Abstract
Sporulation in the fission yeast Schizosaccharomyces pombe is a unique biological process in that the plasma membrane of daughter cells is assembled de novo within the mother cell cytoplasm. A double unit membrane called the forespore membrane (FSM) is constructed dynamically during meiosis. To obtain a dynamic view of FSM formation, we visualized FSM in living cells by using green fluorescent protein fused with Psy1, an FSM-resident protein, together with the nucleus or microtubules. The assembly of FSM initiates in prophase II, and four FSMs in a cell expand in a synchronous manner at the same rate throughout meiosis II. After the meiosis II completes, FSMs continue to expand until closure to form the prespore, a spore precursor. Prespores are initially ellipsoidal, and eventually become spheres. FSM formation was also observed in the sporulation-deficient mutants spo3, spo14, and spo15. In the spo15 mutant, the initiation of FSM formation was completely blocked. In the spo3 mutant, the FSM expanded normally during early meiosis II, but it was severely inhibited during late and postmeiosis, whereas in the spo14 mutant, membrane expansion was more severely inhibited throughout meiosis II. These observations suggest that FSM expansion is composed of two steps, early meiotic FSM expansion and late and post meiotic FSM expansion. Possible regulatory mechanisms of FSM formation in fission yeast are discussed.
INTRODUCTION
Sporulation is an intriguing biological process in that a novel cell is synthesized within the mother cell cytoplasm. Schizosaccharomyces pombe cells initiate a sporulation program when challenged by nutrient starvation, particularly starvation for nitrogen (Egel and Egel-Mitani, 1974; Yamamoto et al., 1997). Sporulation consists of two overlapping processes, meiosis and spore morphogenesis. The most important event of spore morphogenesis is the assembly of double-layered intracellular membranes, termed forespore membranes (FSMs), which develop into the spore plasma membrane (Yoo et al., 1973). During meiosis II, FSMs are assembled by the fusion of membrane vesicles. At this stage, the spindle pole body (SPB), which plays a crucial role in spindle microtubule formation, undergoes a morphological transformation into a multilayered structure as revealed by electron microscopy (Hirata and Tanaka, 1982; Tanaka and Hirata, 1982). As analyzed by fluorescence microscopy using an anti-Sad1 (an SPB component) antibody, compact dots of SPB are transformed into crescent shapes coincident with the initiation of FSM formation (Hagan and Yanagida, 1995; Ikemoto et al., 2000). These morphological alterations of the SPB are called “SPB modification” (Nakase et al., 2008). After SPB modification, membrane vesicles are then recruited to the vicinity of the modified SPBs, and subsequently fuse there to generate forespore membranes (Hirata and Tanaka, 1982; Tanaka and Hirata, 1982). As the nucleus divides in meiosis II, the FSM expands, and eventually it encapsulates a haploid nucleus generated by two rounds of meiosis, producing the membrane-bounded precursor of the spore, the prespore. In the space between the inner and outer membranes of the FSM, spore wall materials are deposited to form layers of spore walls (Yoo et al., 1973; Hirata and Tanaka, 1982). Finally, the inner layer of the FSM becomes the spore plasma membrane, whereas the outer layer of the membrane autolyzes. Mature spores are then liberated from an ascus upon autolysis of the ascus walls (Tanaka and Hirata, 1982). The basic process of sporulation in the budding yeast Saccharomyces cerevisiae (Byers, 1981; Kupiec et al., 1997; Neiman, 2005), is very similar to that of S. pombe as described above.
To address the molecular mechanism of sporulation, we have identified and analyzed a number of sporulation-specific genes (Bresch et al., 1968; Ikemoto et al., 2000; Asakawa et al., 2001; Nakamura et al., 2001, 2002, 2005; Nakase et al., 2001, 2004, 2008; Nakamura-Kubo et al., 2003; Yoshida et al., 2005; Ye et al., 2007). Spo2, Spo13, and Spo15 proteins localize to the SPB. The morphological transformation of the SPB is not detected in either spo2, spo13, or spo15 mutants, suggesting that these proteins are involved in the initiation of FSM formation (Ikemoto et al., 2000; Nakase et al., 2008). spo3+ is a sporulation-specific gene encoding an FSM-integrated transmembrane protein. Anucleated spores are often observed in the spo3 mutant, because FSM formation is impaired (Hirata and Shimoda, 1992; Nakamura et al., 2001). In addition to these genes, the general secretion apparatus is also required for vesicle transport during sporulation. The S. pombe Sec12 homologue, Spo14/Stl1, is necessary for proper construction of the FSM (d'Enfert et al., 1992; Nakamura-Kubo et al., 2003). Sec12 is responsible for vesicle transport from the endoplasmic reticulum (ER) to the Golgi apparatus in S. cerevisiae (Nakano et al., 1988). Spo20 is structurally and functionally related to the major S. cerevisiae phosphatidylinositol/phosphatidylcholine-transfer protein Sec14, which is required for vesicle formation from the Golgi apparatus (Bankaitis et al., 1990; Nakase et al., 2001). Spo20 regulates formation of the FSM, in addition to its known roles in post-Golgi vesicle trafficking (Nakase et al., 2001). Additional sec genes are also involved in FSM formation (Nakamura et al., 2001, 2005).
The spatiotemporal coordination of meiotic nuclear divisions and FSM formation is essential for accurate distribution of the genome into four haploid spores. Cytological observations by electron and fluorescence microscopy described above have been obtained with fixed specimens. To gain a better understanding of sporulation in S. pombe, appreciation for the dynamic aspect of FSM formation is essential. Here, we made time-lapse observations of living cells, which provides information and insights that cannot be obtained from fixed specimens. We report the behavior of FSMs in individual living cells of wild-type and sporulation-deficient mutant strains. Based on these observations, we propose a model for FSM assembly in S. pombe.
