Abstract
Low-cost and high-resolution on-chip microscopes are vital for reducing cost and improving efficiency for modern biomedicine and bioscience. Despite the needs, the conventional microscope design has proven difficult to miniaturize. Here, we report the implementation and application of two high-resolution (≈0.9 μm for the first and ≈0.8 μm for the second), lensless, and fully on-chip microscopes based on the optofluidic microscopy (OFM) method. These systems abandon the conventional microscope design, which requires expensive lenses and large space to magnify images, and instead utilizes microfluidic flow to deliver specimens across array(s) of micrometer-size apertures defined on a metal-coated CMOS sensor to generate direct projection images. The first system utilizes a gravity-driven microfluidic flow for sample scanning and is suited for imaging elongate objects, such as Caenorhabditis elegans; and the second system employs an electrokinetic drive for flow control and is suited for imaging cells and other spherical/ellipsoidal objects. As a demonstration of the OFM for bioscience research, we show that the prototypes can be used to perform automated phenotype characterization of different Caenorhabditis elegans mutant strains, and to image spores and single cellular entities. The optofluidic microscope design, readily fabricable with existing semiconductor and microfluidic technologies, offers low-cost and highly compact imaging solutions. More functionalities, such as on-chip phase and fluorescence imaging, can also be readily adapted into OFM systems. We anticipate that the OFM can significantly address a range of biomedical and bioscience needs, and engender new microscope applications.
Keywords: optofluidic microscopy, phenotype characterization, microfluidic
Optical microscopy pervades almost all aspects of modern biomedicine and bioscience; to name a few key areas, optical microscopes are vital instruments in microorganism studies, cell biology, and clinical pathology. However, despite the long history of microscopy and the remarkable range of optical tools that have been developed since the invention of the first microscope in the early 1600s, the fundamental design of microscopes has undergone little change. A typical microscope still consists of an objective, space for relaying the image, and an eyepiece or an imaging lens to project a magnified image onto a person's retina or a camera. In addition to its relatively high implementation cost (precise and expensive lenses are needed), the conventional microscope design has also proven difficult to miniaturize (1, 2). A relatively modern invention—digital inline holographic microscopy (DIHM) (3)—showed that it is possible to render microscope-resolution images of objects without the use of lenses; however, as a method, DIHM requires significant postmeasurement computation and the use of a coherent light source. In 2005, Lange et al. (4) reported a direct projection method to implement compact and low-cost imaging systems. In Lange's method, the specimen is placed directly on a CMOS image sensor, and the projection image is then recorded by the sensor (Fig. 1A). The resolution in such a system is given by the sensor pixel size. Because the typical pixel size of a commercial CCD or CMOS sensor is >3 μm, this approach is incapable of yielding images that have resolution comparable with conventional microscope images (resolution of 1 μm or better). Despite their low image qualities, recent works (5) showed that these pixelated images are useful for certain high-throughput cell-identification applications.
Fig. 1.
Comparison of on-chip imaging schemes. (A) Direct projection imaging scheme. By placing the specimen directly on top of the sensor grid, we can obtain a projection image with resolution equal to the sensor pixel size. (B) By placing the specimen on a grid of apertures, we can obtain a sparse image. However, for the same grid density, the obtained image will not be much improved over that of A. (C) By raster-scanning the specimen over the aperture grid, we can obtain a “filled-in” image. In this case, the image resolution is limited by the aperture size. Grid density is no longer a factor in resolution consideration. (D) The scanning scheme can be simplified into a single-pass flow of the specimen across the grid by orientating the grid at a small angle (θ) with respect to the flow direction (x axis). (E) The aperture grid can be simplified by substitution with a long linear aperture array. This scheme is the basis for the optofluidic microscopy method.
Our present objective is to determine whether it is possible to modify the direct projection imaging accordingly so that microscope-resolution images can be collected. We believe that if such an approach can be found, it can be a viable low-cost and compact replacement for the conventional microscope system for a range of applications.
