Abstract
U2 small nuclear ribonucleoprotein (snRNP) auxiliary factor 65 kDa (U2AF65) is an essential splicing factor in the recognition of the pre-mRNA 3′ splice sites during the assembly of the splicing commitment complex. We report here that U2AF65 is proteolyzed during apoptosis. This cleavage is group I or III caspase dependent in a noncanonical single site localized around the aspartic acid128 residue and leads to the separation of the N- and C-terminal parts of U2AF65. The U2AF65 N-terminal fragment mainly accumulates in the nucleus within nuclear bodies (nucleoli-like pattern) and to a much lesser extent in the cytoplasm, whereas the C-terminal fragment is found in the cytoplasm, even in localization studies on apoptosis induction. From a functional viewpoint, the N-terminal fragment promotes Fas exon 6 skipping from a reporter minigene, by acting as a dominant-negative version of U2AF65, whereas the C-terminal fragment has no significant effect. The dominant-negative behavior of the U2AF65 N-terminal fragment can be reverted by U2AF35 overexpression. Interestingly, U2AF65 proteolysis in Jurkat cells on induction of early apoptosis correlates with the down-regulation of endogenous Fas exon 6 inclusion. Thus, these results support a functional link among apoptosis induction, U2AF65 cleavage, and the regulation of Fas alternative splicing.
INTRODUCTION
U2 small nuclear ribonucleoprotein (snRNP) auxiliary factors (U2AF65-U2AF35) cooperate in 3′ splice site (3′ss) recognition through direct interactions with the branch point and the branch-binding protein SF1/BBP as well as with the polypyrimidine tract (PT) and the conserved AG dinucleotide of the 3′ss (Valcárcel et al., 1996; Merendino et al., 1999; Wu et al., 1999; Zorio and Blumenthal, 1999; Soares et al., 2006). U2AF65 contains three RNA recognition motifs (RRMs) and an N-terminal region rich in basic residues (RS domain). RRMs 1 and 2 of U2AF65 display a canonical RRM-fold, bind RNA in vitro, and are implicated in high-affinity binding to the PT (Zamore et al., 1992; Banerjee et al., 2003; Sickmier et al., 2006). RRM3 is a noncanonical RRM and mediates the SF1 interaction (Selenko et al., 2003). Recognition of the PT by U2AF65 is key to splice site selection and spliceosome assembly for the major class of introns in higher eukaryotes. Both length and pyrimidine content of this sequence are critical determinants of the relative strength of competing 3′ss and therefore of differential 3′ss use as a major source of proteome diversity (Reed, 1989; Izquierdo and Valcárcel, 2006).
Apoptosis is induced by many different stimuli, including growth factor withdrawal, chemotherapeutic compounds, membrane-bound death receptors, or DNA-damaging agents (Krammer, 2000; Adams, 2003). Whatever the initial signal, common characteristic cytological and molecular changes occur during apoptosis (Adams, 2003). Several signaling components and apoptosis pathways have been identified and elucidated at the molecular level (Adams, 2003). When apoptosis is induced, caspases, a family of aspartyl-specific cysteine proteases, are activated and act as effectors of apoptosis (Earnshaw et al., 1999). The effector caspases then selectively cleave specific target proteins, resulting in the biochemical, morphological, and molecular features associated with imminent cell death (Earnshaw et al., 1999). An understanding of the mechanisms of apoptosis requires the caspase substrates to be identified and the consequences of their proteolytic cleavage to be established. Until now, caspases have been reported to cleave >300 different proteins (Fischer et al., 2003), but, in most cases, the consequences of protein cleavage are poorly understood.
The aim of this work was to examine whether U2AF65 proteolysis during apoptosis has functional consequences on the control of Fas alternative splicing. The results show that the N-terminal fragment down-regulates Fas exon 6 inclusion. Moreover, the results suggest that alternative splicing is modulated by U2AF65 cleavage during early apoptosis.
MATERIALS AND METHODS
Cell Cultures and Apoptosis Induction
HeLa and Jurkat cells were cultured as described previously (Izquierdo and Valcárcel, 2007a). Apoptosis was induced by means of the following agents: 500 ng/ml anti-Fas antibody (immunoglobulin [Ig]M clone CH11) (Millipore, Billerica, MA), 2 μM staurosporine (Sigma-Aldrich, St. Louis, MO), and 10 μg/ml cycloheximide (Sigma-Aldrich). In addition, HeLa cells were irradiated with UV light (100 mJ/cm2). For inhibition of caspases, cells were preincubated for 1 h with 5 μM pan-caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (z-VAD-fmk; BIOMOL Research Laboratories, Plymouth Meeting, PA) before treatment with apoptotic agents.
Plasmids
Fas minigene was described previously (Förch et al., 2000; Izquierdo et al., 2005). The constructs containing human U2AF65 and U2AF35 cDNAs were kindly provided by Dr. J. Valcárcel (Centre de Regulació Genòmica, Barcelona, Spain). The green fluorescent protein (GFP)-U2AF65 derivative plasmids containing either a 128-amino acid deletion at the N terminus, a 347-amino acid deletion at the C terminus, or directed mutagenesis from aspartic128 to alanine were generated with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). Fas ΔI6 minigene was generated from Fas wild-type (wt) minigene by a polymerase chain reaction (PCR)-based method as described above. The sequence of all constructs was verified by sequencing.