MATERIALS AND METHODS
Yeast Strains, Media, and Transformation
The S. pombe strains used in this study are listed in Table 1. The media used have been described previously (Egel and Egel-Mitani, 1974; Gutz et al., 1974; Moreno et al., 1990). S. pombe cells were grown at 30°C and sporulated at 28°C.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YN68 (FY12296)a | h90 leu1<<GFP-psy1 | Nakase et al. (2004) |
| YN104 (FY12332)a | h90 spo15::ura4 ade6-M210 leu1-32 ura4-D18 | This study |
| TN106 (FY13150)a | h90 ade6-M216 leu1-32 | This study |
| TN200 (FY7331)a | h90 spo3::ura4 ade6-M216 leu1-32 ura4-D18 | This study |
| TN240 | h90 spo3::ura4 ade6<<GFP-psy1 leu1-32 ura4-D18 | This study |
| TN241 | h90 spo14-B221 leu1<<GFP-psy1 | This study |
| TN242 | h90 spo15::ura4 leu1<<GFP-psy1 ura4-D18 | This study |
| TN340 | h90 ade6<<YFP-psy1 leu1-32 | This study |
| TN341 | h90 spo3::ura4 ade6<<YFP-psy1 leu1-32 ura4-D18 | This study |
| TN342 | h90 spo14-B221 ade6<<YFP-psy1 leu1-32 ura4-D18 | This study |
| TN344 | h90 ade6<<YFP-psy1 leu1-32 pREP1(CFP-atb2) | This study |
| TN345 | h90 spo3::ura4 ade6<<YFP-psy1 leu1-32 ura4-D18 pREP1(CFP-atb2) | This study |
| TN346 | h90 spo14-B221 ade6<<YFP-psy1 leu1-32 pREP1(CFP-atb2) | This study |
| TN349 | h90 spo15::ura4 ade6<<YFP-psy1 leu1-32 ura4-D18 | This study |
| TN353 | h90 spo15::ura4 ade6<<YFP-psy1 leu1-32 ura4-D18 pREP1(CFP-atb2) | This study |
| MKL216 (FY7259)a | h90 spo14-B221 ade6-M216 leu1-32 | This study |
a Yeast Genetic Resource Center of Japan supported by the National BioResource Project (YGRC/NBRP) (http:/yeast.lab.nig.ac.jp/nig/). The S. pombe strains constructed in this study will be deposited at the YGRC/NBRP.
Plasmid Construction
Plasmid pIA was constructed by inserting a 3-kb fragment containing the ade6+ gene into the SspI site of pBluescript II KS− (Stratagene, La Jolla, CA). Two oligonucleotides were used to amplify the yellow fluorescent protein (YFP) gene by polymerase chain reaction (PCR) by using primer FP1 5′-CAGTCCTCGAG(XhoI)CATGGTGAGCAAGGGCGAGGAGCTG-3′ and FP2 5′-CAGTCGGATCC(BamHI)GTCGACTTGTACAGCTCGTCCATGCCGAG-3′as primers. Underlined sequences indicate the restriction enzyme sites. The PCR product was digested with XhoI and BamHI, and then ligated into the corresponding site of pIA, yielding pIA(YFP). The psy1+ open reading frame (ORF) was then amplified by PCR from genomic DNA, digested with SalI and SacI (sites included in the primers), and ligated into SalI/SacI-digested pIA(YFP), yielding pIA(YFP-psy1ORF). The psy1+ promoter region was then amplified by PCR from genomic DNA, digested with XhoI (sites included in the primers), and ligated into XhoI-digested pIA(YFP-psy1ORF), yielding pIA(YFP-psy1). Finally, pIA(YFP-psy1) was linearized by digestion at the BamHI site in ade6+ and used to transform strains TN106, YN104, TN200, and MKL216 (Table 1), yielding strains TN340, TN349, TN341, and TN342, respectively. The cyan fluorescent protein (CFP) gene was amplified by PCR using primers FP1 and FP2. The PCR product was digested with XhoI and BamHI, and then they were ligated into the corresponding site of pREP1 (Maundrell, 1993), yielding pREP1(CFP). pREP1(CFP-atb2) was constructed as follows. Two oligonucleotides were used to amplify the atb2+ gene by PCR using 5′-CCCGTCGAC(SalI)AATGAGAGAGATCATTTCCAT-3′ and 5′-CCCGAGCTC(SacI)GGAAGAGAAATTAGTTTCAAA-3′ as primers. The PCR product was digested with SalI and SacI, and then they were ligated into the corresponding site of pREP1 (Maundrell, 1993), yielding pREP1(CFP-atb2).
Live Analysis
Time-lapse microscopy was performed in growth chambers kept at 28°C with inverted fluorescence microscopes (IX-71; Olympus, Tokyo, Japan) equipped with a 60×/1.25 numerical aperture (NA- or 100×/1.35 NA oil immersion objective (Plan-Apo; Olympus), a cooled charge-coupled device (CCD) camera (ORCA-ER; Hamamatsu Photonics, Hamamatsu, Japan), and a filter wheel (Mac5000; Ludl Electronic Products, Hawthorne, NY) populated with Chroma filters (Chroma Technology, Rockingham, VT). The CCD camera, a filter wheel, and image acquisition were controlled by AQUACOSMOS software (Hamamatsu Photonics). Digital images were processed with Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA).