It is difficult to conceive or develop a direct projection imaging strategy by which single-time-point images at resolution better than the sensor pixel size can be acquired. However, if we permit ourselves to exploit the time dimension during the image acquisition process, it is possible to develop viable high-resolution direct projection imaging strategies in which resolution and sensor pixel size are independent. As an example, one can imagine covering a sensor grid with a thin metal layer and etching a small aperture onto the layer at the center of each sensor pixel. The sensor pixel will then be sensitive only to light transmitted through the aperture. By placing a target specimen on top of the grid, we can then obtain a sparsely sampled image of the object (Fig. 1B). A “filled-in” image can be generated by raster-scanning the specimen over the grid (or equivalently, raster-scanning the grid under the specimen) and compositing the time varying transmissions through the apertures appropriately (Fig. 1C). We can see that in this case, the resolution is fundamentally determined by the aperture size and not the pixel size. Therefore, by choosing the appropriate aperture size, we can achieve high resolution. The imaging strategy can be simplified by tilting the aperture grid at a small angle (θ) with respect to x axis and replacing the raster-scan pattern with a single linear translation of the specimen across the grid (Fig. 1D). As long as a sufficient number of apertures span across the specimen completely in y axis, and neighboring apertures overlap sufficiently along y axis, a filled-in high-resolution image of the specimen will be achieved. The design can be further simplified by replacing the tilted 2D aperture grid with a long tilted 1D aperture array (Fig. 1E). This imaging strategy (6) forms the basis of the optofluidic microscopy (OFM) method. The OFM method shares a lot of similarities with near-field scanning optical microscopy methods (7). In fact, the OFM aperture array can be interpreted as a series of NSOM apertures. Whereas NSOM sensors are generally raster-scanned over the target objects, the OFM approach uses object translation to accomplish scanning—in the microfluidic system, this is a far simpler and more efficient strategy.
Here, we report the implementation and application of two high-resolution, lensless, and fully on-chip microscope systems based on the OFM method. The first OFM system is customized for Caenorhabditis elegans imaging. It utilizes a gravity-driven-flow mechanism to translate C. elegans across the OFM sensing area. The second system is optimized for cell and other spherical/ellipsoidal object imaging. It utilizes an electrokinetic drive to translate samples in the OFM system. This approach minimizes potential rotations or tumbles of the cells during the scanning process.
In the next section, we will describe the implementation of the gravity-driven-flow-based OFM system, provide details about its characteristics, and report on its use for automated C. elegans imaging and phenotype characterization. Next, we will describe the electrokinetically driven OFM system, and report on its use for imaging Chlamydomonas (single cellular algae), mulberry pollen spores, and polystyrene spheres. We will then discuss the OFM contrast mechanism and the resolution characteristics of OFM systems. Finally, we conclude by briefly discussing the potential applications of the OFM method.
Results
Gravity-Driven-Flow-Based Optofluidic Microscope System.
Design.
This on-chip OFM system was fabricated on a commercially available 2D CMOS image sensor (Micron MT9V403C12STM) with a 9.9-μm pixel size. We planarized the surface of the sensor with a 2-μm-thick SU8 photoresist and coated it with a 300-nm-thick Al layer. We then milled two lines of apertures (1 μm diameter) separated by a single line of sensor pixels onto the Al layer with a focused ion beam (FIB) machine (FEI Nova 200). The apertures were spaced 9.9 μm apart so that each aperture mapped uniquely onto a single sensor pixel (Fig. 2A). Each line consisted of 100 apertures. A 0.2-μm-thick poly(methyl methacrylate) (PMMA) layer was spin-coated on top of the Al film to protect the OFM apertures. Finally, we bonded an optically transparent poly(dimethylsiloxane) (PDMS) microfluidic chip containing a channel (width of 50 μm, height of 15 μm) on top of the sensor with a Karl Suss mask aligner (MJB3). The channel was oriented at θ = 0.05 radians with respect to the aperture arrays. The top of the system was uniformly illuminated with white light (≈20 mW/cm2, approximately the intensity of sunlight) from a halogen lamp.
Fig. 2.
OFM prototype. (A) schematic of the device (top view). The OFM apertures (white circles) are defined on the Al (gray) coated 2D CMOS image sensor (light gray dashed grid) and span across the whole microfluidic channel (blue lines). (B) The actual device compared with a U.S. quarter. (C) Upright operation mode. (D) Flow diagram of the OFM operation. Two OFM images of the same C. elegans are acquired by the two OFM arrays, respectively (red arrows). If the image correlation is <50%, the image pair is rejected. Otherwise, the area and the length of the worms are measured automatically by evaluating the contour (green dashed line) and the midline (yellow dashed line). (E) Cross-section of the fabrication of an electrokinetically driven OFM device.
