Abstract
Background
Fungal and animal mitochondrial genomes typically have one tRNA for each synonymous codon family. The codon-anticodon adaptation hypothesis predicts that the wobble nucleotide of a tRNA anticodon should evolve towards maximizing Watson-Crick base pairing with the most frequently used codon within each synonymous codon family, whereas the wobble versatility hypothesis argues that the nucleotide at the wobble site should be occupied by a nucleotide most versatile in wobble pairing, i.e., the tRNA wobble nucleotide should be G for NNY codon families, and U for NNR and NNN codon families (where Y stands for C or U, R for A or G and N for any nucleotide).
Results
We here integrate these two traditional hypotheses on tRNA anticodons into a unified model based on an analysis of the wobble costs associated with different wobble base pairs. This novel approach allows the relative cost of wobble pairing to be qualitatively evaluated. A comprehensive study of 36 fungal genomes suggests very different costs between two kinds of U:G wobble pairs, i.e., (1) between a G at the wobble site of a tRNA anticodon and a U at the third codon position (designated MU3:G) and (2) between a U at the wobble site of a tRNA anticodon and a G at the third codon position (designated MG3:U).
Conclusion
In general, MU3:G is much smaller than MG3:U, suggesting no selection against U-ending codons in NNY codon families with a wobble G in the tRNA anticodon but strong selection against G-ending codons in NNR codon families with a wobble U at the tRNA anticodon. This finding resolves several puzzling observations in fungal genomics and corroborates previous studies showing that U3:G wobble is energetically more favorable than G3:U wobble.
Background
The wobble versatility hypothesis [1-6], abbreviated as WVH, states that the wobble site of tRNA anticodon should have G for NNY codons (where Y stands for C or U and N for any nucleotide) because G can pair with both C and U in RNA, and should have U for NNR to pair with both A and G. For NNN codon families, the wobble site should be U because U is known to be the most versatile in wobble-pairing [7-12]. In contrast, the codon-anticodon adaptation hypothesis, or CAAH for short, invokes the codon usage bias as a determining factor, i.e., the wobble site of tRNA anticodon should co-evolve with codon usage so that the nucleotide in the wobble site of tRNA anticodon should match the most abundant codon in a synonymous codon family [6,13-15]. The association between the major codon and the anticodon of the most abundant tRNA has been documented in Escherichia coli [16,17], Saccharomyces cerevisiae [18], and other species and organelles [15,19-22].
Here we develop a general hypothesis of codon-anticodon adaptation based on an analysis of wobble costs, and derive its predictions that can be tested by genomic data. The wobble cost may be viewed as reduction in decoding efficiency and accuracy because such reduction would be selected against over evolutionary time. We will refer to this new general hypothesis based on wobble cost as WCH (for wobble cost hypothesis). The two traditional hypotheses, CAAH and WVH, will be shown to be special forms of WCH.
Following the shorthand notation of Ogle et al. [23], I designate the translation cost through wobble base-pairing between nucleotide i at the third codon position of a codon and the nucleotide j at the wobble site of tRNA anticodon as Mi3:j (where M is for wobble cost. The letter C would be more suitable to represent cost but it may confuse with the nucleotide C). We assume Mi3:j = 0 if nucleotides i and j form Watson-Crick base pairing. The reason for this assumption is that C:G and A:U pairs have not been found to contribute to ribosomal stalling (which reduces translation efficiency) or amino acid mis-incorporation (which reduces translation accuracy) although almost all non-Watson-Crick base pairings have been shown to reduce translation efficiency and accuracy. We define MY3:U as the wobble cost between a wobble U at the tRNA anticodon and a C or U at the third codon position. For simplicity, we also assume MA3:A = MC3:A = MG3:A = MA3:C = MC3:C = MU3:C = MA3:G = MG3:G = MO (where the subscript O is for "other", i.e., MO is the wobble cost between base pairs that are not Watson-Crick base pairs, not U3:G or G3:U base pairs and not Y3:U pairs). In general, MU3:G and MG3:U are expected to be smaller than MC3:U, MU3:U, and MO because G and U can base-pair in RNA, MY3:U is expected to be smaller than MO because a wobble U at tRNA anticodon is known to be the most versatile in wobble-pairing [7-12].
A classical study of U:G wobble pairs [24] suggests a preference for the U being at the 3' end rather than at the 5' end, which implies that U3:G wobble pair is energetically more favorable than G3:U wobble pair [25]. Subsequent studies have shown that, while the U3:G wobble pair occurs on the ribosome, the unmodified G3:U wobble pair does not [2,23,26]. These findings suggest that MU3:G may be smaller than MG3:U, although its generality is unknown.
Two-fold NNY and NNR codon families
First consider the NNY codon family where Y is either C or U. Designate the number of C-ending and U-ending synonymous codons by NC and NU, respectively, and the total cost of wobble pairing as MwG when the wobble site of the anticodon is G, and as MwA when the wobble nucleotide is A (we do not need to consider the case when the wobble site is U or C for NNY codon families because such cases have never been observed and because a tRNA with a wobble U or C to translate NNC and NNU codons is against physiochemical reasons). We now express the total cost MwG and MwA as
| (1) |
Note that MC3:G = MU3:A = 0 according to our definition. The dependence of MwG and MwA on the relative frequencies of NC, expressed as proportion of C-ending codons in the NNY codon family (PC), is graphically shown in Fig. 1, with MO assumed to equal 2•;MU3:G. The condition for MwG = MwA, i.e., when the wobble site of the tRNA anticodon can take either G or A without a fitness differential, is
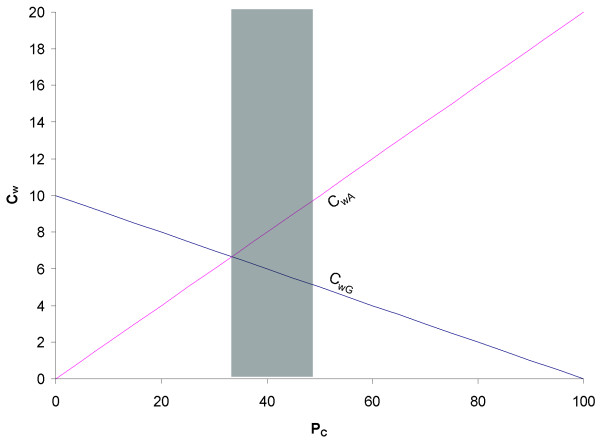
Figure 1.