To observe green fluorescent protein (GFP)-tagged Psy1 and chromatin, living cells scraped from an agar plate were suspended in 1 μg/ml Hoechst 33342, a DNA-specific fluorescence dye, and incubated for 5 min. Cells were collected by centrifugation, suspended in SSL-N medium, and placed onto a thin film of 2% agarose gel. The film was sandwiched between a pair of coverslips and placed on the stage. Live analysis was performed using a Chroma 86013 filter set (Chroma Technology). To observe CFP-Atb2 and YFP-Psy1 simultaneously, samples were prepared as described above without staining by Hoechst 33342. In this case, a Chroma 86006 filter set (Chroma Technology) was used. Both GFP- and YFP-tagged Psy1 are functional, because these fusion genes complemented the lethality of psy1Δ (Nakamura et al., 2001; data not shown).
RESULTS
Live Observation of Forespore Membrane Formation
To our knowledge, the dynamic behavior of FSM in live cells has not yet been documented. We previously showed that the fission yeast syntaxin-like protein Psy1 localizes to the FSM. The use of the GFP-Psy1 fusion construct may allow observation of FSM formation in living cells. Strain YN68, in which the GFP-tagged psy1+ gene was chromosomally integrated and was driven by its own promoter, was cultured on sporulation medium (SSA) for 16 h, stained with Hoechst 33342, and GFP-Psy1 and chromatin were then simultaneously traced by time-lapse fluorescence microscopy. Supplemental Figure 1 and Supplemental Movie 1 show the behavior of FSMs and nuclei observed in a single living cell over time from an early stage of meiosis II to the completion of the prespore. GFP-Psy1 was initially dispersed in the cytoplasm before meiosis II started (0–4 min). A pair of GFP-Psy1 dots seemed at opposite sides of each nucleus (6 min), and then four dots in a cell expanded to form cup-shaped structures in a synchronized manner (8–18 min). Subsequently, as the nuclei separated away from each other during anaphase II, the two cup-shaped structures also separated with each nucleus (22–32 min). After meiosis II ended, the FSMs were still expanding (38 min), and eventually closed into sacs (46 min). Finally, the sacs gradually became spherical (46–64 min). Similar behavior of the FSM was observed with another FSM-localized protein, Spo3-GFP (data not shown).
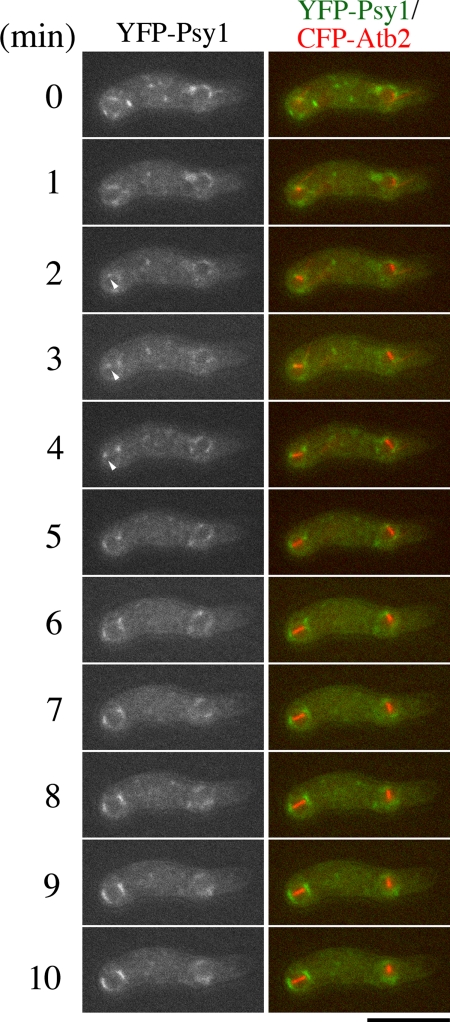
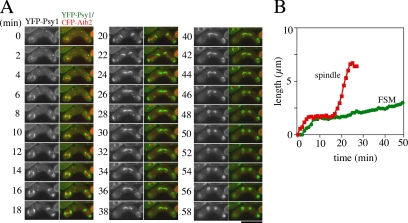
Our observations suggest that FSM formation is highly coordinated with meiosis II. To examine spatiotemporal relationships between FSM formation and meiosis II, we observed both FSMs and meiosis II in more detail. For this purpose, we tagged the psy1+ gene with the YFP gene and the atb2+ gene, encoding an α-tubulin, with the CFP gene and performed two-color imaging. A previous report revealed that, in meiotic cells expressing low levels of GFP-Atb2, meiosis proceeded normally and viable spores were formed (Ding et al., 1998). We confirmed that CFP-Atb2 also behaved like GFP-Atb2 in meiosis (data not shown). The normal spindle dynamics in mitosis consists of three distinct periods: phase I, phase II, and phase III (Nabeshima et al., 1998). In phase I, the spindle elongates to an average length of 2.5 μm. Phase II is characterized by a period of constant spindle length. Phase III is the period at which the spindle elongates rapidly to an average length of 10 μm. Phase I corresponds to a prophase-like stage. Most of phase II is prometaphase and metaphase, whereas sister chromatid separation (anaphase A) occurs at the end of this phase. Phase III begins immediately after or simultaneously with the onset of anaphase B. These three phases have a strong resemblance to the principal events occurring in mitosis in higher eukaryotes (Nabeshima et al., 1998). Essentially the same phases are observed in meiosis (Yamamoto and Hiraoka, unpublished data). Therefore, simultaneous observation of YFP-Psy1 and CFP-Atb2 allows examination of the temporal relationship between FSM formation and meiosis II. The contour length of FSM was measured to monitor its expansion (Figure 1B). As shown in Figure 1A and in Supplemental Movie 2, shortly after meiosis II started (when spindle microtubules were formed), the YFP-Psy1 signal showed up as four dots at both ends of the meiotic spindles (6 min), confirming that FSM formation initiates at SPB. Both electron microscopy and fluorescence microscopy by using fixed specimens reveal that FSM formation initiates during meiosis II. However, the stage of meiosis II at which the assembly of FSM initiates remains unclear. We next examined the appearance of YFP-Psy1 dots in greater detail. Because the FSM is initially assembled on the SPB by the fusion of membrane vesicles, we defined the initiation of FSM formation as the appearance of GFP- or YFP-Psy1 dots at the end of the spindle microtubules. As shown in Figure 2, soon after the spindle microtubules seemed during meiosis II, two dots of YFP-Psy1 were observed at both ends of each spindle microtubule. Table 2 shows the timing of appearance of the YFP-Psy1 dot at the ends of the spindle microtubules. The duration from the onset of meiosis II (appearance of spindle microtubules) to the appearance of YFP-Psy1 dots at the ends of the microtubule spindles (Table 2, dot) was apparently shorter than phase I, indicating that FSM formation initiates during phase I. Because phase I corresponds to prophase II, FSM formation initiates at prophase II. The YFP-Psy1 signal then expanded at a constant rate. After meiosis II ended (the spindle microtubules collapsed at 34 min), the FSM continued to expand for ∼20 min until closing of the FSM to form a prespore (Figure 1A). Finally, the sacs became spherical. The duration from closure of FSM to becoming spherical was ∼18 min (Table 2, to sphere).