Each line of apertures represents an OFM scanning array. The metal layer blocks light from the underlying pixels; light can only be transmitted through the apertures. The imaging process involves uniformly flowing the specimen through the channel and recording the time varying light transmission through each aperture as the specimen passes. Each time scan represents a line profile across the specimen. Because the specimen passes the apertures sequentially, if the speed of the specimen is uniform, there will be a constant time delay between adjacent line scans. By shifting the line scans with this delay, we can obtain an accurate projection image of the specimen. Specifically, unlike the physical sensing grid in a CCD or CMOS image sensor, the OFM sampling scheme effectively establishes a virtual sensing grid. The grid density is adjustable by changing the number of apertures spanning the channel, the flow speed of the specimen, and the OFM readout rate. Higher pixel density allows the OFM to oversample the specimen, thereby preventing undesirable aliasing artifacts in the images. Finally, as mentioned previously, the resolution of such system is fundamentally limited by the aperture size.
This on-chip OFM prototype (Fig. 2 B and C) utilizes two parallel OFM arrays for two reasons. First, by measuring the time difference between when the specimen first crosses each array and knowing the separation between the two arrays along the channel axis x, we can determine the flow speed of the specimen v, which is important for correct OFM image construction. (Note that the flow speed is determined for each specimen independently. As such, speed variations between specimens have no impact on our ability to perform correct OFM image reconstruction.) Second, significant differences between the two OFM images acquired by the two OFM arrays for the same specimen will indicate shape changes, flow speed variations, and/or rotations of the specimen during the data acquisition. Because accurate OFM imaging requires the absence of these variations, discrepancy between the images is a possible criterion for rejecting that image pair (Fig. 2D). We set our rejection criterion at the image-pair correlation threshold of <50%. During our experiments, ≈50% of the specimens were rejected based on this criterion. We note that this processing approach was highly conservative in that it also rejected a large proportion of acceptable images in which image-pair correlation was low because of small variations in flow speed and slight sample shifts. We believe that better flow controls (such as smoother channels and better speed tracking) and better image-processing algorithms can significantly lower the rejection rate. This is an area that is worth further study.
C. elegans imaging demonstration.
We demonstrated the proper functioning of this on-chip microscope system by employing it to image C. elegans larvae. To facilitate efficient flow of the specimens through the system, we took the following steps in preparing the microfluidic channel.
The PDMS microfluidic channel was designed with two smooth funnels at both ends, and oxygen plasma was used to render the inner surface of the PDMS channel hydrophilic. Before use, we conducted a surface treatment process (detailed in Materials and Methods) to reduce sample adhesion to the channel walls. We operated the prototype in the upright mode (Fig. 2C), so that gravity could drive the flow; this eliminated the need for bulky external pumps. When the specimen solution (newly hatched C. elegans L1 larvae in S-basal buffer, ≈20 worms per microliter) was injected into the top funnel, the solution wetted the channel and the specimens were continuously pulled into the channel by the gravity-driven microfluidic flow. To prevent the worms from wiggling, we immobilized them by subjecting them to a 70°C heat bath for 3 min. Because of the sedimentation of the worms in the solution, the throughput of OFM imaging was not constant. The maximum observed throughput was approximately five worms per minute. However, the flow speed of worms v in the channel was fairly uniform (≈500 μm/sec). The data readout rate f of each OFM array was 1,000 frames/sec, and imaging of each worm required ≈2.5 sec. The spacing of the OFM virtual grid along the x and y axes are both 0.5 μm (less than the 1-μm aperture size).
Fig. 3A shows a pair of OFM images acquired by the two OFM arrays from the same wild-type C. elegans. The image correlation between them was 56%. Consistent internal structures were found in both OFM images. Fig. 3B shows an image collected from a similar worm that was placed directly onto an unprocessed CMOS sensor (note that the pixel size is 9.9 μm); the worm was barely distinguishable in this low-resolution direct projection image. Fig. 3C shows a conventional microscope image of a similar worm acquired through a ×20 Olympus objective lens (650-nm resolution for 555-nm wavelength under Sparrow's criterion). Similar internal structures of C. elegans appeared in both the microscope and the OFM images. This confirmed that the OFM can render images comparable in quality to those of a conventional microscope with similar resolution.
Fig. 3.
