppression of IIB. However, neither experimental condition is able to completely block Transwell migration. By contrast, blebbistatin and Y27632, both of which inhibit myosins IIA and IIB, completely block Transwell migration at concentrations predicted from the dose–response relationship to produce >95% inhibition (Figure 7, B and C). Together, these results suggest that myosins IIA and IIB both contribute to glioma migration. Further studies will be needed to determine the respective roles for these two myosin II isoforms, and the degree to which they play distinct versus overlapping roles.
Finally, the results of our study speak to the importance of studying glioma cell motility in an assay that faithfully recapitulates the extracellular milieu found within brain, because these tumor cells are capable of adapting their motility to the particular environment through which they are moving. They also suggest that glioma cells have a repertoire of motility “tools” that includes myosin II and that they use selectively, depending on the nature of the extracellular environment they are invading. What other components are part of this “tool box”, and how glioma cells use them, will be investigated in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Harald Sontheimer and James Goldman for helpful discussions. We particularly thank Trine Giaever for creating Figure 8. This work was supported in part by NIH grant NS045070 (P.C.) and NIH grant HDO 40182 (RV).
Abbreviations used:
- ECM
extracellular matrix
- GFP
green fluorescent protein
- PDGF
platelet-derived growth factor
- RLC
regulatory light chain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0319) on May 21, 2008.
REFERENCES
- Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J. Biol. Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Assanah M., Lochhead R., Ogden A., Bruce J., Goldman J., Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A., Baudoin J. P., Alvarez C., Bornens M., Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betapudi V., Licate L. S., Egelhoff T. T. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- Bruehlmeier M., Roelcke U., Blauenstein P., Missimer J., Schubiger P. A., Locher J. T., Pellikka R., Ametamey S. M. Measurement of the extracellular space in brain tumors using 76Br-bromide and PET. J. Nucl. Med. 2003;44:1210–1218. [PubMed] [Google Scholar]
- Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L., Korn E. D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Buckner J. C., Brown P. D., O'Neill B. P., Meyer F. B, Wetmore C. J., Uhm J. H. Central nervous system tumors. Mayo Clin. Proc. 2007;82:1271–1286. doi: 10.4065/82.10.1271. [DOI] [PubMed] [Google Scholar]
- Burger P. C., Kleihues P. Cytologic composition of the untreated glioblastoma with implications for evaluation of needle biopsies. Cancer. 1989;63:2014–2023. doi: 10.1002/1097-0142(19890515)63:10<2014::aid-cncr2820631025>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Conti M. A., Adelstein R. S. Non-muscle myosin II moves in new directions. J. Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- Dahl K. N., Kahn S. M., Wilson K. L., Discher D. E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- Ernest N. J., Weaver A. K., Van Duyn L. B., Sontheimer H. W. Relative contribution of chloride channels and transporters to regulatory volume decrease in human glioma cells. Am. J. Physiol. Cell Physiol. 2005;288:C1451–C1461. doi: 10.1152/ajpcell.00503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin A., Suzuki S. O., Weiker M., Goldman J. E., Bruce J. N., Canoll P. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia. 2006;53:799–808. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- Fillmore H. L., VanMeter T. E., Broaddus W. C. Membrane-type matrix metalloproteinases (MT-MMPs): expression and function during glioma invasion. J. Neurooncol. 2001;53:187–202. doi: 10.1023/a:1012213604731. [DOI] [PubMed] [Google Scholar]
- Gillespie G. Y., Soroceanu L., Manning T., Gladson C. L., Rosenfeld S. S. Glioma migration can be blocked by non-toxic inhibitors of myosin II. Cancer Res. 1999;59:2076–2082. [PubMed] [Google Scholar]
- Goldbrunner R. H., Bernstein J. J., Tonn J. C. Cell-extracellular matrix interaction in glioma invasion. Acta Neurochir. 1999;141:295–305. doi: 10.1007/s007010050301. [DOI] [PubMed] [Google Scholar]
- Guo P., Hu B., Gu W., Xu L., Wang D., Huang H. J., Cavenee W. K., Cheng S. Y. Platelet-derived growth factor-B enhances glioma angiogenesis by stimulating vascular endothelial growth factor expression in tumor endothelia and by promoting pericyte recruitment. Am. J. Pathol. 2003;162:1083–1093. doi: 10.1016/S0002-9440(10)63905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzinger D. B., Demuth T., Berens M. E. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J. Natl. Cancer Inst. 2007;99:1583–1593. doi: 10.1093/jnci/djm187. [DOI] [PubMed] [Google Scholar]
- Kakita A., Goldman J. E. Patterns and dynamics of SVZ cell migration in the postnatal forebrain: monitoring living progenitors in slice preparations. Neuron. 1999;23:461–472. doi: 10.1016/s0896-6273(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Kolega J. The role of myosin II motor activity in distributing myosin asymmetrically and coupling protrusive activity to cell translocation. Mol. Biol. Cell. 2006;17:4435–4445. doi: 10.1091/mbc.E06-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. H., Bresnick A. R. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- Lim D. A., Cha S., Mayo M. C., Chen M-H, Keles E., VandenBerg S., Berger M. S. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neurooncology. 2007;9:424–429. doi: 10.1215/15228517-2007-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limouze J., Straight A. F., Mitchison T., Sellers J. R. Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- Ma X., Kawamoto S., Uribe J., Adelstein R. S. Function of the neuron-specific alternatively spliced isoforms of nonmuscle myosin IIB during mouse brain development. Mol. Biol. Cell. 2006;17:2138–2149. doi: 10.1091/mbc.E05-10-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelak A. N., et al. Phase I single-dose study of intracavitary-administered iodine-131-TM-601 in adults with recurrent high-grade glioma. J. Clin. Oncol. 2006;24:3644–3650. doi: 10.1200/JCO.2005.05.4569. [DOI] [PubMed] [Google Scholar]