Figure 1.
Simultaneous observation of FSM and microtubules. (A) Two color live imaging of YFP-Psy1 (FSM) and CFP-Atb2 (microtubules) in wild-type strain TN344. In merged images, YFP-Psy1 (green) and CFP-Atb2 (red) are shown. Frames were taken every 2 min. Bar, 10 μm. (B) Kinetics of FSM expansion and spindle extension during meiosis II. The contour length of one out of four FSMs (green line) and one of two spindle microtubules (red line) was measured by Aquacosmos software and plotted.
Figure 2.
FSM initiation during meiosis II. Two-color live imaging of YFP-Psy1 (green) and CFP-Atb2 (red) in wild-type strain TN344. The arrowheads indicate the position of YFP-Psy1 dots at the ends of the spindle microtubule. Frames were taken every 1 min. Bar, 10 μm.
Table 2.
Temporal relationship between FSM formation and meiosis II
| Duration of meiosis II (min) |
Duration of events of FSM formation (min) |
n | |||||
|---|---|---|---|---|---|---|---|
| Phase I | Phase II | Phase III | Dota | Closureb | To spherec | ||
| Wild type | 7.89 ± 1.45 | 14.78 ± 2.28 | 10.52 ± 1.48 | 4.73 ± 1.68 | 19.16 ± 2.91 | 17.76 ± 3.68 | 10 |
| spo3Δ | 7.57 ± 1.40 | 13.00 ± 2.17 | 9.91 ± 1.20 | 5.07 ± 1.00 | 24.67 ± 3.55 | N.D. | 15 |
| spo14 | 7.64 ± 1.45 | 15.00 ± 3.11 | 10.68 ± 1.25 | 4.64 ± 0.93 | 26.96 ± 4.29 | N.D. | 14 |
| spo15Δ | 8.41 ± 1.62 | 16.17 ± 6.34 | 9.57 ± 1.28 | 12 | |||
N.D., not determined because the prespores in these mutants were too small for an accurate determination of shape.
a From initiation of phase I to the appearance of YFP-Psy1 dots at the edge of spindle microtubules.
b From completion of meiosis II to closure of the FSM.
c From closure until the prespore became spherical.
Observations of fixed specimens (Nakamura et al., 2001) and time-lapse observation of living cells (Figures 1 and 2) suggest that the four FSMs expand synchronously. To confirm this, the contour length of each of the FSMs was measured. As shown in Figure 3(wild type), four FSMs expanded in a synchronous manner.
Figure 3.
Synchrony of the expansion of four FSMs. The contour length of four FSMs was measured by Aquacosmos software and plotted. Wild type (YN68), spo3Δ (TN240), and spo14 (TN241).
FSM Formation Is Completely Blocked in spo15 Mutant Cells
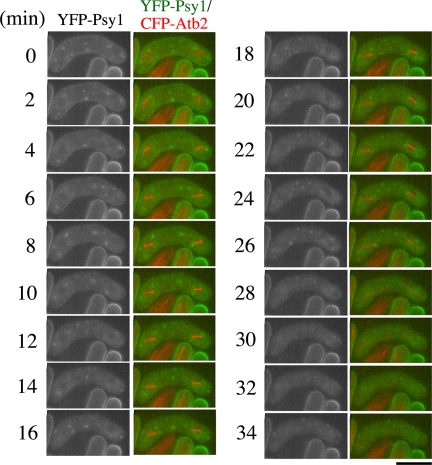
To gain further insight into the molecular mechanism of FSM formation, we examined FSM formation in sporulation-deficient mutants (spo). We have isolated several spo+ genes that are involved in sporulation, and we have determined their molecular functions. Most spo mutants are defective in FSM formation. Therefore, we presumed that live observation of the FSM in spo mutants would allow dissection of the events occurring during FSM formation. Spo15 is a large protein with a predicted molecular mass of 220 kDa containing coiled-coil regions. Spo15 is associated with SPB throughout the life cycle. spo15 mutants are defective in morphological changes that occur in SPBs during meiosis. These results suggest that Spo15 plays a key role in FSM initiation (Ikemoto et al., 2000). However, we have no direct evidence that Spo15 is involved in the initiation of FSM formation. Therefore, we examined the behavior of FSM by time-lapse observation in spo15 mutant cells. Strain, TN242, which expresses GFP-Psy1, was cultured in sporulation medium, stained with Hoechst 33342, and then GFP-Psy1 was traced by time-lapse fluorescence microscopy. Like wild-type cells, GFP-Psy1 was dispersed in the cytoplasm of the spo15 zygote before meiosis II (Supplemental Figure 2, 0–4 min). However, no FSM-like structure was observed when meiosis II progressed (20–44 min; Supplemental Movie 3). Next, we observed the behavior of FSMs and microtubules simultaneously in the spo15Δ null mutant. In wild-type cells, YFP-Psy1 dots showed up at the ends of the spindle microtubules during prophase II (Figure 2). When meiosis II initiated, YFP-Psy1 dots were not detected at the ends of spindle microtubules. The FSM-like structures were not formed even after meiosis II ended (Figure 4). Given its localization to SPB, these results demonstrate that Spo15 plays an essential role in initiation of FSM formation.