Images of wild-type C. elegans L1 larvae. (A) Duplicate OFM images acquired by the two OFM arrays for the same C. elegans. (B) Direct projection image on a CMOS sensor with 9.9-μm pixel size. (C) Conventional microscope image acquired with a ×20 objective.
Quantitative phenotype characterization of C. elegans.
The function of a gene must manifest itself in a certain phenotype to be observed. However, the formidable number of genes and their combinations imposes a difficult challenge to systematic phenotype characterization (8, 9). Inexpensive, automated, and quantitative phenotype characterization devices are critical to comprehensive biology studies. Motivated by the extensive use of phenotype characterization, especially morphology, in the genetic studies of microorganisms and cells, we used the OFM prototype to image and analyze phenotypes of C. elegans.
To compare body sizes of the wild-type (N2), sma-3 (e491), and dpy-7 (e88) C. Elegans, we imaged 25 specimens of each strain. The sma-3 (e491) gene is part of a family of transforming growth factor β pathway components (10). The dpy-7 gene encodes a cuticular collagen required for proper body form (11). The typical OFM images of the three strains (Fig. 4 A–C) show that the sma-3 worm is smaller and thinner than the wild-type worm, and that the dpy-7 worm is fatter and shorter than the wild-type worm. These observations are consistent with those made under a conventional microscope.
Fig. 4.
Phenotype characterization of C. elegans L1 larvae. (A–C) Typical OFM images of wild-type, sma-3, and dpy-7 worms, respectively. (D and E) The length (D) and effective width (E) of wild-type, sma-3, and dpy-7 worms, respectively (color-coded). The columns represent the mean values in the population; the hatched areas correspond to the confidence intervals of the mean values; and the error bars are the standard deviations indicating the variation between individuals in the population. Twenty-five worms were evaluated for each phenotype.
Because OFM images are naturally digitalized, we can perform large volume and automatic quantitative information extraction by computer assisted postprocessing. We developed a MATLAB program (the algorithm is described in Materials and Methods) to determine the area and length of the worms in batches (Fig. 2D). From those two quantities, we then computed an effective width for each worm by dividing the area by the length. In Fig. 4 D and E the columns represent the mean length and width of the three C. elegans strains; the hatched areas correspond to the confidence intervals of our mean length and width estimates. The standard deviations (blue error bars) of the measurement indicate the variation between individuals within the strain. The measured mean length and width were 252.9 ± 3.1 μm and 11.7 ± 0.1 μm for wild-type, 214.3 ± 2.9 μm and 11.5 ± 0.1 μm for sma-3, and 199.1 ± 4.3 μm and 12.1 ± 0.1 μm for dpy-7. They were consistent with reported data (12). The three strains have distinct length (P < 0.01 for each pair; Student's t test). dpy-7 mutants are significantly wider than N2 and sma-3 (P < 0.05 and P < 0.01, respectively), but we observed no statistically significant width difference between sma-3 and N2 for the sample size used.
Electrokinetic-Drive-Based OFM System.
Flow dynamic difference between pressure and electrokinetic drive.
This next OFM system was customized for imaging cells and other spherical/ellipsoidal objects. Pressure-driven liquid flow in microfluidic channel typically develops a parabolic velocity profile (Poiseuille flow) due to the nonslip boundary condition on the channel side walls. An object flowing in the channel will receive a torque from this nonuniform velocity profile and start to tumble if it is slightly off-center or if it is nonsymmetric. This nonuniform translational movement can prevent the OFM system from acquiring an accurate image of the object.
We prepared the following experiment to observe this effect. A PDMS microfluidic channel, of dimension 3 mm in length, 40 μm in width, and 13 μm in height was bonded with a PMMA-coated glass slide. The channel was then passivated by using the process described in Materials and Methods. We then injected heat-killed (70°C water bath for 30 min) Chlamydomonas cells into the channel by a syringe. A difference in fluid column height between the channel inlet and outlet induced a pressure differential in the channel and actuated the flow (13). The parabolic velocity profile exerted an unsymmetrical distribution of drag force on the objects flowing along the microfluidic channel and caused the cells to rotate.