Protein Analysis
Whole-cell extracts were prepared by resuspending an aliquot of untransfected and transfected HeLa cells in lysis buffer (50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.1% Nonidet P-40 plus a cocktail of protease inhibitors), freeze-thawing three times, and centrifuging at 10,000 rpm for 5 min. Postnuclear fractions were isolated from aliquots of HeLa cells suspended in lysis buffer, incubated for 5 min at 4°C, and centrifuged at 4000 rpm for 5 min. The supernatants were collected and stored as cytoplasmic fractions. The resulting pellets were resuspended in lysis buffer, freeze-thawed three times and centrifuged at 10,000 rpm for 5 min. The supernatants were collected as enriched nuclear fractions. All centrifugations were carried out in a microfuge at 4°C. The different fractions were stored at −70°C. Protein concentration was determined with the Bradford reagent (Bio-Rad protein assay; Bio-Rad, Hercules, CA) using bovine serum albumin as standard. For Western blot analyses, equal amounts of protein from the fractions were loaded on 10% SDS-PAGE and Western blots were performed using nylon membranes and the corresponding antibodies. Immunoblots were carried out using the following antibodies: anti-U2AF65 (MC3; provided by Dr. J. Valcárcel); anti-T cell intracellular antigen (TIA)-1 (C-20), anti-TIAR (C-18), and anti-caspase-3 (H-277) from Santa Cruz Biotechnology (Santa Cruz, CA); anti-polypyrimidine tract-binding protein (PTB) (BB7; provided by Dr. C. W. Smith, University of Cambridge, United Kingdom); anti-α-tubulin (B-5-1-2; Sigma-Aldrich); anti-poly(ADP-ribose) polymerase (PARP) (C-2-10; BIOMOL Research Laboratories); and anti-GFP (JL-8; Clontech, Mountain View, CA). HeLa extracts were analyzed for caspase-3 or caspase-3–like activity with peptide substrate DEVD-para-nitroanilide (DEVD-p-NA) (ApoAlert caspase-3 colorimetric assay kit; Clontech). In vitro transcription and translation assays were performed with a transcription-translation–coupled system (Promega, Madison, WI). HeLa nuclear extracts (Dignam et al., 1983) or TIA-1b/U2AF65 translated in vitro was incubated with recombinant caspase-1–10 (MultiCasPak-4, Active Caspases 1-10, AK-010; BIOMOL Research Laboratories). Immobilon INSTA-Blot membrane containing samples of several human cell lines (HeLa, Jurkat, Daudi, 293, Rh 30, A375, T98G, HCT-116, and Hep-G2) were obtained from Calbiochem (San Diego, CA).
Fluorescence Microscopy Analysis
HeLa cells were grown for 24 h on coverslips, and then they were washed three times with phosphate-buffered saline (PBS), fixed in absolute methanol at −20°C for 2 min, washed three times with PBS, and processed. For immunofluorescence experiments, the coverslips were incubated for 1 h at room temperature with the primary antibody diluted in PBS solution (anti-U2AF65 MC3 antibody [dilution 1:200]). The samples were then washed three times with PBS and incubated for 1 h with the secondary antibody (Alexa 594-coupled donkey anti-mouse IgG [dilution 1:500]) plus Hoechst 33258 (0.1 μM). The samples were then washed three times in PBS and mounted with Mowiol (Calbiochem) before fluorescence analysis. The subcellular distribution of green fluorescent protein (GFP)-tagged proteins was analyzed 24 h after transfection, and Hoechst 33258 (0.1 μM) was added to the cell medium for 15 min before cell fixation. The microscopic observations of endogenous U2AF65-antibody complexes, Hoechst, or GFP-fused proteins were carried out by epifluorescence using a fluorescence microscope (DM 4000B; Leica, Wetzlar, Germany) equipped with corresponding excitation filters. Digital images were acquired with a Leica camera and processed with the Leica software package.
DNA Transfections, RNA Isolation, and Reverse Transcription (RT)-PCR Analysis
DNA transfections, total and cytoplasmic RNA isolations, and RT-PCR analyses were carried out as described previously (Izquierdo and Valcárcel, 2007b). Control experiments with different input amounts of RNA indicated that the amplification was quantitative under these conditions (Izquierdo et al., 2005). The oligonucleotide primers used were TRAIL.sense (5′-ATGATTTTGAGAACCTCTGAGGAAA-3′) and TRAIL.antisense (5′-TTTGTTTGTCGTTC-TTTGTGTTTTC-3′) for human TRAIL gene; caspase-2.sense (5′-ATGGACGAAGCGGAT-CGG-3′) and caspase-2.antisense (5′-CCCTGGCCTTAT-GATGTT-3′) for human caspase-2 gene. The primers PT1 and PT2, β-actin.sense and β-actin.antisense, as well as Fas exon5.sense and Fas exon7.antisense used to analyze alternatively spliced products from wild type/mutant Fas minigene, endogenous β-actin and Fas genes, respectively, were described previously (Förch et al., 2000; Izquierdo and Valcárcel, 2007b).
siRNA Duplex Preparation and Transfection of siRNAs
The sequences of the 21-nt siRNA oligonucleotides used for targeting TIA-1, TIAR, and PTB were synthesized and annealed as described previously (Elbashir et al., 2001; Izquierdo et al., 2005). HeLa cells were transfected at 30% confluence with 2 μg of small interfering RNAs (siRNAs) by using Lipofectin (Invitrogen). Cells were incubated for 72 h before protein extract preparation.
RESULTS
U2AF65 Is Cleaved during Apoptosis in Jurkat Cells
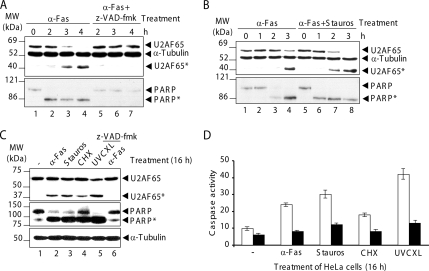
Jurkat cells were incubated with the anti-Fas antibody, a well-established apoptotic agent. After 2 h of treatment, a fragment of U2AF65 with an apparent molecular mass of 42 kDa was identified by anti-U2AF65 antibody MC3. This fragment became more prominent 3–4 h after treatment (Figure 1A, lanes 2–4, identified as U2AF65*). The cleavage of U2AF65 was inhibited when apoptosis was blocked by pretreatment for 1 h with the z-VAD-fmk peptide, a broad-spectrum caspase inhibitor (Figure 1A, lanes 5–7). It is therefore very likely that U2AF65 cleavage is a consequence of apoptosis induction. The cleavage of the well-characterized caspase-3 and -7 target protein PARP was also determined as a control marker of caspase activity (Figure 1A, bottom). In agreement with published data (Earnshaw et al., 1999), PARP cleavage leads to the appearance of an 85-kDa product as early as 2 h after antibody treatment (Figure 1A, bottom, lanes 2–4). PARP cleavage was inhibited in the presence of 5 μM z-VAD-fmk (Figure 1A, bottom, lanes 5–7). Together, these results suggest that U2AF65 is cleaved by caspases during apoptosis. However, U2AF65 cleavage was less efficient and slightly delayed compared with cleavage of PARP (Figure 1A, top and bottom, lanes 2–4).