Figure 4.
Behavior of YFP-tagged Psy1 during sporulation in the spo15 mutant. Two-color live imaging of YFP-Psy1 and CFP-Atb2 in spo15Δ mutant strain TN353. In merged images, YFP-Psy1 (green) and CFP-Atb2 (red) are shown. Frames were taken every 2 min. Bar, 10 μm.
FSM Expansion during Late Meiosis II and Afterward Is Impaired in spo3 Mutant Cells
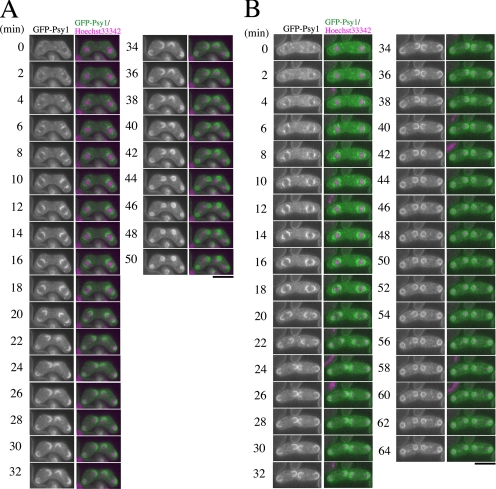
The expansion of the FSM is mediated by fusion of vesicles. The spo3+ gene encodes a protein that has a transmembrane region at its N terminus. Indeed, Spo3 localizes to the FSM and is essential for its expansion (Nakamura et al., 2001). Electron microscopy revealed that membrane vesicles accumulated in the cytoplasm of immature spo3Δ asci, suggesting that Spo3 is essential for the assembly of the FSM. In most wild-type cells, the FSM encapsulates each haploid nucleus. About 70% of the spo3Δ zygotes form four aggregates of GFP-Psy1 near nuclei (class I). These aberrant structures may represent remnants of collapsed membranes. The rest (∼30%) of the spo3Δ zygotes contains four, albeit remarkably small, nucleated prespores (class II) (Nakamura et al., 2001). These phenotypes have also been observed in electron microscopic studies showing aberrant prespores called spore-like bodies in spo3 mutants (Hirata and Shimoda, 1992). To clarify these defects in FSM formation in more detail, we made time-lapse observations of FSM formation in spo3 mutant cells. Figure 5, A and B, shows an example of GFP-Psy1 behavior in class I and class II phenotypes, respectively. In class I mutant cells (Figure 5A and Supplemental Movie 4), the FSMs seemed to initiate and to expand normally at an early stage of meiosis II (−18 min). Interestingly, at late meiosis II, expansion of the FSMs was severely impaired (22–32 min). After the meiosis II completed, the FSMs closed without engulfing the nuclei. Finally, four small sacs formed near the nuclei (34–50 min). In class II phenotype cells, the FSMs behaved essentially as the class I phenotype cells. However, despite the apparent defect in FSM expansion, the FSM somehow engulfed the nucleus, resulting in formation of prespores that were remarkably small (Figure 5B and Supplemental Movie 5). These data suggest that the defect in FSM formation between class I and class II cells is identical, but that the difference in phenotype is due to the degree of FSM expansion; the defect in class I being more severe than in class II.
Figure 5.
Behavior of GFP-tagged Psy1 during sporulation in the spo3 mutant. spo3Δ strain TN240 which expresses GFP-Psy1, was cultured on sporulation medium (SSA) for 16 h, and stained with Hoechst 33342 to allow simultaneous observation of chromatin by time-lapse fluorescence microscopy. Frames were taken every 2 min. In merged images, GFP-Psy1 (green) and Hoechst 33342 (magenta) are shown. (A) Class I mutant: four aggregates of GFP-Psy1 are formed near nuclei. (B) Class II mutant: four nucleated prespores are formed but they are remarkably small. Bar, 10 μm.
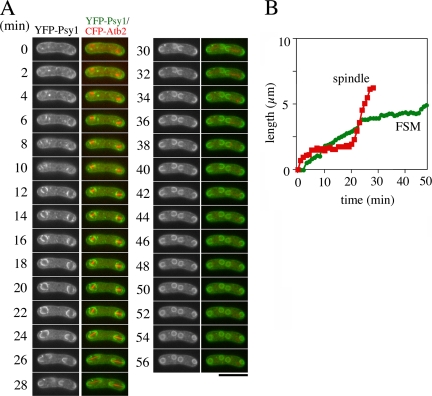
To address the relationship between FSM expansion and meiosis II progression in spo3 mutants, we simultaneously observed the FSM and microtubules by time-lapse fluorescence microscopy as described above. As in wild-type cells, the YFP-Psy1 dots in the spo3Δ cells first occurred at the both sides of the spindle microtubule at phase I (Figure 6A, Table 2, and Supplemental Movie 6). The FSMs expanded at a rate similar to that observed in wild-type cells until phase II ended (Figure 6B, Table 3, and Supplemental Movie 6). Interestingly, the expansion of the FSMs was severely inhibited at about the time of onset of phase III (Table 3). After completion of meiosis II, the FSMs closed. These results indicate that FSM expansion during late and postmeiosis II is impaired in the spo3 mutant. The duration of phase I, II, and III in spo3Δ cells was similar to that in wild-type cells (Table 2), supporting our previous observation that meiosis II proceeds normally in spo3Δ cells (Nakamura et al., 2001).