Conceptual illustration of the dependence of wobbling cost involving a G or an A at the wobble site of tRNA anticodon (MwG and MwA) on the proportion of C-ending codons (PC) in an NNY codon family, with MC3:A = 2MU3:G. The shaded area corresponds to PC smaller than 1/2 but larger than 1/3.
| (2) |
In the scenario in Fig. 1 with MO = 2MU3:G, the condition for MwG = MwA is PC = 1/3. Thus, when PC drifts above 1/3, it would become more beneficial (less costly) to have a G at the wobble nucleotide of tRNA anticodon. However, when PC drifts below 1/3, then a wobble A would be more advantageous and should be favored by natural selection. Naturally, if MU3:G = 0, then we should always have a wobble G at the tRNA anticodon regardless of codon usage.
Now we consider three special cases with reference to Fig. 1 and the two traditional hypotheses, CAAH and WVH. First, if NC = NU = N/2, then CAAH, which predicts that the wobble nucleotide of tRNA anticodon should form Watson-Crick base pair with the most abundant codon, has no prediction at all because the two codons are equally abundant. In contrast, if we assume MU3:G < MO as is depicted in Fig. 1, we have MwG < MwA, and WCH predicts that the anticodon wobble site should be a G. This is the same prediction as WVH.
Second, if NC > NU, then MwG < MwA assuming MU3:G ≤ MO, and WCH predicts that the anticodon wobble site should be occupied by a G. This prediction is shared by both CAAH and WVH.
Third, when NC < NU but NC > NU·MU3:G/MO, then WCH will still predict a G at the wobble nucleotide of tRNA anticodon because MwG is still smaller than MwA. Take the scenario in Fig. 1 for example, when NC < NU but PC > 1/3 (which correspond to the shaded area in Fig. 1), we have MwG < MwA (Fig. 1), so natural selection should favor a G at the wobble site of tRNA anticodon. WVH happens to have the same prediction. However, CAAH will predict an A at the wobble site of tRNA anticodon if U-ending codons are more abundant than C-ending codons. This is in contrast to WVH, i.e. CAAH is inapplicable when PC is within the shaded range in Fig. 1.
Fourth, when NC << NU, especially in the extreme case when NC = 0, then Eq. (1) is reduced to MwG = NU• MU3:G and MwA = 0. Because now MwG > MwA, WCH predicts an A at the anticodon wobble site. In this case, WVH would still predict a G at the wobble site because it ignores the codon frequencies (i.e., it ignores the relative magnitude of NC and NU), but CAAH would predict an A at the wobble site, which is the same prediction as WCH. Only in this particular case when NC << NU can CAAH and WVH be clearly differentiated. As is depicted in Fig. 1, the chance for CAAH to be supported decreases as MU3:G decreases relative to MO. CAAH would have little chance to be unambiguously supported if MU3:G is close to 0. Thus, as long as MU3:G is small relative to MO, we should expect CAAH to be less supported than WVH. A recent study shows that CAAH indeed receives much less support than WVH when both were tested against fungal mitochondrial genomic data [27].
The simple Eq. (1) offers a way to assess relative costs of different wobble pairs. For example, if nature has chosen a G at the wobble site of the tRNA anticodon, then we may infer that MwG < MwA. So, from Eq. (1), we have
| (3) |
Suppose we have an NNY codon family translated by a tRNA with a G at the wobble site of the anticodon. If MU3:G is large, then there would be strong selection favoring C-ending codons against U ending codons. So the ratio of NC/NU may be very large. In such a case the estimated MU3:G/MO ratio, being smaller than a very large value, is not informative. However, in different codon families, mutation pressure may allow NU to drift up relative to NC. Suppose we have three NNY codon families each translated by a tRNA with a G at the anticodon wobble site. If the NC/NU ratio for the three NNY codon families are 100/2, 80/100 and 200/20, respectively, we may infer that MU3:G/MO < 80/100 (because it is the most precise). Alternatively, with many NC/NU ratios from many NNY codon families translated by tRNA with a G at its anticodon wobble site, we may compute the lower 95% confidence limit of the NC/NU ratio (LCL95.G where the subscript G indicates tRNA with a G at the anticodon wobble site) and infer that MU3:G/MC3:A < LCL95.G.
If we always have very large NC/NU ratios, we may infer that selection against U-ending codons must be strong, with little chance for mutation to elevate NU. This is a strong indication of a large MU3:G. Along the same line of reasoning, we may infer that MU3:G is very small if NU can often as large as, or even larger than, NC.
Similarly, if nature has chosen an A at the wobble site of tRNA anticodon, then we may infer MwG > MwA, so
| (4) |
We can apply exactly the same rationale for the NNR codon family leading to parallel conclusions. For example, if nature has chosen a U at the wobble site of the tRNA anticodon, then we may infer that MwU < MwC, so that
| (5) |
Similarly, if MG3:U is very large, then there should be strong selection against G-ending codons in favor of A-ending codons. This will produce large NA/NG ratios. In contrast, a large NG comparable to NA indicates a very small MG3:U cost.