Figure 1.
U2AF65 cleavage occurs during apoptosis. (A) Jurkat cells in the absence (lanes 1–4) or in the presence (lanes 5–7) of z-VAD-fmk were incubated with the anti-Fas antibody. (B) Jurkat cells were incubated with anti-Fas antibody alone (lanes 1–4) or with this antibody plus staurosporine (lanes 5–8). (C) HeLa cells were treated with the indicated apoptotic agents for 16 h. Lane 6, HeLa cells pretreated with z-VAD-fmk as described in A. In A–C, protein extracts were immunoblotted with the indicated antibodies. The identities of molecular mass markers and protein bands are shown. (D) Protein extracts from HeLa cells treated as in described in C were harvested to test caspase-3 or caspase-3-like activity. Where indicated (black bars), 5 μM DEVD-fmk was incubated before the addition of the substrate DEVD-p-NA. Caspase activity was displayed as the number of nanomoles of p-NA released per hour per total milligram of protein as calculated from a standard curve by using free p-NA. The values of caspase activity are means ± SEM for at least two independent samples.
U2AF65 Cleavage Is a General Apoptotic Event
To rule out whether U2AF65 cleavage was restricted to anti-Fas antibody-mediated apoptosis, cleavage was analyzed in Jurkat cells treated with anti-Fas antibody alone (Figure 1B, lanes 1–4); anti-Fas antibody together with a potent and general kinase inhibitor, staurosporine (Figure 1B, lanes 5–8); or with a strong translational elongation inhibitor, cycloheximide (data not shown). Both apoptotic agents increased the efficiency of U2AF65 cleavage (Figure 1B, top, lanes 4 and 7–8; data not shown). The extent of proteolysis seems to correlate well with the potency of the apoptotic agent used. To reinforce this point, HeLa cells were treated for 6 h (data not shown) or 16 h (Figure 1C) with the following apoptotic agents: anti-Fas antibody, staurosporine, cycloheximide, and UV light. As shown in Figure 1C (lanes 2–5), similar profiles of U2AF65 cleavage were observed in all cases. PARP cleavage and caspase-3/caspase-3–like activity were used as control markers of apoptosis (Figure 1C, middle, lanes 2–5; and D, white bars). α-Tubulin was used both as a loading control and as a protein resistant to apoptotic stimuli (Figure 1C, bottom). Inhibition of U2AF65 and PARP cleavage and caspase-3/caspase-3–like activity were observed when the HeLa cells were preincubated with z-VAD-fmk and DEVD-fmk peptides, respectively (Figure 1C, top and middle, lanes 6; and D, black bars). These data confirm that U2AF65 cleavage is a general molecular event during apoptosis.
Subcellular Localization of U2AF65 during Apoptosis
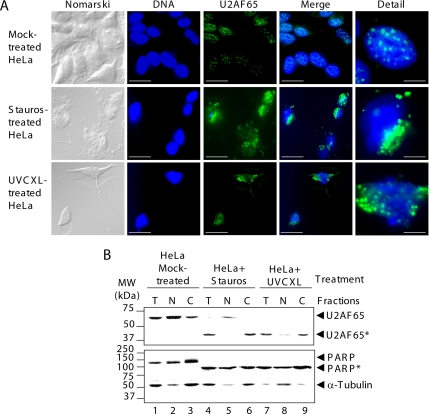
In nonapoptotic cells, U2AF65 is localized mainly in the nucleus, with accumulation in speckles (Figure 2A, top images; Gama-Carvalho et al., 1997). However, the U2AF65-related fluorescence was found in the cytoplasm of apoptotic HeLa cells treated with staurosporine or irradiated with UV light (Figure 2A, middle and bottom), suggesting changes in the subcellular distribution of U2AF65 and/or the resulting U2AF65 proteolytic fragments. To confirm this notion, the distribution pattern of the U2AF65 proteolytic fragment (42 kDa) generated upon apoptosis induction and recognized by monoclonal anti-U2AF65 antibody MC3 was investigated. This fragment was localized mainly in the cytoplasmic fraction (Figure 2B, top, lanes 4–9). Apoptosis did not change the subcellular distribution of the 85-kDa proteolytic fragment of PARP with respect to the native protein (Figure 2B, bottom). However, apoptosis induction by UV light irradiation alters nuclear pore and transport systems (Buendia et al., 1999) as shown by the distribution of α-tubulin (Figure 2B, bottom, lane 8).
Figure 2.
Subcellular localization of U2AF65 in apoptotic cells. (A) HeLa cells were mock treated or treated with staurosporine or irradiated with UV light and then stained with anti-U2AF65 mAb MC3. From left to right, images show cells examined using Nomarski method (Nomarski), stained with Hoechst through a 4,6-diamidino-2-phenylindole (DAPI) filter (DNA), or stained with anti-U2AF65 antibody and examined with a tetramethylrhodamine B isothiocyanate filter (U2AF65). The two right-hand images (Merge and Detail) are the sum of DNA and U2AF65 images as well as detailed images. In DNA, U2AF65 and Merge images, bars represent 30 μm. In detailed images, bars represents represent 5 μm. (B) Fractions containing either total protein (T), nuclei (N), or postnuclear supernatant (C) from HeLa cells mock treated (lanes 1–3), treated with staurosporine (lanes 4–6), or irradiated with UV light (lanes 7–9) were analyzed with the indicated antibodies. The migrations of molecular mass markers and protein bands are indicated.
In Vitro Cleavage of U2AF65 by Group I or III Caspases
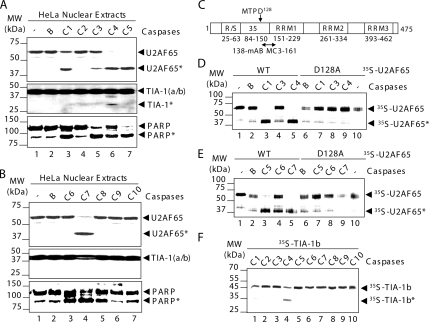
To prove that caspases are directly involved in U2AF65 cleavage and to identify the responsible caspases, in vitro cleavage assays were carried out. TIA-1 and U2AF65- enriched nuclear extracts (Dignam et al., 1983) were incubated with recombinant caspases from group I (caspase-1, -4, and -5), group II (caspase-2, -8, -9, and -10), or group III (caspase-3, -6, and -7) (Figure 3). The cleavage patterns showed the appearance of the 42-kDa proteolytic fragment of U2AF65 in the samples containing caspase-1, -3, -4, -5, and -7 (Figure 3A, lanes 3 and 5–7; and B, lane 4). Nevertheless, TIA-1—used as a specificity control—was only cleaved by caspase-4 (Figure 3A, middle, lane 6). PARP cleavage was used as a positive control of caspase activity (Figure 3, A and B, bottom, lanes 3–7). These findings demonstrate that group I or III caspases mainly cleave U2AF65 in vitro, giving rise to the same proteolytic product obtained in Jurkat and HeLa cells after apoptosis induction.