We found that the use of dc electrokinetics provides a simple and direct way to control the motion of biological cells in the on-chip OFM system to suppress object rotation. By imposing a uniform electric field in a PDMS channel (3 mm in length, 40 μm in width, and 13 μm in height, bonded with a PMMA-coated glass slide), a dipole can be induced on target ellipsoidal Chlamydomonas cell (heat-killed by 70°C water bath for 30 min). The dispersed dipole moment can only be stabilized when its major axis is aligned along the electric field lines. In other words, the cell will experience an electroorientation force (14). At the same time, because the cell Chlamydomonas cell is likely to carry a net electrical charge, the external electric field will also exert an electrophoretic force on the cell.(15). This induces the cell to move along the channel. The velocity-dependent viscous Stokes drag will eventually match this force and result in a constant rotation-free translational motion of the cell. The application of external electric field also causes the translation of the electric double layer (EDL) at the channel walls; this phenomenon is known as electroosmosis (16). Under the thin EDL assumption, the electroosmotic plug-like velocity profile will exert a symmetrical shear stress distribution on the cells. In steady-state situations, this movement is also nonrotational. At a constant voltage of 25 V to a pair of platinum electrodes at the channel inlet and outlet, we found that an average translational speed of 270 μm/sec was achieved for the cells.
In addition, we observed that the cells typically settled into their steady-state orientation via electroorientation within a flow distance of 200 μm. This information was useful in informing the OFM system design because it indicated that we need to allow for an extra flow channel length before the OFM aperture array for the specimens to reach steady state before the image acquisition. Finally, we note that we did not encounter any cell lysis issues over the voltage range (10–65 V for a 3-mm-long microfluidic channel) we tested.
To compare the performance of pressure drive and electrokinetic drive for creating rotation-free sample translation, we performed the following experiment. We flowed 100 Chlamydomonas cells, with size ranged from 8 to 16 μm in width, through the channel by pressure drive and electrokinetic drive. We chose a region of interest that is 500 μm in length and 1.25 mm from the inlet for observation. This region of interest matches with the location and length of the OFM array in our second OFM system. A statistical distribution of cell rotation events in that region observed through a conventional microscope is recorded. We define a cell as experiencing rotation if it has rotated by >3° during its passage through the region of interest. In total, 83 cells experienced rotation under pressure drive whereas only 6 rotating cells were observed under electrokinetic drive. In fact, these errant movements were mainly observed after a prolonged experiment period, typically ≈30 min, and were likely caused by the presence of debris on the channel floor after extended channel usage.
Design.
This system was similar to the gravity-driven OFM system in its basic layout with a couple key differences and a few minor noncritical design variations (Fig. 2E). For brevity, we shall presently enumerate only the significant differences. This system was fabricated on a 2D CMOS imaging sensor (Micron MT9M001C12STM). The CMOS chip comprised of a grid lattice of 1,280 × 1,024 square pixels with the pixel size at 5.2 μm. We planarized the sensor surface with PMMA and coated it with a 15-nm-thick chromium seed layer and a 300-nm-thick gold layer. The aperture array consisted of 120 holes of diameter 0.5 μm and separation 10.4 μm. We next deposited a second layer of PMMA (0.4 μm thick) on the gold layer to insulate the metal layer from the electric field that would be subsequently applied (17). Then, we emplaced a PDMS chip containing a microfluidic channel of dimension 2.4 mm in length, 40 μm in width, and 13 μm in height onto the chip. Finally, we inserted a pair of platinum electrodes into the inlet/outlet of the channel.
Chlamydomonas, mulberry pollen spores, and polystyrene sphere imaging demonstration.
We imaged three different samples, namely, Chlamydomonas cells (8–16 μm; Carolina Scientific), mulberry pollen spores (11 μm to 16 μm; Duke Scientific), and polystyrene microspheres (10 μm; PolyScience) with this system.
Before the experiment, we treated the channel surface using the procedure detailed in Materials and Methods. The Chlamydomonas cells were heat treated (70°C water bath for 30 min) before the experiment. The mulberry pollen spores and polystyrene microspheres were dissolved in deionized water and sonicated for 5 min before use.
During imaging acquisitions, the potential difference between the electrodes was set at ≈20 V. The typical translation speed of the objects in the OFM system was ≈1.5 mm/sec. The data readout rate was 2,000 frames/sec. For an object of dimension 15 μm, this implied a typical OFM image acquisition time of 0.3 sec. The higher-than-expected translation speed was attributable to residual pressure differential in the channel. Nevertheless, the electroorientation force was sufficiently strong to suppress sample rotations.
Several OFM images of the three different samples are shown respectively in Fig. 5 A–E in comparison with images acquired with an inverted microscope (Olympus IX-71) under a ×20 objective in Fig. 5 F–J.