Given previous studies indicating that U3:G is energetically much more favorable than G3:U [2,23-26], we should expect MU3:G < MG3:U. The reasoning above paves the way for us to test whether this is generally true among the genetically diverse fungal species.
Three-fold AUH (Isoleucine) codons
Designate codons ending with A, C, and U as NA, NC and NU, respectively. The wobble cost of having an A, G, C, or U at the wobble site of tRNA anticodon is
| (6) |
It is obvious that MwC is always greater than MwA, so we should never find a C at the wobble site of a tRNAIle anticodon, i.e., we can disregard MwC. If nature has chosen G at the wobble site of tRNAIle anticodon, then we may infer that MwG is the smallest. From MwG < MwA and MwG < MwU, we have
| (7) |
where the assumption is made on the basis of previous observations that U is generally the most versatile in wobble-pairing among the four nucleotides [7-12].
Four-fold NNN codons
Designate the number of codons ending with A, C, G, and U as NA, NC, NG, and NU, respectively. The wobble costs involving an A, C, G or U at the wobble site of tRNA anticodon are, respectively,
| (8) |
If nature has chosen a U at the wobble site of the tRNA anticodon, then Eq. (8) does not give us any simple inequality to estimate the cost ratios. However, if nature has chosen an A at the wobble site of the tRNA anticodon, then from MwA < MwG and MwA < MwC, we can infer
| (9) |
In what follows, we estimate MU3:G/MO, MG3:U/MO, and MU3:C/MO by using fungal mitochondrial genomic data. Cells in fungal species are generally rapid-replicating which necessitates efficient translation. Rapidly replicating unicellular organisms are theoretically expected to be under strong selection to increase the rate of biosynthesis [15,28] and they typically exhibit strong codon-anticodon adaptation [29]. Thus, fungal species should be ideal for evaluating evolutionary hypothesis on codon-anticodon adaptation.
Methods
We retrieved 36 fungal mitochondrial genomes (Table 1) by using NCBI Entrez. Three different genetic codes are used in different fungal genomes. Among the 36 fungal mitochondrial genomes, seven genomes use translation table 3, 27 genomes use translation table 4, and two genomes use translation table 16 (Table 1). When results are similar among genomes using the same translation table, only results from a representative genome are presented. The number of codon families supporting CAAH (NCAAH) and WVH (NWVH) is compiled following the following rationale [27]. Suppose a lysine (Lys) codon family has 20 AAA and 60 AAG codons. WVH would ignore the codon usage bias and predict a wobble U in the tRNALys anticodon because U can pair with both A and G, whereas CAAH would predict a wobble C in the tRNALys anticodon to maximize the Watson-Crick match with the more frequent G-ending codons. If the tRNALys anticodon is found to have a wobble U, then WVH is supported; if a wobble C is found, then CAAH is supported. If we have 60 AAA codons and 20 AAG codons and if tRNALys anticodon has a wobble U, then both hypotheses are supported, i.e., they are indistinguishable and are not included in Table 1. The methionine codon families are not included in Table 1 but discussed in detail elsewhere [27,30].
Table 1.
Number of codon families unambiguously supporting the codon-anticodon adaptation hypothesis (NCAAH) and the wobble versatility hypothesis (NWVH) in each fungal species.
| Species | Accession* | Code† | NCAAH | NWVH |
|---|---|---|---|---|
| Allomyces macrogynus | NC_001715 | 4 | 1 | 13 |
| Ashbya gossypii ATCC 10895 | NC_005789 | 3 | 0 | 10 |
| Aspergillus niger | NC_007445 | 4 | 0 | 12 |
| Aspergillus tubingensis | NC_007597 | 4 | 0 | 12 |
| Candida albicans SC5314 | NC_002653 | 4 | 1 | 11 |
| Candida glabrata | NC_004691 | 3 | 0 | 12 |
| Candida metapsilosis | NC_006971 | 4 | 1 | 15 |
| Candida orthopsilosis | NC_006972 | 4 | 1 | 15 |
| Candida parapsilosis | NC_005253 | 4 | 1 | 15 |
| Candida stellata | NC_005972 | 4 | 1 | 13 |
| Moniliophthora perniciosa | NC_005927 | 4 | 0 | 13 |
| Epidermophyton floccosum | NC_007394 | 4 | 2 | 12 |
| Harpochytrium sp. JEL94 | NC_004760 | 4 | 1 | 3 |
| Harpochytrium sp. JEL105 | NC_004623 | 4 | 1 | 4 |
| Hyaloraphidium curvatum | NC_003048 | 4 | 1 | 4 |
| Hypocrea jecorina | NC_003388 | 4 | 1 | 13 |
| Kluyveromyces lactis | NC_006077 | 4 | 0 | 13 |
| Kluyveromyces thermotolerans | NC_006626 | 3 | 0 | 11 |
| Lecanicillium muscarium | NC_004514 | 4 | 0 | 10 |
| Monoblepharella sp. JEL 15 | NC_004624 | 4 | 0 | 15 |
| Mortierella verticillate | NC_006838 | 4 | 1 | 13 |
| Penicillium marneffei | NC_005256 | 4 | 1 | 12 |
| Pichia Canadensis | NC_001762 | 4 | 2 | 12 |
| Podospora anserine | NC_001329 | 4 | 0 | 13 |
| Rhizophydium sp.136 | NC_003053 | 16 | 0 | 2 |
| Rhizopus oryzae | NC_006836 | 4 | 1 | 14 |
| Saccharomyces cerevisiae | NC_001224 | 3 | 1 | 12 |
| Saccharomyces castellii | NC_003920 | 3 | 0 | 14 |
| Saccharomyces servazzii | NC_004918 | 3 | 0 | 9 |
| Schizophyllum commune | NC_003049 | 4 | 1 | 13 |
| Schizosaccharomyces japonicus | NC_004332 | 4 | 1 | 16 |
| Schizosaccharomyces octosporus | NC_004312 | 4 | 1 | 14 |
| Schizosaccharomyces pombe | NC_001326 | 4 | 1 | 14 |
| Smittium culisetae | NC_006837 | 4 | 1 | 15 |
| Spizellomyces punctatus1 | NC_003052 | 16 | 0 | 4 |
| Yarrowia lipolytica | NC_002659 | 3 | 0 | 11 |
| Sum | 23 | 414 |
* GenBank accession number.