Figure 3.
U2AF65 is mainly cleaved by group I or III caspases at the aspartate128. (A and B) HeLa nuclear extracts were incubated with either no additional agents (lane 1), buffer alone (lane 2), or buffer plus each of the caspases (lanes 3–7) and analyzed by Western blotting using the antibodies against the indicated proteins. (C) Schematic structure of human U2AF65 protein. The amino acid sequence around the caspase cleavage site is shown in single-letter symbol of amino acids. The RS-rich sequence, U2AF35 binding site and RRM functional domains are indicated by open boxes, and the amino acid number is shown. The epitope recognized by anti-U2AF65 mAb MC3 is indicated. (D and E) 35S-labeled U2AF65 wt (lanes 1–5) or mutant D128A (lanes 6–10) was incubated with either none (lanes 1 and 10), buffer alone (lanes 2), or buffer plus each of the caspases indicated (lanes 3–5 and 7–9). (F) 35S-labeled TIA-1b was incubated with each of the caspases (lanes 1–10) as indicated. In panels, the positions of U2AF65, TIA-1 or PARP and molecular mass markers are shown.
Van Damme et al. (2005) suggested that the aspartic acid128 of U2AF65 protein, in the MTPD sequence context (Figure 3C), defines a noncanonical target site for caspases (Van Damme et al., 2005). The size of the U2AF65 proteolytic fragment recognized by the monoclonal antibody (mAb) MC3 (Figure 3C) and the high degree of conservation of this aspartate residue in the animal kingdom from Homo sapiens to Drosophila melanogaster (Supplemental Figure 1) are consistent with this prediction. To test this possibility, in vitro cleavage reactions were carried out using 35S-labeled U2AF65 and recombinant caspases. An efficient cleavage was obtained on incubation with caspases-1, -4, -5, and -7 (Figure 3, D and E, lanes 3 and 5). This cleavage decreased markedly when an U2AF65 mutant, in which the aspartate128 was substituted by alanine (D128A), was used (Figure 3, D and E, lanes 7 and 9). The less efficient cleavage observed with caspase-3 or -6 (Figure 3, D and E, lanes 4) was blocked when the D128A mutant protein was used as well (Figure 3, D and E, lanes 8). Efficient cleavage of 35S-labeled TIA-1 isoform b by caspase-4 was used as a specificity control (Figure 3F, lane 4). As a whole, these results confirm that aspartate128 is the caspase target site on human U2AF65 protein. An intriguing observation was that U2AF65 cleavage by caspase-6 is different in HeLa cells compared with reticulocyte lysates (compare Figure 3, B and E). This fact was also observed with TIA-1 cleavage by caspase-4 (Figure 3, A and F). The results suggest that the recombinant caspases used in our study are more effective at cleaving proteins expressed in reticulocyte lysates than proteins from HeLa nuclear extracts. The simplest explanations are that the relative quantities of target proteins for caspases are different, and/or there are additional requirements for optimal caspase activity that vary between sample sources.
The U2AF65 N-Terminal Fragment Seems to Be Localized in Nuclear Bodies
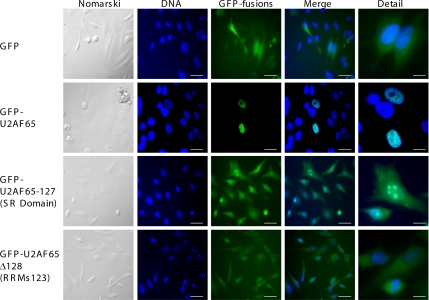
The above-mentioned results suggest that U2AF65 cleavage by caspases gives rise to an N-terminal fragment (U2AF65-127) containing the arginine and serine (RS)-rich domain, which functions as a nuclear localization signal (amino acids [aa]23-aa63) and the U2AF35 binding site (Figure 3C; Gama-Carvalho et al., 1997; Gama-Carvalho et al., 2001), and a C-terminal fragment (U2AF65-Δ128) that contains the three RRMs (Figure 3C). Therefore, it seems likely that resulting proteolytic fragments should have a different subcellular distribution (Gama-Carvalho et al., 1997, 2001). To test this hypothesis, GFP was fused to the N termini of both fragments, and HeLa cells were transfected with the corresponding constructs (Figure 4). GFP alone had a cytoplasmic and nuclear localization (Figure 4, first row), whereas GFP-U2AF65 localized mainly to speckles in the nucleus (Figure 4, second row). However, GFP-U2AF65-127 fusion protein showed a heterogeneous distribution in nuclear bodies with nucleolus-like pattern and diffuse fluorescence around the cytoplasm (Figure 4, third row). The GFP-U2AF65-Δ128 fusion protein was mainly found in the cytoplasm (Figure 4, fourth row). The distribution patterns of the GFP-tagged U2AF65 fragments were further investigated and confirmed by subcellular fractionation (data not shown). These findings were confirmed by performing localization studies in HeLa cells transfected with plasmids expressing either GFP-U2AF65 or U2AF65-GFP fusion proteins on apoptosis induction. In nonapoptotic cells, GFP-U2AF65 or U2AF65-GFP recombinant proteins were localized in nuclear speckles (Supplemental Figure 2, first and fourth rows). However, in apoptotic cells treated with staurosporine or irradiated with UV light, GFP-related fluorescence from GFP-tagged U2AF65 fragments was preferentially detected in cell clusters (N-terminal fragment) (Supplemental Figure 2, second and third rows) and cytoplasm (C-terminal fragment) (Supplemental Figure 2, fifth and sixth rows).
Figure 4.