Fig. 5.
Cell and microsphere images. (A–E) Images taken from the on-chip OFM driven by dc electrokinetics of Chlamydomonas (A and B), mulberry pollen (C and D), and a 10-μm polystyrene microsphere. (F–J) Images taken from a conventional light transmission microscope with a ×20 objective of Chlamydomonas (F and G), mulberry pollen (H and I), and a 10-μm polystyrene microsphere (J). (Scale bars: 10 μm.)
Contrast and Resolution.
OFM contrast mechanism.
The OFM image contrast shares similar origins with the contrast in conventional microscopy images. The OFM achieves its highest resolution in the plane that is just above the aperture array. In effect, the OFM is similar to a conventional microscope in which the focal plane is locked at the plane that is just below the target object. The light field at that plane consists of the combination of the unscattered component of the illumination and the light fields that are scattered by scattering sites in the object. The presence of a scattering site immediately above a specific point in that plane will typically result in a dark patch in the image as the illumination light is scattered away by the scatterer. At other locations, the constructive interference of scattered light and the illumination field can result in a higher-than-average light field brightness. The dark boundary in Fig. 5D is attributable to a diminished light field from the presence of the pollen boundary scattering light away. The bright boundary is attributable to the constructive interference of that scatter light component with the illumination at those locations.
However, we note that the above-mentioned similarity between the OFM and a conventional microscope with a fixed focal plane only holds if near-field components are insignificant. Because the OFM samples the wavefront without resorting to propagative projection, it is also sensitive to near-field light components. Therefore, it is possible for an OFM system to achieve better resolution by using smaller apertures.
Object thickness limit.
It is worth noting that, similar to a conventional transmission microscope, there is, in principle, no upper limit to the sample thickness that the OFM can process. In practice, the OFM will fail to acquire an image if the sample is too optically scattering or absorptive to permit sufficient light to be transmitted through the OFM apertures. This practical limit exists for the conventional transmission microscope as well.
OFM resolution.
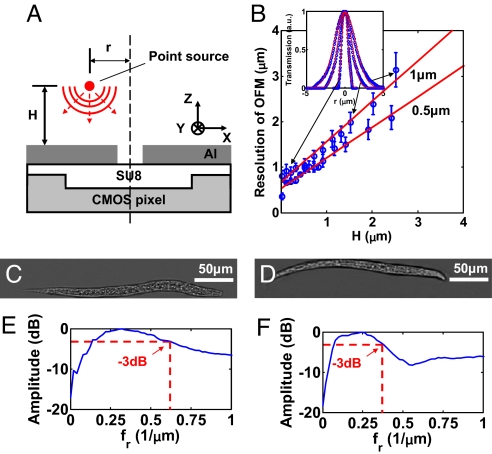
The resolution characteristics of the OFM are useful for informing on system design and image interpretation. We began by characterizing the point spread functions (PSF) associated with individual OFM apertures. We measured the PSF of our prototypes by laterally scanning a near-field scanning optical microscope (NSOM) (Alpha-SNOM; WITec) tip across the apertures (1 and 0.5 μm in diameter) at various heights H and measuring the signal detected by the underlying pixel (Fig. 6A). We approximated the NSOM tip, which was <100 nm in diameter, as a point source. Fig. 6B Inset shows representative OFM PSF plots at H = 0.1, 1.5, and 2.5 μm for the 1-μm-diameter aperture. The PSF broadened as a function of H. We quantified the height-dependent resolution of our prototype by the PSF's width. Fig. 6B shows the resolution [Sparrow's criterion (18)] as a function of H. From the plot, we can see that the ultimate resolution of the gravity-driven-flow-based OFM system was 0.9 μm (with 0.2-μm-thick PMMA above the metal layer accounted for) and the resolution degraded to 3 μm at H = 2.5 μm. The ultimate resolution of the electrokinetic drive based OFM system was 0.8 μm (with the 0.4-μm-thick PMMA above the metal layer accounted for) and the resolution degraded to 2 μm at H = 2.5 μm. The result was consistent with our more detailed study on the light collection characteristics of small apertures (19).
Fig. 6.