† Genetic code.
The tRNA and CDS sequences were extracted and analyzed by using DAMBE [31,32]. The CDS-derived codon usage is also computed with DAMBE. The anticodon in almost all tRNA sequences from all species share the regular feature of being flanked by two nucleotides on either side to form a loop that is held together by a stem. For example, the anticodon loop (AC loop) of the tRNAArg genes translating CGN codons in Epidermophyton floccosum is 28CGUGUUACGGCCACG42, where the starting and ending numbers indicate the position of the AC loop in the tRNA sequence, with the anticodon 5'-ACG-3' (matching codon CGU) flanked by two nucleotides on either side (in bold) to form a loop that is held together by a stem made of the first and the last four nucleotides. Similarly, the other tRNAArg translating AGR codons is 25AAAAUACUUCUAAUAUUUU43, with the AC loop held together by a six-base stem. However, some tRNA sequences have a suspicious AC loop and DAMBE will flag them out. The AC loop is then identified by aligning the tRNA sequences against other isoaccepting tRNA sequences with a regular AC loop [6]. Some tRNA anticodon loop has the anticodon flanked by three instead of two nucleotides. For example, the anticodon loop in tRNALeu in the mitochondrial genome of Kluyveromyces thermotolerans is GAUACUCUUAAGAUGUAUU, with the anticodon UAA flanked by three nucleotides (in bold) on both sides. There are a few tRNA sequences in which anticodon loop cannot be identified.
Some mitochondrial genomes in GenBank are annotated incorrectly. For example, tRNAPro in the mitochondrial genome of Ashbya gossypii ATCC 10895 has an anticodon of UGG matching codon CCA (the most frequently used proline codon), but the GenBank file (NC_005789) annotated the anticodon to match codon CCU.
A few fungal mitochondrial genomes do not have a complete set of tRNA genes. For example, the mitochondrial genomes of Hyaloraphidium curvatum and Harpochytrium sp. JEL94 have seven and eight tRNA genes, respectively, and consequently will need tRNA import from the nuclear genome. This may cause complication in analyzing codon-anticodon adaptation. However, removing such genomes does not alter the conclusions.
Some species exhibit extreme avoidance of certain codon families. For example, Ashbya gossypii ATCC 10895 codes Arg with only AGR codons without using any CGN codons. In contrast, Hyaloraphidium curvatum codes Arg with only CGN codons without using any AGR codons. Such avoidance of certain codon families would facilitate the evolutionary loss of the associated tRNA [33-35], although it is not always clear whether the avoidance is the cause or the consequence of the loss of the associated tRNA.
We computed relative synonymous codon usage, or RSCU [36], as a measure of codon usage bias within a codon family by using DAMBE [31,32]. Some coding sequences are incomplete. For example, the cox1 CDS in Aspergillus niger is annotated as "join(<19768..20614,21640..22495)". The first two nucleotides (i.e., at positions 19768 and 19769) represent a partial codon and are discarded in computing codon frequencies.
Results and discussion
Wobble cost between G and U: MU3:G and MG3:U
Recall that the two inequalities
| (10) |
are, respectively, for NNY codons translated by tRNA with a G at the wobble site of tRNA anticodon, and for NNR codons translated by tRNA with a U at the wobble site of tRNA anticodon. The observed NC/NU ratios for Allomyces macrogynus (representing fungal mitochondrial genomes with translation table 4) are much smaller than NA/NG ratios (Table 2). The smallest NC/NU value is 0.279 whereas the smallest NA/NG value is 2.372 (Table 2). We have mentioned before that, if MU3:G is very small, then a wobble G at the tRNA anticodon will not impose strong selection against U-ending codons, and NU may drift up and down with mutation relative to NC. This will lead to relatively small NC/NU ratios. From the minimum NC/NU value of 0.279, we may infer that MU3:G < 0.279•MO, i.e., MU3:G is quite small relative to MO.
Table 2.
NC/NU ratios for NNY codons and NA/NG ratios for NNR codons in Allomyces macrogynus (representing fungal mitochondrial genomes with translation table 4).
| CF* | AA† | AC‡ | NCod § | NC/NU | CF* | AA† | AC‡ | NCod § | NA/NG |
|---|---|---|---|---|---|---|---|---|---|
| AAY | N | GUU | 360 | 0.295 | AAR | K | UUU | 411 | 2.543 |
| AGY | S | GCU | 230 | 0.314 | AGR | R | UCU | 202 | 6.769 |
| CAY | H | GUG | 213 | 0.357 | CAR | Q | UUG | 173 | 3.806 |
| GAY | D | GUC | 356 | 0.299 | GAR | E | UUC | 263 | 2.372 |
| UAY | Y | GUA | 390 | 0.279 | UUR | L | UAA | 742 | 5.870 |
| UGY | C | GCA | 94 | 0.382 | |||||
| UUY | F | GAA | 592 | 0.726 | |||||
| Minimum | 0.279 | 2.372 | |||||||
| Mean | 0.379 | 4.272 | |||||||
| Std Dev | 0.157 | 1.975 | |||||||
* CF: Codon family
† AA: One-letter code of amino acid
‡ AC: Anticodon
§ NCod: Number of codons in the codon family
The AUH codon family coding for amino acid Ile in A. macrogynus mitochondrial genome is translated by a tRNA with a GAU anticodon. According to Eq. (7), the MU3:G/Co ratio should also be smaller than the NC/NU ratio. The observed NC/NU ratio is 0.3605 (= 159/441). This is similar to the NC/NU ratio in NNY codon families (Table 2). Thus, the wobble cost of MU3:G relative to MO from the AUH codon family is similar to that derived from NNY codon families.