Subcellular distribution of the U2AF65 N- and C-terminal fragments as GFP-tagged fusions. HeLa cells were transfected with plasmids expressing GFP (top images), GFP-fused with either U2AF65 (top middle images), the U2AF65 N-terminal fragment (SR domain) (lower middle images), or the U2AF65 C-terminal fragment (RRM123) (bottom images). The left-hand images show cells examined using Nomarski method. Fluorescence from GFP was observed using a fluorescein filter. To visualize nuclei, DNA was stained with Hoechst and examined using a DAPI filter. The two right-hand columns are merged and detailed images, respectively. In DNA, GFP-fusions and Merge images, bars represent 30 μm. In detailed images, bars represent 5 μm.
Functional Impact of the U2AF65 Proteolytic Fragments on Fas Splicing
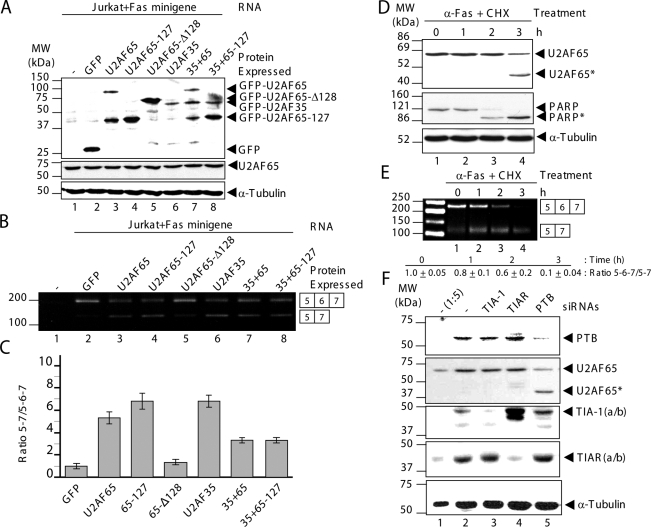
The major function of U2AF65 is the recognition of the 3′ss of pre-mRNAs during RNA splicing taking place in the nucleus (Zamore et al., 1992). Because the U2AF65 fragments generated during apoptosis were relocalized in the nucleolus and cytoplasm, it was deemed relevant to study their effects on U2AF65-dependent splicing by using a human minigene. This minigene comprises human Fas genomic sequences from exons 5–7 (Förch et al., 2000). The features of consensus sequences at the 3′ss of introns 5 and 6 are very interesting from a mechanistic viewpoint because intron 5 3′ss can be defined as weak, and thus highly U2AF65 dependent, whereas intron 6 3′ss can be defined as strong, and subsequently less U2AF65 dependent (Izquierdo et al., 2005; Pacheco et al., 2006a). U2AF65 overexpression (Figure 5A, lane 3) versus GFP (Figure 5A, lane 2) promoted partial skipping of exon 6 in Jurkat cells (Figure 5B, lane 3, and C). This was evident also when the N-terminal fragment was overexpressed (Figure 5, A and B, lanes 4; and C). Given the fact that overexpression of U2AF65 leads by itself to the appearance of a truncated fragment that comigrates with U2AF65-127 (Figure 5A, lane 3), the simplest interpretation of these results is that the effect observed is due to this truncated fragment. In contrast, overexpression of the U2AF65 C-terminal fragment (Figure 5A, lane 5) did not significantly affect Fas splicing (Figure 5B, lane 5; and C), indicating that the U2AF65 N-terminal fragment down-regulates exon 6 inclusion. This effect could be explained by assuming that overexpression of the U2AF65 N-terminal fragment sequesters endogenous U2AF35 and therefore causes dilution of functional endogenous U2AF65-U2AF35 heterodimer, which will not favor the selection of the weak 3′ss of intron 5. If this were true, U2AF35 transfection should have similar consequences. As expected, U2AF35 overexpression (Figure 5A, lane 6) promoted significant exon 6 skipping (Figure 5B, lane 6; and C). This idea was further supported in that overexpression of both U2AF subunits (Figure 5A, lane 7) can partially rescue exon 6 inclusion (Figure 5B, lane 7; and C). In the same vein, overexpression of the U2AF65 N-terminal fragment compared with U2AF35 (Figure 5A, lane 8) promotes inclusion of Fas exon 6 (Figure 5B, lane 8; and C). Similar results were also obtained using HeLa cells (data not shown). Together, these findings support the idea of dominant-negative behavior of the U2AF65 N-terminal fragment, suggesting that apoptosis-associated caspase activity disrupts U2AF65 interaction with U2AF35, which might contribute to down-regulation of Fas exon 6 splicing.
Figure 5.
U2AF65 cleavage modulates endogenous Fas alternative splicing in Jurkat cells. (A) Protein extracts from untransfected and transfected Jurkat cells with the plasmids indicated were analyzed by Western blotting using anti-GFP, anti-U2AF65, or α-tubulin antibodies. Ectopic GFP expression from GFP-tagged U2AF65 plasmids and its derivatives is shown in the top panel. Endogenous U2AF65 and α-tubulin expression and equal loading are shown in the bottom panels. (B) Cytoplasmic RNAs from Jurkat cells processed in A were analyzed by RT-PCR using primers PT1 and PT2. (C) The values of ratios between 5–7 and 5-6-7 amplification products are means ± SEM for at least two to three independent experiments. These values were expressed relative to GFP expression (B, lane 2), whose value was fixed arbitrarily to 1. (D and E) Jurkat cells were incubated with the apoptosis-inducers anti-Fas antibody (500 ng/ml) and cycloheximide (10 μg/ml) for the indicated times (0–3 h). (D) Total cell extracts were analyzed by immunoblotting using antibodies against U2AF65, PARP, and α-tubulin. (E) Cytoplasmic RNAs were isolated from apoptotic Jurkat cells in D and analyzed by RT-PCR. The products of amplification were separated by agarose-gel electrophoresis. The values of ratios between 5-6-7 and 5–7 amplification products are means ± SEM for at least two independent experiments. These values were normalized relative to lane 1, whose value was fixed arbitrarily to 1. In D and E, positions of molecular mass markers, protein bands and predicted alternatively spliced products are indicated. (F) The siRNA-mediated down-expression of PTB promotes the U2AF65 cleavage. Western blot analysis of HeLa cell lysates (5 μg [1:5] or 25 μg, lanes 1 and 2–5, respectively) prepared 72 h after transfection with siRNAs against GFP (lanes 1 and 2), TIA-1 (lane 3), TIAR (lane 4), and PTB (lane 5). The blot was probed with antibodies against PTB, U2AF65, TIA-1, TIAR, and α-tubulin proteins, as indicated. Molecular weight markers and the identities of protein bands are indicated.