Resolution of the OFM prototype. (A) Schematic of the PSF measurement. (B) Resolution of the prototype at various heights H above a 0.5- and a 1-μm-diameter aperture under Sparrow's criterion. (Inset) Representative OFM PSF plots at H = 0.1, 1.5, and 2.5 μm for the 1-μm-diameter aperture. (C and D) OFM images of wild-type C. elegans L1 larvae in a 15- and 25-μm-tall microfluidic channels, respectively. (E and F) Radial frequency spectra of OFM images in C and D, respectively. The −3-dB bandwidths (dashed red lines) are at 0.62 and 0.38 μm−1, respectively.
We note that, given the OFM's contrast mechanism, a better approach for resolution characterization will be to translate a point scatterer across the aperture under a uniform illumination field and measure the light collected by the aperture. However, we further note that the point source and point scatterer configurations are optically similar in the context of resolution considerations. Under Sparrow's criterion (18) and in the small scatterer limit, the point source resolution computation is directly translatable for point scatter consideration.
We also verified the degradation of the OFM resolution with height through a C. elegans imaging experiment in which we varied the channel height. Fig. 6 C and D shows OFM images of wild-type C. elegans in 15- and 25-μm-tall channels, respectively. A shallow channel was able to better confine the specimen close to the aperture array and thus was able to provide better resolved images (Fig. 6C). Fig. 6 E and F shows the radial frequency spectrums of the OFM images, which revealed that the −3-dB bandwidths were at 0.62 and 0.38 μm−1, respectively, for the 15- and 25-μm channels.
Discussion
The application of OFM for cell imaging is a particularly promising area. As an automatic on-chip cell microscopy method, OFM can potentially be used in applications such as blood fraction analysis (20), urine screening for infection (21, 22), stem cell screening and sorting (23, 24), tumor cell counting (25, 26), and drug screening (27).
The compact, simple, and lensless OFM can significantly benefit a broad spectrum of biomedicine applications and bioscience researches, and also change the ways we conduct certain experiments. For example, the availability of tens or even hundreds of microscopes on a chip can allow automated and massively parallel imaging of large populations of cells or microorganisms. An on-chip microscope system can also potentially provide low cross-contamination risk (by being cost-effective enough to be disposable) point-of-care analysis in the clinical settings. In a Third World environment, a complete, low-cost, and compact microscope system suitable for malaria diagnosis can be a boon for a health worker who needs to travel from village to village.
Materials and Methods
Culture of C. elegans for Imaging.
The alleles used were dpy-7(e88), sma-3(e491), and wild-type (N2). C. elegans were maintained and handled as described in ref. 28. Briefly, all strains were cultured at 20°C. Bleaching was used to synchronize the development of C. elegans L1 larvae.
PEG Grafting Process to Promote the Flow of Samples in OFM Imaging.
The microfluidic channel was filled up and flushed with a 10% poly(ethylene glycol) (PEG) solution, 0.5 mM NAIO4, and 0.5% (by weight) benzyl alcohol. Under the activation of UV light for 1 h, the channel surface was conjugated with the PEG molecules. The process is similar to the one described in ref. 29. The PEG grafted surface prevented nonspecific adsorption with biological entities and lubricated the object flow. The chip can be rinsed with deionized water, dried, and stored under ambient condition because the PEG grafted surface has a long-term stability.
The Algorithm for Automatic Determination of C. elegans Length and Width.
(i) Delineate the boundary around each C. elegans from the OFM images. (ii) Calculate the area occupied by each C. elegans based on the boundary. (iii) Segment the C. elegans image along its length and calculate the centroid for each segment. (iv) Connect the centroids by a continuous line. The length of the C. elegans is given by the length of this line. (v) The width of the C. elegans is calculated by dividing the area occupied by the nematode with its length.
Acknowledgments.