The CUN codon family coding for amino acid Leu in A. macrogynus mitochondrial genome is translated by a tRNA with an AAG anticodon. Note that no A→I conversion has been observed in mitochondria [37,38] so Eq. (9) is applicable. According to Eq. (9), the MU3:G/Co ratio should be greater than the NC/NU ratio. The observed NC/NU ratio is 0.1905 (= 48/252). Thus we have MU3:G > 0.1905•MO. This inequality, together with the previous inequality of MU3:G < 0.279•MO, leads to 0.1905•MO < MU3:G < 0.279•MO.
Given that MU3:G > 0.1905•MO, we can infer that MwG > MwA when NC/NU < 0.1905, i.e., when U-ending codons are more than five times as frequent as C-ending codons. In other words, when NC/NU < 0.1905, the wobble cost of having an A at the wobble site of the tRNA anticodon is smaller than that of having a G at the wobble site, and should be favored by natural selection. Among the 36 fungal mitochondrial genomes, only two genomes have a NNY codon family with a correspondent tRNA that has an A at the wobble site of its anticodon. In the mitochondrial genome of Penicillium marneffei, the tRNA translating the AAY codon family has a wobble A in its anticodon. The NC/NU ratio is 0.087 (= 41/473) which is much smaller than 0.1905. Similarly, in the mitochondrial genome of Pichia Canadensis, the tRNA translating the AGY codon family has a wobble A in its anticodon. The NC/NU ratio is 0.0083 (= 1/120) which is also much smaller than 0.1905. Thus, the prediction that a wobble A at the tRNA anticodon is advantageous over a wobble G when NC/NU < 0.1905 is consistent with empirical data.
In contrast to the small NC/NU ratios in NNY, AUH and CUN codon families, all NA/NG ratios in NNR codon families are substantially larger (Table 2). We have argued before that, if MG3:U is very small, then a U at the wobble site of tRNA anticodon would impose little selection against G-ending codons in NNR codon families, and mutation may allow NG to drift up, leading to large NG values relative to NA. However, if MG3:U is large, then G-ending codons should be strongly selected against and NG would be small relative to NA, leading to large NA/NG ratios. The much larger NA/NG ratios than NC/NU ratios (t = 5.2967, DF = 10, p = 0.0003, two-tailed test) suggest that MG3:U is much greater than MU3:G.
There is a caveat in evaluating the relative magnitude of MU3:G/MO, and MG3:U/MO by the NC/NU and NA/NG ratios because these ratios can be affected by AT-biased mutations. The mitochondrial genome of A. macrogynus is 57472 bp, with the number of C+G being 22700 and that of A+U being 34772. If we exclude those nucleotides in coding sequences, then the numbers of C+G and A+U are 14136 and 17656, respectively. This may be considered as the background frequencies maintained by mutation bias, which leads to the expected NC/NU ratio of 0.8001 (=NC+G/NA+U) and that of NA/NG ratio of 1.2490 (=NA+U/NC+G). Thus, we expect NC/NU to be smaller than NA/NG even when there is no difference between MU3:G and MG3:U. To establish the argument that MU3:G is indeed smaller than MG3:U, we need to demonstrate that (1) there is no selection against U-ending codons in NNY codon families by showing that the observed NC/NU ratio is not greater than 0.8001, and (2) there is selection against G-ending codons in NNR codon families by showing that NA/NG is significantly greater than 1.2490. It is not enough to show that NC/NU < NA/NG.
We note that the observed NC/NU values for the seven NNY codon families in Table 2 are all smaller than the expected 0.8001, suggesting no selection against U-ending codons in NNY codon families (i.e., small MU3:G). In contrast the observed NA/NG ratios for the five NNR codon families are all much greater than the expected 1.2490 (Table 2), consistent with the interpretation of selection against G-ending codons in NNR codon families (i.e., large MG3:U). This is consistent with the interpretation that MU3:G < MG3:U. One can perform a χ2-test for each of the NNR codon families to see if G-ending codons are underused. The tests are all highly significant, with p < 0.00001.
The results are similar for fungal genomes with translation table 3, with the result from a representative species (Saccharomyces cerevisiae) presented in Table 3. Again the NC/NU ratios in NNY codon families are much smaller than NA/NG ratios in NNR codon families. We should note that the S. cerevisiae mitochondrial genome is much more AT-biased than the A. macrogynus mitochondrial genome, with the proportion of (G+C) in non-coding sequences being only 0.1484. The reason for the GC deficiency in yeast is not clear, but it may be caused either by mutation bias or by the low abundance of C in living cells [39-41]. In any case, the expected NC/NU and NA/NG ratios, given the biased genomic AT content, are 0.1742 and 5.7405, respectively. We note that the observed NC/NU ratios among the NNY codon families are all smaller than the expected value of 0.1742 except for the UUY (Phe) codon family (Table 3), against suggesting little selection against U-ending codons (i.e., small MU3:G). In contrast, the NA/NG ratios are all significantly greater than the expected 5.7405 except for the AUR (Met) codon family, suggesting selection against G-ending codons (i.e., large MG3:U). The exceptional AUR (Met) codon family has a tRNA with a CAU anticodon which would favor G-ending codons and is expected to be different.
Table 3.