To provide more evidence of the functional relevance of U2AF65 cleavage for Fas alternative splicing in a cellular context, Jurkat cells were incubated with the apoptosis inducers anti-Fas antibody and cycloheximide for the indicated times (0–3 h), and endogenous Fas exon 6 splicing event was examined (Figure 5, D and E). Cell extracts were processed by immunoblotting with antibodies against U2AF65, PARP, and α-tubulin proteins (Figure 5D) to confirm apoptosis induction and U2AF65 cleavage. Additionally, cytoplasmic RNAs were analyzed by RT-PCR (Figure 5E) to determine the alternatively spliced products of the Fas gene. The results show that U2AF65 proteolysis during early apoptosis induction for 3 h favors Fas exon 6 skipping (Figure 5E). In the same vein, the consequences of the complete U2AF65 cleavage on splicing events of endogenous pre-mRNAs during apoptosis were also investigated. In vivo splicing events of proapoptotic (Fas and caspase-2), anti-apoptotic (tumor necrosis factor-related apoptosis-inducing ligand [TRAIL]), and control (β-actin) pre-mRNAs were tested. Jurkat cells treated with anti-Fas antibody together with staurosporine for 4 h showed that full endogenous U2AF65 proteolysis occurred (Supplemental Figure 3A). In these conditions, the results indicated that the events of alternative splicing were affected differently by apoptosis than the events of constitutive splicing. This affirmation is based on the fact that inhibition of all alternative splicing events examined was observed, whereas constitutive splicing corresponding to the β-actin exons 5 and 6 seemed to be only partially affected (Supplemental Figure 3C), although in the putative contribution of the half-life of β-actin mRNA cannot be ruled out. In contrast, the “disappearance” of unspliced products might be a consequence of experimental approach used (semiquantitative RT-PCR), which does not allow amplification of unspliced endogenous mRNA precursors given that these are too large. To directly address this, a mutant Fas minigene reporter—identified as Fas ΔI6, which contains a partial deletion (1000 nt) of intron 6—was used to visualize whether Fas pre-mRNA was spliced or unspliced in mock cells and treated Jurkat cells with apoptotic agents (Supplemental Figure 3D). The results showed that in nonapoptotic Jurkat cells, the Fas ΔI6 pre-mRNA was spliced with strong exon 6 skipping, whereas the Fas pre-mRNA was unspliced on apoptosis induction (Supplemental Figure 3D). The accumulation of unspliced Fas pre-mRNA correlates well with the reduction of intact U2AF65 and the appearance of proteolyzed U2AF65. However, the constitutive splicing of endogenous β-actin pre-mRNA was partially affected in the same experimental conditions (Supplemental Figure 3D). Together, these observations are consistent with the idea that U2AF65 cleavage might represent an epigenetic event that induces drastic changes in the regulation of gene expression on apoptosis induction. The observations also reinforce the idea that proteolytic cleavage of U2AF65 may represent a potential mechanism for increasing alternative splicing relative to constitutive splicing during early apoptosis, through the imbalance between free and heterodimeric forms of U2snRNP auxiliary factors.
PTB siRNA-mediated Knockdown Promotes U2AF65 Cleavage
Finally, to illustrate whether cleaved U2AF65 is present in human cells under normal growing conditions, we screened several well-established human cell lines (HeLa, Jurkat, Daudi, 293, Rh 30, A375, T98G, HCT-116, and Hep-G2). The results indicate that cleaved U2AF65 is not found in cell lines tested (data not shown). However, HeLa cells transfected with a double-stranded siRNA against PTB showed a significant reduction in the relative U2AF65 level (Figure 5F, lane 5). Interestingly, this reduction occurred in parallel with the appearance of a 42-kDa fragment of U2AF65 (indicated as U2AF65*) in this cell extract (Figure 5F, lane 5). Normal levels of intact U2AF65 protein were observed in HeLa cells treated with siRNAs against TIA-1 or TIAR in the same depletion conditions (80%) (Figure 5F, lanes 3 and 4). The levels of α-tubulin were used as loading control (Figure 5F, lanes 1–5). Given that PTB-siRNA transfection led to the altered morphology in which a significant number of the cells were round and lost adherence (data not shown; He et al., 2007), the appearance of the 42-kDa proteolytic fragment of U2AF65 was considered a consequence of genotoxic response/stress to PTB depletion.
DISCUSSION
We report here that human U2AF65 cleavage is a general apoptotic event dependent on groups I or III caspase activity, generating two U2AF65 fragments with a prevalent subcellular distribution in nuclear bodies that seem to be nucleoli (N-terminal fragment) and cytoplasm (C-terminal fragment). The relevant biological consequences for the regulation of Fas alternative splicing are derived from this observation.
Cellular Consequences of U2AF65 Cleavage by Caspases
The caspase recognition of Asp128 in U2AF65 is preceded by a noncanonical caspase target sequence containing the amino acids MTPD. This sequence represents a nonconventional cleavage site for caspases from groups I—with conventional cleavage site including WEHD and (W/L)EHD—and III—with conventional cleavage site including DEVD and VEHD (Lavrik et al., 2005). However, the sequence together with its context is highly conserved from Drosophila to human (Supplemental Figure 1), implying that U2AF65 cleavage during apoptosis may play an important physiological role in a large group of animals. In addition, this finding may suggest that not only the primary sequence but also the conformation of U2AF65 may contribute to caspase recognition. Our results also suggest that the effector caspases from groups I and III are the caspases that specifically cleave this target protein during apoptosis.