Figure 6.
Simultaneous observation of FSM and microtubules in the spo3 mutant. Two-color live imaging of YFP-Psy1 and CFP-Atb2 in spo3Δ mutant strain TN345. In merged images, YFP-Psy1 (green) and CFP-Atb2 (red) are shown. Frames were taken every 2 min. Bar, 10 μm. (B) Time course changes in the FSM and spindle microtubules during meiosis II. The contour lengths of FSMs (green line) and spindle microtubules (red line) are also shown.
Table 3.
Development of the FSM in spo mutants
| Length of forespore membrane (μm) at |
||||||
|---|---|---|---|---|---|---|
| End of phase I | End of phase II | End of phase III | 15 min after completion of meiosis II | FSM closure | n | |
| Wild type | 0.73 ± 0.18 | 3.06 ± 0.35 | 5.51 ± 0.62 | 8.39 ± 1.13 | 8.56 ± 0.71 | 10 |
| spo3Δ | 0.81 ± 0.31 | 2.88 ± 0.53 | 3.83 ± 0.38 | 4.47 ± 0.60 | 5.30 ± 0.55 | 16 |
| spo14 | 0.90 ± 0.48 | 2.29 ± 0.77 | 2.99 ± 1.08 | 3.84 ± 1.51 | 4.81 ± 1.50 | 10 |
The FSM Formation Is Impaired from Initiation to Closure in spo14 Mutant Cells
spo14+ encodes a budding yeast Sec12-like protein, necessary for budding of vesicles from the endoplasmic reticulum. Our previous study revealed that Spo14 is responsible for the assembly of the FSM by supplying membrane vesicles (Nakamura-Kubo et al., 2003). Therefore, we examined FSM formation in the spo14-B221 mutant. As shown in Supplemental Figure 3 and Supplemental Movie 7, GFP-Psy1 dots occurred at opposite sides of nuclei as in wild-type cells (0–4 min), but expansion occurred to be inhibited at early meiosis (0–18 min). The GFP-Psy1 signal in spo14 mutant cells had a smaller curvature than in wild-type and spo3 mutant cells. Anaphase II progressed without further expansion of FSMs (20–24 min), resulting in formation of four tiny aggregates adjacent to the nuclei (44–54 min).
To examine FSM formation in a spo14 mutant in more detail, the FSM and microtubules were visualized by YFP-Psy1 and CFP-Atb2, respectively. FSM formation initiated at phase I as in wild-type cells (Table 2, 2–4 min), but the expansion of the FSMs was severely impaired during phase II (8–16 min). Indeed, the length of the FSMs at the end of phase I in spo14 mutant cells was not different from in wild-type cells (Table 3). At late and postmeiosis, the FSMs expanded very slowly (Figure 7, A and B, Supplemental Movie 8, and Table 3). This is in marked contrast to the spo3 mutant, where FSM formation seems to proceed normally until phase III. These data suggest that FSM formation is impaired from initiation to closure in spo14 mutants.
Figure 7.
Simultaneous observation of FSM and microtubules in the spo14 mutant. Two-color live imaging of YFP-Psy1 and CFP-Atb2 in spo14 mutant strain TN346. YFP-Psy1 (green) and CFP-Atb2 (red) are shown. Frames were taken every 2 min. Bar, 10 μm. (B) Kinetics of FSM formation and spindle microtubule movements during meiosis II. The contour lengths of FSMs (green line) and spindle microtubules (red line) are also shown.
In a previous study, we distinguished two classes of phenotype in spo14-B221 cells. In class I, FSM formation was arrested halfway, whereas in class II, four aggregates of GFP-Psy1 were observed near the SPB (Nakamura-Kubo et al., 2003). However, our live observations in the present study show that the form of the FSMs in the class I phenotype seems to be similar to that observed during meiosis (Supplemental Figure 3, 28–42 min). Eventually, the FSMs closed to form small aggregates, corresponding to the class II phenotype. Thus, we conclude that the FSM-like structure in the class I phenotype is not a terminal phenotype, but rather an intermediate form of the FSM. Thus, the class II phenotype is the terminal phenotype uniformly observed in spo14 mutants.
DISCUSSION
The FSM formation is one of the most morphologically dramatic events in the S. pombe life cycle, in that membrane is synthesized de novo within mother cells. To our knowledge, until now, electron and fluorescence microscopic observations of the FSM have been restricted to fixed cells. Although the current study with live cells has yielded results that are consistent with previous data based on fixed cells, the time-lapse analysis has provided more information about dynamic features of FSM assembly.
Initiation of FSM Formation
Because FSM formation takes place coincident with meiosis II, these two events must be highly coordinated. We observed FSM and microtubules simultaneously and analyzed the spatiotemporal relationship between FSM formation and meiosis II. The precise stage at which FSM formation begins was not determined, although previous studies have shown that initiation of FSM formation occurs during meiosis II. Our time-lapse analysis revealed a pair of YFP-Psy1 dots sandwiching a nucleus, which corresponds to the initiation of FSM formation, at the SPBs seemed during phase I of meiosis II. Given that phase I corresponds to prophase, these data indicate that FSM formation begins at prophase II.