NC/NU ratios for NNY codons and NA/NG ratios for NNR codons in Saccharomyces cerevisiae (representing fungal mitochondrial genomes with translation table 3).
| CF* | AA† | AC‡ | NCod § | NC/NU | CF* | AA† | AC‡ | NCod § | NA/NG |
|---|---|---|---|---|---|---|---|---|---|
| AAY | N | GUU | 786 | 0.065 | AAR | K | UUU | 599 | 16.114 |
| AGY | S | GCU | 119 | 0.035 | AGR | R | UCU | 225 | 36.500 |
| AUY | I | GAU | 728 | 0.093 | AUR | M | CAU | 428 | 1.326 |
| CAY | H | GUG | 183 | 0.070 | CAR | Q | UUG | 165 | 8.706 |
| GAY | D | GUC | 264 | 0.052 | GAR | E | UUC | 220 | 9.000 |
| UAY | Y | GUA | 495 | 0.076 | UGR | W | UCA | 130 | 20.667 |
| UGY | C | GCA | 92 | 0.070 | UUR | L | UAA | 911 | 64.071 |
| UUY | F | GAA | 445 | 0.369 | |||||
| Minimum | 0.035 | 1.326 | |||||||
| Mean | 0.104 | 22.341 | |||||||
| Std Dev | 0.109 | 21.560 | |||||||
* CF: Codon family
† AA: One-letter code of amino acid
‡ AC: Anticodon
§ NCod: Number of codons in the codon family
The NC/NU ratio for the UUY (Phe) codon family is consistently greater than those of other NNY codon families (Tables 2, 3). One may suspect whether, for this particular codon family, there is a significant MU3:G. The rate of tRNAPhe with anticodon 3'-AAG-5' dissociating from the UUU codon is about twice as high as that from the fully matched UUC codon [42]. Also, the tRNAPhe misread CUC codons more than twice as often as CUU codons [42]. These two lines of evidence suggest that C3:G pair is much more favorable than U3:G pair, i.e., MU3:G for the UUY (Phe) codon family may indeed be substantially greater than MC3:G. Unfortunately, there has been no other similar studies on codon-anticodon pairing for other NNY codon families. One should also note that the tRNAPhe in this study comes from Escherichia coli, and the result may not be applicable to fungal species.
The two fungal species using genetic table 16, i.e., Spizellomyces punctatus and Rhizophydium sp. 136 each have only a partial set of tRNA genes. Among NNY codon families in S. punctatus, only the GAY (coding for Asp) and UAY codon family (coding for Tyr) have an identifiable tRNA, with anticodons being GUC and GUA, respectively. The expected NC/NU and NA/NG ratios are 0.4536 and 2.2048, respectively, based on the nucleotide frequencies of non-coding sequences in the mitochondrial genome. The observed NC/NU ratios for GAY and UAY codon families are 0.1667 (= 40/240) and 0.2411 (= 81/336), respectively, suggesting no selection against U-ending codons (i.e., small MU3:G). In contrast, the NA/NG ratios are much larger, being 19.2308 (= 250/13) for the AAR codon family (coding for amino acid Lys) translated by a tRNA with a UUU anticodon and 11.1538 (= 145/13) for the CAR codon family (coding for amino acid Gln) translated by a tRNA with a UUG anticodon. These results suggest selection against G-ending codons (i.e., large MG3:U) There is no other NNR codon family with identifiable tRNAs in S. punctatus. The other species using translation table 16, Rhizophydium sp. 136, exhibits a similar pattern.
In short, results from fungal mitochondrial genomes are consistent with no selection against U-ending codons in NNY codon families but significant selection against G-ending codons in NNR codon families, indicating that MU3:G is smaller than MG3:U. These findings corroborate previous biochemical studies demonstrating that U3:G is energetically much more favorable than G3:U [2,23-26]. However, G3:U can be almost as good as A3:U when U is modified to xm5U [43].
The finding of a small MU3:G can explain puzzling observations in codon usage in fungal mitochondrial genomes. Take the tRNASer translating the AGY codon family in the mitochondrial genome of Ashbya gossypii ATCC 10895 for example. The genome contains 31 AGU codons and no AGC codon. CAAH would have predicted an ACU anticodon with perfect base pair with AGU codons, but the observed anticodon is GCU consistent with WVH. Such an anticodon makes sense only if MU3:G is equal to MC3:G, i.e., a wobble G in a tRNA anticodon can pair with U just as good as with C in the third codon position.
Can this finding be generalized to nuclear genomes and the translation machinery in the cytoplasm? If MU3:G is generally small, then we have an answer to a puzzling observation that has long baffled molecular evolutionary geneticists. The UGY codon family (coding for Cys) in the nuclear genome of S. cerevisiae is translated by tRNA coded in four tRNACys genes all with a wobble G at the tRNA anticodon. One would have predicted that UGC codons should be strongly preferred over UGU codons to avoid the wobble cost MU3:G. However, the observed numbers of UGC and UGU codons in S. cerevisiae CDSs is 13802 and 22873, respectively, opposite to the predicted trend. The bias is even stronger in highly expressed genes. For example, in the codon usage table of highly expressed genes distributed with the EMBOSS package [[44], Eyeastcai.cut], the numbers of UGC and UGU codons are 3 and 39, respectively. The unexpected codon usage bias (unexpected because the bias is in the wrong direction) becomes easy to understand if MU3:G is equal to MC3:G, i.e., there is no need to overuse the UGC codons to avoid MU3:G so AT-biased mutation in yeast (whose genome is highly AT-biased), which increases the frequency of U-ending codons, is not checked by counteracting selection.