U2AF65 is a protein that resides in the nucleus, mainly in speckles (Gama-Carvalho et al., 1997), which are nuclear bodies where transcription and splicing factors accumulate. The results presented suggest that after apoptosis induction and caspase activation, U2AF65 is cleaved into two parts: the N-terminal fragment, containing the R/S domain and U2AF35 binding motif; and the C-terminal fragment, which contains the three RRMs. By transfecting HeLa cells with the GFP/U2AF65-RS-35 and GFP/U2AF65-RRM1, -2 and -3 derivative constructs, which produce GFP-fused to N-/C-terminal fragments of U2AF65, we have demonstrated that these U2AF65 fragments seem to be localized in the nucleolus and cytoplasm, even when the localization studies are performed on induction of apoptosis. Subcellular redistribution of U2AF65 fragments may be attributed to several phenomena occurring during apoptosis. First, the N-terminal region of U2AF65 (up to RRM1) containing a stretch of basic amino acids followed by RS dipeptides and a putative bipartite nuclear localization signal (NLS) (Gama-Carvalho et al., 2001) is separated from the C-terminal region (RRM1, -2, and -3). Therefore, the RS domain on U2AF65 can act as an NLS, and it is sufficient to direct a heterologous protein to the nucleus. Moreover, the fragment containing the RS-rich domain is sufficient to relocalize a heterologous protein to the nucleolus. Several experimental results suggest that RS domains can interact with both single- and double-strand RNAs such as intron/exon RNA sequences or the ribosomal RNAs localized to the nucleolus (Shen and Green, 2006). Given that GFP-U2AF65-127 fusion protein is small it probably enters the nucleolus by diffusion and is retained by nonspecific binding of the RS domain to rRNA (Gama-Carvalho et al., 2001; Shen and Green, 2006). Second, with regard to the cytoplasmic localization of this protein, modification of the nuclear pore complex occurs during apoptosis. Several reports have suggested that the activation of caspases during apoptosis increases nuclear pore permeability by cleaving components of both the nuclear pore and the nuclear transport system (Buendia et al., 1999; Ferrando-May, 2005). These modifications result in leakage of proteins restricted to the nucleus into the cytoplasm and vice versa (Figure 2B; see α-tubulin in lane 8). Third, inhibition of transcription during apoptosis may partly contribute to the relocalization of U2AF65 fragments, because inhibition of cellular transcription by actinomycin D induces the partial redistribution of U2AF65 to the cytoplasm (Gama-Carvalho et al., 1997).
Early recognition of the 3′ ends of introns is achieved in higher eukaryotes by the U2AF, which is made up of a large subunit (U2AF65) and a small subunit (U2AF35) that interact to form a stable heterodimer. U2AF65 binds to the polypyrimidine tract upstream from the 3′ splice site and promotes U2 snRNP binding to the pre-mRNA; therefore, it is an essential factor for splicing. When apoptosis is induced and U2AF65 is cleaved, the nuclear functions of complete U2AF65, such as the control of Fas alternative splicing, are compromised. Our findings (Figure 5, A–C) suggest that apoptosis-associated caspase activity disrupts U2AF65 functional interaction with U2AF35, which might contribute to splicing modulation. This observation could be explained by assuming that the U2AF65 N-terminal fragment—the U2AF65 C-terminal fragment generated by caspases was mainly localized in the cytoplasm and did not affect Fas splicing—promotes dilution of functional endogenous U2AF65-U2AF35 heterodimer, which will not favor the selection of the 3′ splice sites. These findings agree with previous results that strongly suggest that U2AF regulates the splicing of a subset of endogenous pre-mRNAs containing alternative 3′ss associated with PTs of different strengths (Pacheco et al., 2006a,b). Therefore, it is tempting to speculate that a fine-tuning regulation is operating to control the amount of each factor to compensate the cellular balance of free U2AF65 or U2AF35 and U2AF65-U2AF35 heterodimer. Additionally, U2AF65 has been implicated in the regulation of 3′ end processing (Millevoi et al., 2006; Danckwardt et al., 2007). In this regard, we speculate that the coupling of splicing and 3′ end processing of mammalian pre-mRNAs should be altered as well. A very striking common feature of the apoptosis-targeted proteins is the presence of an RNA binding motif, which is likely to play a role in RNA splicing (Fischer et al., 2003). The physiological significance of such selective changes on RNA-binding protein family members and/or spliceosome machinery-associated factors during apoptosis remains unknown. It will certainly be a challenge to understand a process as complex as splicing/3′ end processing in the context of apoptosis, where multiple factors are processed and may be functionally modified. In contrast, U2AF65 cleavage and the resulting fragments could also have functional consequences for the metabolism of the cytoplasmic mRNAs. In our experiments, the U2AF65 N- and C-terminal portions had no effect on the translational activity of the β-galactosidase reporter mRNA (data not shown). However, U2AF65 cleavage during apoptosis may modulate translation/stability of particular species of mRNAs given that a role for U2AF65 in the cytoplasmic metabolism of specific mRNAs has been reported (Gama-Carvalho et al., 2006).
Fas Splicing Regulation during Apoptosis
Fas is a quintessential death receptor, and cells undergo apoptosis on ligation. Alternative Fas pre-mRNA splicing can regulate the sensitivity of Fas-expressing cells to Fas-induced apoptosis. The protein isoform of the mRNA lacking exon 6 encodes the soluble form of the receptor able to inhibit Fas signaling (Cheng et al., 1994). This process is regulated in a number of physiological situations, including activation-mediated T cell death and autoimmune lymphoproliferative disorders (Bidere et al., 2006). However, other studies have shown that soluble human Fas protein can induce death of transformed cells by “reverse” apoptotic signaling via transmembrane Fas ligand (Proussakova et al., 2003). Its cytotoxicity seems to be linked to the oligomerization of soluble Fas antigen (Proussakova et al., 2003). Thus, several questions need to be answered about this issue in futures studies: What is the role of altered splicing of Fas protein? Would this inactivate Fas production? Is this a protective response, that is, could it prevent apoptosis, or is it more likely to accelerate cell death by apoptosis? Greater understanding of the mechanisms that regulate function and expression of the Fas receptor as well as its mediation in other processes, including cell proliferation or differentiation and their signaling pathways, will provide relevant insight into the regulatory circuits of apoptosis.