Expansion of the FSM
The FSM expands continuously during meiosis. The FSM continued to expand even after meiosis II ended. In spo3Δ cells, the FSM expanded normally until the onset of anaphase IIB, after which the expansion is severely impaired (Figure 6 and Table 3). In contrast, a spo14 mutation, which causes a defect in ER-to-Golgi membrane vesicle transport, compromised FSM formation throughout meiosis II. From these results, we suggest that FSM expansion is composed of at least two steps: “early meiotic FSM expansion” and “late and postmeiotic FSM expansion.” Early meiotic FSM expansion takes place coincident with the initiation of meiosis II. In this step, the FSM expands by fusion with vesicles that are likely derived from the ER through the Golgi. In contrast, late and postmeiotic FSM expansion requires Spo3 function in addition to the general secretory machinery. The form of the FSM at anaphase II may be considerably different in spo3 and spo14 mutants. The FSM seems to be more rigid in the spo14 mutant than in wild type or in the spo3 mutant, possibly due to differences in composition.
The coordination of cell cycle events during cell proliferation is attained in part through surveillance systems called “checkpoint controls.” When there are defects in critical cell cycle events, checkpoint mechanisms arrest or delay progression of the cell cycle to prevent aneuploidy and cell lethality (Hartwell and Weinert, 1989). In addition, S. pombe has a cytokinesis or contractile ring checkpoint, which monitors formation and integrity of the medial actomyosin ring, and thereby delays subsequent nuclear division (Le Goff et al., 1999; Liu et al., 2000). We examined whether progression of meiosis II is delayed when FSM formation is inhibited. Inhibition of FSM formation by either spo3, spo14, or spo15 mutations was found not to affect the progression of meiosis II (Table 2). Conversely, some mutants, such as rec mutants, which undergo abnormal meiotic nuclear divisions are able to form spores, though they are apparently misshapen (Ponticelli and Smith, 1989). Thus, unlike mitosis, no checkpoint-like control coordinating meiosis and FSM formation seems to exist.
Closure of the FSM
Live analysis also showed that FSM closure takes place ∼20 min after meiosis II ends. In spo3 and spo14 mutants, FSM closure was also observed, suggesting that the degree of FSM expansion does not directly control its closure. By electron microscopy, the leading edge of FSM is observed as an electron dense structure, which is apparently distinct from the FSM (Hirata, unpublished data). Several proteins have been identified as leading edge components both in S. cerevisiae and S. pombe (Knop and Strasser, 2000; Moreno-Borchart et al., 2001; Nickas and Neiman, 2002; Okuzaki et al., 2003; Itadani et al., 2006). In S. pombe, Meu14 is one of the leading edge components. Meu14 possesses a coiled-coil domain and localizes to the leading edge of the FSM during meiosis II. A meu14Δ mutant consequently produces abnormal spores at a high frequency, suggesting that Meu14 is responsible for spore morphogenesis, especially determination of FSM closure. The ring structure of Meu14 forms in both spo3Δ or spo15Δ cells, supporting the notion that closure of the FSM occurs normally in these mutants (Okuzaki et al., 2003). Therefore, leading edge proteins such as Meu14, but not proteins related to FSM expansion (e.g., Spo3 and Spo14) might control FSM closure.
The expansion and closure of four FSMs in a zygote was found to occur in a synchronous manner. The mechanism responsible for this synchrony is not clear. Our live analysis revealed that the four FSMs expanded synchronously in both spo3 and spo14 mutants (Figure 3). Moreover, our preliminary data indicate that uncontrolled FSM expansion and closure occurred in the meu14Δ mutant (Ito, Shimoda, and Nakamura, unpublished data). The leading edge of the FSM may control the synchrony of FSM expansion as well as closure itself.
Spore Morphogenesis
Live analysis also revealed that the prespore became spherical in shape after FSM closure. A previous study by electron microscopy showed that spore wall materials are deposited between the inner and outer membranes of the prespore, after which the spore wall was formed (Yoo et al., 1973). Prespores might become spherical concomitantly with formation of the spore wall. Alternatively, at this stage, actin patches are dispersed at the periphery of the prespores (Petersen et al., 1998; Itadani et al., 2006; Ohtaka et al., 2007). Actin relocalization may be required for spherical shaping of spores. Simultaneous observation of actin and FSM will allow us to address this question.
Possible Model for the FSM Formation
Based on the present work and on previous electron and fluorescence microscopic observations using fixed specimens, we propose a new model for the morphogenesis of FSM relative to meiosis (Figure 8). 1) Initiation of FSM formation. During prophase I, outer plaques form on the cytoplasmic side of the SPB, where membrane vesicles accumulate and begin to fuse with one another. Although the molecular mechanism that triggers FSM formation is still unknown, Spo15 is involved in this step. 2) Early meiotic FSM expansion. Membrane vesicles derived from the ER through the Golgi apparatus fuse to the FSM. The general secretory machinery including Spo14, Psy1, and Sec9, a synaptosomal-associated protein of 25 kDa orthologue, are responsible for the membrane fusion. 3) Late and postmeiotic FSM expansion. At this stage, Spo3 is responsible for membrane fusion. 4) FSM closure. About 20 min after completion of meiosis II, the FSM closes. Leading edge components such as Meu14 are involved in this process. Based on the observation that the FSM can close in FSM formation-defective mutants (e.g., spo3 and spo14), FSM closure can occur independently of expansion. 5) Spherical shaping of prespores. After FSM closure, the resulting prespores becomes spherical as spore wall material accumulates in the space between the inner and outer membranes of the prespores leading to spore wall formation.
Figure 8.
A model for assembly of the FSM.
Supplementary Material
ACKNOWLEDGMENTS
Some of the S. pombe strains were provided by the Yeast Genetic Resource Center Japan supported by the National BioResource Project (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig/). This study was supported by Grant-in-Aid for Scientific Research on Priority Areas “Genome Biology” to C. S., and on “Cell Cycle Control” and ”Life of Proteins” to T. N. from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and NOVARTIS Foundation (Japan) for the Promotion of Science to T. N. J. K. is a recipient of the research fellowship for young scientist from the Japan Society for the Promotion of Science.