- Manning T. J., Jr., Rosenfeld S. S., Sontheimer H. Lysophosphatidic acid stimulates actomyosin contraction in astrocytes. J. Neurosci. Res. 1998;53:343–352. doi: 10.1002/(SICI)1097-4547(19980801)53:3<343::AID-JNR8>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Manning T. J., Jr., Parker J. C., Sontheimer H. Role of lysophosphatidic acid and rho in glioma cell motility. Cell Motil. Cytoskeleton. 2000;45:185–199. doi: 10.1002/(SICI)1097-0169(200003)45:3<185::AID-CM2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Mohanam S., Sawaya R. E., Yamamoto M., Bruner J. M., Nicholson G. L., Rao J. S. Proteolysis and invasiveness of brain tumors: role of urokinase-type plasminogen activator receptor. J. Neurooncol. 1994;22:153–160. doi: 10.1007/BF01052890. [DOI] [PubMed] [Google Scholar]
- Nabors L. B., et al. Phase I and correlative biology study of cilengitide in patients with recurrent malignant glioma. J. Clin. Oncol. 2007;25:1651–1657. doi: 10.1200/JCO.2006.06.6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi F., Fukui R., Hoshiga M., Negoro N., Ii M., Nakakohji T., Kohbayashi E., Ishihara T., Hanafusa T. Different migratory and proliferative properties of smooth muscle cells of coronary and femoral artery. Atherosclerosis. 2003;171:39–47. doi: 10.1016/j.atherosclerosis.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Prados M. D., et al. Phase 1 study of erlotinib HCl alone and combined with temozolomide in patients with stable or recurrent malignant glioma. Neurooncology. 2006;8:67–78. doi: 10.1215/S1522851705000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Limouze J., Combs C. A., Straight A. F., Sellers J. R. Blebbistatin, a myosin II inhibitor, Is photoinactivated by blue light. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C. J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- Salhia B., Hwang J. H., Smith C. A., Mitsutoshi N., Rutka F., Symons M., Rutka J. T. Role of myosin II activity and the regulation of myosin light chain phosphorylation in astrocytomas. Cell Motil. Cytoskeleton. 2008;65:2–24. doi: 10.1002/cm.20240. [DOI] [PubMed] [Google Scholar]
- Schaar B. T., McConnell S. K. Cytoskeletal coordination during neuronal migration. Proc. Nat. Acad. Sci. USA. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer H. J. The forms of growth in glioma and their practical significance. Brain. 1940;63:1–35. [Google Scholar]
- Shih A. H., Dai C., Hu X., Rosenblum M. K., Koutcher J. A., Holland E. C. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64:4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- Steward W. P., Thomas A. L. Marimastat: the clinical development of a matrix metalloproteinase inhibitor. Expert Opin. Investig. Drugs. 2000;9:2913–2922. doi: 10.1517/13543784.9.12.2913. [DOI] [PubMed] [Google Scholar]
- Stupp R., Hegi M. E., Gilbert M. R., Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J. Clin. Oncol. 2007;25:4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- Suzuki S. O., Goldman J. E. Multiple cell populations in the early postnatal subventricular zone take distinct migratory pathways: a dynamic study of glial and neuronal progenitor migration. J. Neurosci. 2003;23:4240–4250. doi: 10.1523/JNEUROSCI.23-10-04240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Verkhovsky A. B., McQuade K. M., Borisy G. G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. L., Ravid S., Spudich J. A. Control of non-muscle myosins by phosphorylation. Annu. Rev. Biochem. 1992;81:721–759. doi: 10.1146/annurev.bi.61.070192.003445. [DOI] [PubMed] [Google Scholar]
- Thorne R. G., Nicholson C. In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc. Natl. Acad. Sci. USA. 2006;103:5567–5572. doi: 10.1073/pnas.0509425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G., Wu Y., Sasaki Y., Hartshorne D. J., Yamakita Y., Yamashiro S., Matsumura F. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 2004;164:427–439. doi: 10.1083/jcb.200306172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J-W., Bremner K. H., Vallee R. B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Uehita M., et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- Vincente-Manzanares M., Zareno J., Whitmore L., Choi C. K., Horwitz A. F. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. H., Qin X., Zhao T., Ye L., Tamaka H., Nakamura A. Blebbistatin blocks PDGF-induced migration of vascular smooth muscle cell through inhibiting the ATPase activity of smooth muscle myosin II. J. Pharmacol. Sci. 2006;100(Supplement):264. [Google Scholar]
- Wick W., Platten M., Weller M. Glioma cell invasion: regulation of metalloproteinase activity by TGF-beta. J. Neurooncol. 2001;53:177–185. doi: 10.1023/a:1012209518843. [DOI] [PubMed] [Google Scholar]
- Zou M., Famulski K. S., Parhar R. S., Baitei E., Al-Mohanna F. A., Farid N. R., Shi Y. Microarray analysis of metastasis-associated gene expression profiling in a murine model of thyroid carcinoma pulmonary metastasis: identification of S100A4 (Mts1) gene overexpression as a poor prognostic marker for thyroid carcinoma. J. Clin. Endocrinol. Metab. 2004;89:6146–6154. doi: 10.1210/jc.2004-0418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.