Apoptosis is a crucial event in many biological processes (Earnshaw et al., 1999; Krammer, 2000; Adams, 2003). The essential splicing factor U2AF65 participates in spliceosome recruitment and regulation of constitutive and alternative splicing. The present study provides important new information on cleavage of U2AF65 during apoptosis. Cleaved U2AF65 may influence the progression of apoptosis, particularly through effects on alternative splicing control. According to the current understanding of apoptosis, caspase activation can be considered the point of no return, that is, the cell seems to be definitively committed to die. However, situations might arise in which caspases are activated but cells do not enter apoptosis due to overexpression of inhibitors of apoptosis and/or survival factors. It is feasible that regulation of splicing under these conditions influences the apoptotic process in a long-term manner, as a second wave, by producing inhibitors or activators of survival. There are many examples of alternatively spliced factors with pro- or anti-apoptotic activities (for review, see Schwerk and Schulze-Osthoff, 2005). Why should a cell programmed to die modify its RNA splicing metabolism? The regulation of apoptosis might require a sudden response, which would be obtained by posttranscriptional RNA processing rather than by the biosynthesis of RNA. For example, it might be important to remove ribonucleoprotein complexes, perhaps to quickly mount a comprehensive assault on most of the cell's vital systems and to allow hierarchical and irreversible destruction of the cell (Keene and Tenenbaum, 2002; Mata et al., 2005; Moore, 2005; Leung and Sharp, 2006; Prasanth and Spector, 2007). In summary, U2AF65 undergoes proteolytic processing by caspases. This processing is associated with a reduction in U2AF65 expression, the formation of two discrete cleavage fragments, and alterations in U2AF65 splicing activity. As such, this caspase-mediated processing may represent an additional mechanism to modulate U2AF65 functionality and Fas alternative splicing under apoptotic conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Valcárcel, J. M. Sierra, and L. M. Soares for assistance, help, and encouragement. We thank J. Alcalde for technical assistance. We are grateful for the generosity of A. Castelló and C. W. Smith, who kindly providing caspase-3 colorimetric assay kit/anti-caspase 3 antibody and anti-PTB BB7 antibody, respectively. This work was supported by a grant from Fondo de Investigaciones Sanitarias (PI051605). The Centro de Biología Molecular “Severo Ochoa” receives an institutional grant from Fundación Ramón Areces.
Abbreviations used:
- NLS
nuclear localization signal
- RRM
RNA recognition motif
- RS
arginine-serine
- SF1/BBP
branch-point binding protein
- U2AF65
U2 snRNP auxiliary factor 65 kDa
- U2AF35
U2 snRNP auxiliary factor 35 kDa.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1125) on May 28, 2008.
REFERENCES
- Adams J. M. Ways of dying: multiple pathways to apoptosis. Genes Dev. 2003;17:2481–2495. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- Banerjee H., Rahn A., Davis W., Singh R. Sex-lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA. 2003;9:88–99. doi: 10.1261/rna.2131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidere N., Su H. C., Lenardo M. J. Genetic disorders of programmed cell death in the immune system. Annu. Rev. Immunol. 2006;24:321–352. doi: 10.1146/annurev.immunol.24.021605.090513. [DOI] [PubMed] [Google Scholar]
- Buendia B., Santa-Maria A., Courvalin J. C. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 1999;112:1743–1753. doi: 10.1242/jcs.112.11.1743. [DOI] [PubMed] [Google Scholar]
- Cheng J., Zhou T., Liu C., Shapiro J. P., Brauer M. J., Kiefer M. C., Barr P. J., Mountz J. D. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- Danckwardt S., et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Martins L. M., Kaufmann S. H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E. Nucleocytoplasmic transport in apoptosis. Cell Death Differ. 2005;12:1263–1276. doi: 10.1038/sj.cdd.4401626. [DOI] [PubMed] [Google Scholar]
- Fischer U., Janicke R. U., Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P., Puig O., Kedersha N., Martínez C., Granneman S., Seraphin B., Anderson P., Valcárcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M., Carvalho M. P., Kehlenbach A., Valcárcel J., Carmo-Fonseca M. Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J. Biol. Chem. 2001;276:13104–13112. doi: 10.1074/jbc.M008759200. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss R. D., Chiang L., Valcárcel J., Green M. R., Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M., Barbosa-Morais N. L., Brodsky A. S., Silver P. A., Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Pool M., Darcy K.M., Lim S.B., Auersperg N., Coon J.S., Beck W.T. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo J. M., Majós N., Bonnal S., Martínez C., Castelo R., Guigo R., Bilbao D., Valcárcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Izquierdo J. M., Valcárcel J. A simple principle to explain the evolution of pre-mRNA splicing. Genes Dev. 2006;20:1679–1684. doi: 10.1101/gad.1449106. [DOI] [PubMed] [Google Scholar]
- Izquierdo J. M., Valcárcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007a;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- Izquierdo J. M., Valcárcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 2007b;282:19410–19417. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- Keene J. D., Tenenbaum S. A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Krammer P. H. CD95's deadly mission in the immune system. Nature. 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- Lavrik I. N., Golks A., Krammer P. H. Caspases: pharmacological manipulation of cell death. J. Clin. Invest. 2005;115:2665–2672. doi: 10.1172/JCI26252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K.L., Sharp P. A. Function and localization of microRNAs in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- Mata J., Marguerat S., Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Merendino L., Guth S., Bilbao D., Martínez C., Valcárcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Millevoi S., Loulergue C., Dettwiler S., Karaa S. Z., Keller W., Antoniou M., Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Pacheco T. R., Coelho M. B., Desterro J.M.P., Mollet I., Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 2006a;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco T. R., Moita L. F., Gomes A. Q., Hacohen N., Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell. 2006b;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Spector D. L. Eukaryotic regulatory RNAs: an answer to the “genome complexity” conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Proussakova O. V., Rabaya N. A., Moshnikova A. B., Telegina E. S., Turanov A., Nanazashvili M. G., Beletsky I. P. Oligomerization of soluble Fas antigen induces its cytotoxicity. J. Biol. Chem. 2003;278:36236–36241. doi: 10.1074/jbc.M305896200. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Schwerk C., Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Selenko P., Gregorovic G., Sprangers R., Stier G., Rhani Z., Kramer A., Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- Shen H., Green M. R. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier E. A., Frato K. E., Shen H., Paranawithana S. R., Green M. R., Kielkopf C. L. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares L. M., Zanier K., Mackereth C., Sattler M., Valcárcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- Valcárcel J., Gaur R. K., Singh R., Green M. R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- Wu S., Romfo C. M., Nilsen T. W., Green M. R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Patton J. G., Green M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zorio D., Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.