Abbreviations used:
- CFP
cyan fluorescent protein
- ER
endoplasmic reticulum
- FSM
forespore membrane
- GFP
green fluorescent protein
- SPB
spindle pole body
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0414) on June 11, 2008.
REFERENCES
- Asakawa H., Kitamura K., Shimoda C. Novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol. Genet. Genomics. 2001;265:424–435. doi: 10.1007/s004380000429. [DOI] [PubMed] [Google Scholar]
- Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bresch C., Muller G., Egel R. Genes involved in meiosis and sporulation of a yeast. Mol. Gen. Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern J. N., Jones E. W., Broach J. R., editors. The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 59–96. [Google Scholar]
- d'Enfert C., Gensse M., Gaillardin C. Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. 1992;11:4205–4211. doi: 10.1002/j.1460-2075.1992.tb05514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D. Q., Chikashige Y., Haraguchi T., Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Egel R., Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- Gutz H., Heslot H., Leupold U., Loprieno N. Schizosaccharomyces pombe. Handb. Genet. 1974;1:395–446. [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hirata A., Shimoda C. Electron microscopic examination of sporulation-deficient mutants of the fission yeast Schizosaccharomyces pombe. Arch. Microbiol. 1992;158:249–255. doi: 10.1007/BF00245240. [DOI] [PubMed] [Google Scholar]
- Hirata A., Tanaka K. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 1982;28:263–274. [Google Scholar]
- Ikemoto S., Nakamura T., Kubo M., Shimoda C. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 2000;113:545–554. doi: 10.1242/jcs.113.3.545. [DOI] [PubMed] [Google Scholar]
- Itadani A., Nakamura T., Shimoda C. Localization of type I myosin and F-actin to the leading edge region of the forespore membrane in Schizosaccharomyces pombe. Cell Struct. Funct. 2006;31:181–195. doi: 10.1247/csf.06027. [DOI] [PubMed] [Google Scholar]
- Knop M., Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., Byers B., Esposito R. E, Mitchell A. P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Jones E. W., Pringle J. R., Broach J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- Le Goff X., Utzig S., Simanis V. Controlling septation in fission yeast: finding the middle, and timing it right. Curr. Genet. 1999;262:163–172. doi: 10.1007/s002940050455. [DOI] [PubMed] [Google Scholar]
- Liu J., Wang H., Balasubramanian M. K. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 2000. 2000;113:1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1990;194:793–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart A. C., Strasser K., Finkbeiner M. G., Shevchenko A., Shevchenko A., Knop M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 2001;20:6946–6957. doi: 10.1093/emboj/20.24.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A. F., Murray A., Chikashige Y., Yamashita Y. M., Hiraoka Y., Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell. 2001;12:3955–3972. doi: 10.1091/mbc.12.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo M., Nakamura T., Shimoda C. Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol. Cell. Biol. 2002;22:309–320. doi: 10.1128/MCB.22.1.309-320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Kashiwazaki J., Shimoda C. A fission yeast SNAP-25 homologue, SpSec9, is essential for cytokinesis and sporulation. Cell Struct. Funct. 2005;30:15–24. doi: 10.1247/csf.30.15. [DOI] [PubMed] [Google Scholar]
- Nakamura-Kubo M., Nakamura T., Hirata A., Shimoda C. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell. 2003;14:1109–1124. doi: 10.1091/mbc.E02-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A., Brada D., Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Nakamura T., Hirata A., Routt S. M., Skinner H. B., Bankaitis V. A., Shimoda C. The Schizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell. 2001;12:901–917. doi: 10.1091/mbc.12.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Nakamura T., Okazaki K., Hirata A, Shimoda C. The Sec14 family glycerophospholipid-transfer protein is required for structural integrity of the spindle pole body during meiosis in fission yeast. Genes Cells. 2004;9:1275–1286. doi: 10.1111/j.1365-2443.2004.00806.x. [DOI] [PubMed] [Google Scholar]
- Nakase Y., Nakamura-Kubo M., Ye Y., Hirata A., Shimoda C., Nakamura T. Meiotic spindle pole bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol. Biol. Cell. 2008;19:2476–2487. doi: 10.1091/mbc.E08-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Neiman A. M. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics. 2002;160:1439–1450. doi: 10.1093/genetics/160.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka A., Okuzaki D., Saito T. T., Nojima H. Mcp4, a meiotic coiled-coil protein, plays a role in F-actin positioning during Schizosaccharomyces pombe meiosis. Eukaryot. Cell. 2007;6:971–983. doi: 10.1128/EC.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki D., Satake W., Hirata A., Nojima H. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 2003;116:2721–2735. doi: 10.1242/jcs.00496. [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I. M. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 1998;111:867–876. doi: 10.1242/jcs.111.7.867. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Smith G. R. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Hirata A. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J. Cell Sci. 1982;56:263–279. doi: 10.1242/jcs.56.1.263. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Imai Y., Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J. R., Broach J. B., Jones E. W., editors. Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- Ye Y., Fujii M., Hirata A., Kawamukai M., Shimoda C., Nakamura T. Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of farnesyl diphosphate synthase (FPS), Fps1, and an FPS-like protein, Spo9, essential for sporulation. Mol. Biol. Cell. 2007;18:3568–3581. doi: 10.1091/mbc.E07-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B. Y., Calleja G. B., Johnson B. F. Ultrastructural changes of the fission yeast (Schizosaccharomyces pombe) during ascospore formation. Arch. Mikrobiol. 1973;91:1–10. doi: 10.1007/BF00409533. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Nakamura T, Shimoda C. The cation-transporting P-type ATPase Cta4 is required for assembly of the forespore membrane in fission yeast. Genes Genet. Syst. 2005;80:317–324. doi: 10.1266/ggs.80.317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.