If the finding of MU3:G << MG3:U is applicable to nuclear genomes, then we predict that NNY codon families need only one type of tRNA (i.e., tRNA with a wobble G at the tRNA anticodon) for translation. In contrast, NNR codon families should ideally be translated by two different types of tRNAs, one with a wobble U for NNA codons and the other with a wobble C for NNG codons (to avoid the relatively high wobble cost of MG3:U). A corollary is that, if a NNY codon family is translated by only tRNA with a wobble G and NNR codon family by tRNA with a wobble U, then codon usage bias should be smaller in the NNY codon family (in which selection against G-ending codon is weak because of the small MU3:G) than in the NNR codon family (in which selection against U-ending codon is strong because of the relatively large MG3:U). Below we test these predictions with genomic data.
NNY codon families are translated by one type of tRNA with a wobble G and NNR codon families are translated by two types of tRNA with a wobble U and a wobble C, respectively, in fungal nuclear genomes
We have inferred that a tRNA with a wobble G at its anticodon should be efficient not only in translating C-ending codons, but also in translating U-ending codons because of the small MU3:G. For this reason, only one type of tRNA with a wobble G should generally be sufficient in translating NNY codon families. In contrast, in NNR codon families, tRNA with a wobble U at its anticodon should be poor in translating G-ending codons because of the large MG3:U. Thus, the presence of G-ending codons in NNR codon families should favour the use of two types of tRNAs, one with a wobble U for translating NNA codons and another with a wobble C for translating NNG codons. In mitochondrial genomes with limited gene content, each codon family is generally translated by a single tRNA species. So this prediction cannot be tested. However, this prediction can be tested with nuclear genomes where the number of tRNA genes is not so limited as in mitochondrial genomes.
The prediction is strongly supported by results from the nuclear genome of Saccharomyces cerevisiae (Table 4). All NNR codon families are translated by two types of tRNAs with anticodons matching NNA and NNG codons, respectively, whereas all NNY codon families are translated by one type of tRNA with a wobble G at the tRNA anticodon (Table 4). This is consistent with the interpretation that the inference of MU3:G << MG3:U derived from fungal mitochondrial genome is also applicable to fungal nuclear genomes.
Table 4.
tRNAs translating two-fold codon families in the nuclear genome of Saccharomyces cerevisiae.
| AA* | Codon† | T‡ | F§ |
|---|---|---|---|
| Arg | AGA | 11 | 314 |
| Arg | AGG | 1 | 1 |
| Gln | CAA | 9 | 153 |
| Gln | CAG | 1 | 1 |
| Glu | GAA | 14 | 305 |
| Glu | GAG | 2 | 5 |
| Leu | UUA | 7 | 42 |
| Leu | UUG | 10 | 359 |
| Lys | AAA | 7 | 65 |
| Lys | AAG | 14 | 483 |
| Asn | AAC | 10 | 208 |
| Asn | AAU | 0 | 11 |
| Asp | GAC | 16 | 202 |
| Asp | GAU | 0 | 112 |
| Cys | UGC | 4 | 3 |
| Cys | UGU | 0 | 39 |
| His | CAC | 7 | 102 |
| His | CAU | 0 | 25 |
| Phe | UUC | 10 | 168 |
| Phe | UUU | 0 | 19 |
| Ser | AGC | 2 | 6 |
| Ser | AGU | 0 | 4 |
| Tyr | UAC | 8 | 141 |
| Tyr | UAU | 0 | 10 |
* Amion acid carried by tRNA
† Codons forming Watson-Crick base pair with the anticodon of tRNA
‡ Copy number of tRNA gene with anticodon matching the corresponding codon.
§ Codon frequencies of highly expressed yeast protein-coding genes compiled in the Eyeastcai.cut file distributed with EMBOSS [44].
One may propose an alternative hypothesis for the observation that NNY codon families are translated by tRNAs with a wobble G whereas NNR codon families translated by tRNAs with a wobble C and a wobble U, respectively. The wobble A in some nuclear tRNAs is known to be converted to inosine or I for short [45-47] which may pair with A, C or U. If a tRNA translating an NNY codon family has its wobble G mutated to wobble A, then the wobble A may undergo the A→I conversion and misread NNA codons. For this reason, the wobble G→A mutation should be strongly selected against, which would explain the lack of wobble A in tRNA translating NNY codons.
This alternative hypothesis invoking the A→I conversion, while logically sound, was dismissed quite early after the discovery [45,47] that the A→I conversion is quite restrictive and occurs mainly at ACG or ARV anticodons (where R is the IUB code for A or G, and V is for A, G or C). Among tRNAs translating NNY codons, only tRNAPhe has a middle A in the anticodon, and the rest do not have an R in the middle of the anticodon. This means that, for all NNY-translating tRNAs except for tRNAPhe, even if their regular wobble G mutates to A, the resulting wobble A will NOT be converted to inosine. Thus, the alternative hypothesis can only explain the avoidance of a wobble A in tRNAPhe but cannot explain the avoidance of a wobble A in other NNY-translating tRNAs.
Conclusion
In summary, our general hypothesis based on wobble costs allows the integration of the two conventional hypotheses (i.e., CAAH and WVH) on codon-anticodon wobble pairing. The integration leads to new ways of evaluating relative wobble cost of different wobble pairings. In particular, the finding that MU3:G is much smaller than MG3:U corroborates previous structural studies showing that U3:G is energetically more favourable than G3:U and leads to a better understanding of the translation efficiency mediated by codon and anticodon wobble pairing.
Acknowledgements
This study is supported by the CAS/SAFEA International Partnership Program for Creative Research Teams and by NSERC's Discovery and Strategic Grants. I thank S. Aris-Brosou, P. Higgs and E. Rocha for discussion and comments and E. Westhof for references. Four anonymous reviewers provided helpful comments leading to substantial improvement of the paper.