We are grateful for the constructive discussions with and the generous help from Professor Axel Scherer, Jigang Wu, Dr. Zahid Yaqoob, Dr. Claudiu Giurumescu, Oren Schaedel and Dr. Xiaoyan Robert Bao. We appreciate the assistance of the Caltech Watson clean-room. This work is funded by DARPA's Center of Optofluidic Integration, the Wallace Coulter Foundation, National Science Foundation Career Award BES-0547657, and National Institutes of Health Grant R21 PA03-107. L.M.L. thanks the Croucher Scholarship for financial support. P.W.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 2.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Sucerquia J, et al. Digital in-line holographic microscopy. Appl Opt. 2006;45:836–850. doi: 10.1364/ao.45.000836. [DOI] [PubMed] [Google Scholar]
- 4.Lange D, Storment CW, Conley CA, Kovacs GTA. A microfluidic shadow imaging system for the study of the nematode Caenorhabditis elegans in space. Sensors Actuators B. 2005;107:904–914. [Google Scholar]
- 5.Ozcan A, Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 6.Heng X, et al. Optofluidic microscopy—A method for implementing a high resolution optical microscope on a chip. Lab Chip. 2006;6:1274–1276. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- 7.Courjon D. Near-Field Microscopy and Near-Field Optics. London: Imperial College Press; 2003. [Google Scholar]
- 8.Bochner BR. New technologies to assess genotype-phenotype relationships. Nat Rev Genet. 2003;4:309–314. doi: 10.1038/nrg1046. [DOI] [PubMed] [Google Scholar]
- 9.Zhong WW, Sternberg PW. Genome-wide prediction of C-elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 10.Savage C, et al. Caenorhabditis elegans genes sma2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilleard JS, Barry JD, Johnstone IL. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol Cell Biol. 1997;17:2301–2311. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage-Dunn C, et al. SMA-3 Smad has specific and critical functions in DBL-1/SMA-6 TGF β-related signaling. Dev Biol. 2000;223:70–76. doi: 10.1006/dbio.2000.9713. [DOI] [PubMed] [Google Scholar]
- 13.Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 14.Hughes MP. Nanoelectromechanics in Engineering and Biology. 1st Ed. Boca Raton, FL: CRC Press; 2003. [Google Scholar]
- 15.Grossman PD, Colburn JC. Capilliary Electrophoresis. 1st Ed. London: Academic; 1992. [Google Scholar]
- 16.Probstein RF. Physicochemical Hydrodynamics. 2nd Ed. New York: Wiley InterScience; 2003. [Google Scholar]
- 17.Park JH, Hwang DK, Lee J, Im S, Kim E. Studies on poly(methyl methacrylate) dielectric layer for field effect transistor: Influence of polymer tacticity. Thin Solid Films. 2007;515:4041–4044. [Google Scholar]
- 18.Jones AW, Bland-Hawthorn J. Towards a general definition for spectroscopic resolution. Astronomical Data Analysis Software and Systems IV; Astronomical Society of the Pacific Conference Series; Provo, UT: Astronomical Society of the Pacific; 1995. pp. 503–507. [Google Scholar]
- 19.Heng X, et al. Characterization of light collection through a subwavelength aperture from a point source. Opt Express. 2006;14:10410–10425. doi: 10.1364/oe.14.010410. [DOI] [PubMed] [Google Scholar]
- 20.Cui W, et al. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrazzo JM, et al. Community-based urine screening for Chlamydia trachomatis with a ligase chain reaction assay. Ann Intern Med. 1997;127:796–803. doi: 10.7326/0003-4819-127-9-199711010-00004. [DOI] [PubMed] [Google Scholar]
- 22.Carroll KC, et al. Laboratory evaluation of urinary-tract infections in an ambulatory clinic. Am J Clinical Pathol. 1994;101:100–103. doi: 10.1093/ajcp/101.1.100. [DOI] [PubMed] [Google Scholar]
- 23.McCowage GB, et al. Multiparameter-fluorescence activated cell sorting analysis of retroviral vector gene transfer into primitive umbilical cord blood cells. Exp Hematol. 1998;26:288–298. [PubMed] [Google Scholar]
- 24.Buchstaller J, et al. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujiwara T, et al. Therapeutic effect of a retroviral wild-type P53 expression vector in an orthotopic lung-cancer model. J Natl Cancer Inst. 1994;86:1458–1462. doi: 10.1093/jnci/86.19.1458. [DOI] [PubMed] [Google Scholar]
- 26.Fox SB, et al. Relationship of endothelial-cell proliferation to tumor vascularity in human breast-cancer. Cancer Res. 1993;53:4161–4163. [PubMed] [Google Scholar]
- 27.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 28.Sulston JE, Hodgkin JA. The Nematode Caenorhabditis elegans. Cold spring Harbor, NY: Cold Spring Harbor Lab Press; 1988. [DOI] [PubMed] [Google Scholar]
- 29.Hu S, et al. Cross-linked coatings for electrophoretic separations in poly(dimethylsiloxane) microchannels. Electrophoresis. 2003;24:3679–3688. doi: 10.1002/elps.200305592. [DOI] [PubMed] [Google Scholar]