References
- Bonitz SG, Berlani R, Coruzzi G, Li M, Macino G, Nobrega FG, Nobrega MP, Thalenfeld BE, Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32(1):223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KL, Wong JT. Anticodon and wobble evolution. Gene. 2004;333:169–177. doi: 10.1016/j.gene.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RP, Sibler AP, Gehrke CW, Kuo K, Edmonds CG, McCloskey JA, Dirheimer G. 5-[[(carboxymethyl)amino]methyl]uridine is found in the anticodon of yeast mitochondrial tRNAs recognizing two-codon families ending in a purine. Biochemistry (Mosc) 1990;29(4):956–959. doi: 10.1021/bi00456a016. [DOI] [PubMed] [Google Scholar]
- Xia X. Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes. Gene. 2005;345(1):13–20. doi: 10.1016/j.gene.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Nishimura S. In: tRNA: structure, biosynthesis and function. Soll D, RajBhandary U, editor. Washington , ASM Press; 1995. Modified nucleotides and codon recognition. pp. 207–223. [Google Scholar]
- Sibler AP, Dirheimer G, Martin RP. Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett. 1986;194(1):131–138. doi: 10.1016/0014-5793(86)80064-3. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Kojima A, Bessho Y, Hori H, Ohama T, Osawa S. Translation of synonymous codons in family boxes by Mycoplasma capricolum tRNAs with unmodified uridine or adenosine at the first anticodon position. J Mol Biol. 1995;251(4):486–492. doi: 10.1006/jmbi.1995.0450. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Suzuki T, Watanabe K. Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J Mol Evol. 2001;53(4-5):314–326. doi: 10.1007/s002390010221. [DOI] [PubMed] [Google Scholar]
- Andachi Y, Yamao F, Iwami M, Muto A, Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1987;84(21):7398–7402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell BG, Anderson S, Bankier AT, de Bruijn MH, Chen E, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325(6106):728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics. 1998;149(1):37–44. doi: 10.1093/genetics/149.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10: 7055–7064. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257(6):3026–3031. [PubMed] [Google Scholar]
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164(4):1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol Biol Evol. 1996;13(6):864–872. doi: 10.1093/oxfordjournals.molbev.a025646. [DOI] [PubMed] [Google Scholar]
- Xia X. Maximizing transcription efficiency causes codon usage bias. Genetics. 1996;144:1309–1320. doi: 10.1093/genetics/144.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Higgs PG. Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol Biol Evol. 2008;25(2):339–351. doi: 10.1093/molbev/msm259. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Sundaralingam M. Stacking of Crick Wobble pair and Watson-Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon Wobble interaction. Nucleic Acids Res. 1978;5(11):4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol. 2007;14(6):498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6(1):9–15. doi: 10.1017/S1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carullo M, Xia X. An Extensive Study of Mutation and Selection on the Wobble Nucleotide in tRNA Anticodons in Fungal Mitochondrial Genomes. J Mol Evol. 2008;66(5):484–493. doi: 10.1007/s00239-008-9102-8. [DOI] [PubMed] [Google Scholar]
- Xia X. Body temperature, rate of biosynthesis and evolution of genome size. Mol Biol Evol. 1995;12:834–842. doi: 10.1093/oxfordjournals.molbev.a040260. [DOI] [PubMed] [Google Scholar]
- Rocha EP. Codon usage bias from tRNA's point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;12:12. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Huang H, Carullo M, Betran E, Moriyama EN. Conflict between Translation Initiation and Elongation in Vertebrate Mitochondrial Genomes. PLoS ONE. 2007;2:e227. doi: 10.1371/journal.pone.0000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J Hered. 2001;92(4):371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xia X. Data analysis in molecular biology and evolution. Boston , Kluwer Academic Publishers; 2001. p. 277. [Google Scholar]
- Higgs PG, Jameson D, Jow H, Rattray M. The evolution of tRNA-Leu genes in animal mitochondrial genomes. J Mol Evol. 2003;57(4):435–445. doi: 10.1007/s00239-003-2494-6. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Higgs PG. A unified model of codon reassignment in alternative genetic codes. Genetics. 2005;170(2):831–840. doi: 10.1534/genetics.104.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol. 2007;64(6):662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Roy H, Qin D, Rubio MA, Alfonzo JD, Fredrick K, Ibba M. Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc Natl Acad Sci U S A. 2007;104(39):15299–15304. doi: 10.1073/pnas.0704441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Sprinzl M, Steinberg S. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995;77(1-2):139–141. doi: 10.1016/0300-9084(96)88117-X. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18(6):291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- Xia X, Palidwor G. Genomic Adaptation to Acidic Environment: Evidence from Helicobacter pylori. Am Nat. 2005;166(6):776–784. doi: 10.1086/497400. [DOI] [PubMed] [Google Scholar]
- Xia X, Wang HC, Xie Z, Carullo M, Huang H, Hickey DA. Cytosine usage modulates the correlation between CDS length and CG content in prokaryotic genomes. Mol Biol Evol. 2006;23(7):1450–1454. doi: 10.1093/molbev/msl012. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell. 2006;21(3):369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. Modified Uridines with C5-methylene Substituents at the First Position of the tRNA Anticodon Stabilize U{middle dot}G Wobble Pairing during Decoding. J Biol Chem. 2008;283(27):18801–18811. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Haumont E, Fournier M, de Henau S, Grosjean H. Enzymatic conversion of adenosine to inosine in the wobble position of yeast tRNAAsp: the dependence on the anticodon sequence. Nucleic Acids Res. 1984;12(6):2705–2715. doi: 10.1093/nar/12.6.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker HF, Foiret D, Morin A, Jin YX, Fournier M, Fourrey JL. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78(6):488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- Auxilien S, Crain PF, Trewyn RW, Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol. 1996;262(4):437–458. doi: 10.1006/jmbi.1996.0527. [DOI] [PubMed] [Google Scholar]