SUMMARY
PICKLE (PKL) codes for a CHD3 chromatin remodeling factor that plays multiple roles in Arabidopsis growth and development. Previous analysis of the expression of genes that exhibit PKL-dependent regulation suggested that PKL acts during germination to repress expression of embryonic traits. In this study, we examined the expression of PKL protein to investigate when and where PKL acts to regulate development. A PKL:eGFP translational fusion is preferentially localized in the nucleus of cells, consistent with the proposed role for PKL as a chromatin remodeling factor. A steroid-inducible version of PKL - a fusion of PKL to the glucocorticoid receptor (PKL:GR) - was used to examine when PKL acts to repress expression of embryonic traits. We found that activation of PKL:GR during germination was sufficient to repress expression of embryonic traits in the primary roots of pkl seedlings whereas activation of PKL:GR after germination had little effect. In contrast, we observed that PKL is required continuously after germination to repress expression of PHERES1, a type I MADS box gene that is normally expressed during early embryogenesis in wild-type plants. Thus PKL acts at multiple points during development to regulate patterns of gene expression in Arabidopsis.
Keywords: CHD3, chromatin remodeling factor, embryo, seedling, germination, developmental transition
INTRODUCTION
Germination of seeds marks a dramatic developmental transition in the life cycle of the plant. Significantly different developmental programs are expressed by the young plant prior to and following germination (Baud et al., 2002; Bewley and Black, 1994; Goldberg et al., 1994; Holdsworth et al., 1999). The latter phase of seed development is referred to as the maturation phase and includes processes such as the accumulation of storage reserves and the acquisition of desiccation tolerance. A young growing seedling, in contrast, undergoes cell expansion and division, mobilizes seed storage reserves, and initiates photosynthesis. Consistent with the distinct developmental characteristics exhibited by maturing seeds and developing seedlings, microarray analysis and proteomics have revealed that vast repertoires of genes are differentially expressed at each of these two stages of development (Gallardo et al., 2001; Gallardo et al., 2002; Girke et al., 2000).
PICKLE (PKL) is necessary to ensure that traits expressed during embryogenesis and seed formation are not expressed after germination (Ogas et al., 1997). pkl seedlings are capable of expressing embryo-associated traits throughout the plant. In particular, the primary roots of pkl seedlings have been demonstrated to express many embryo specific traits after germination, including the accumulation of seed storage reserves (oil, protein, and phytate) and the ability to undergo somatic embryogenesis (Henderson et al., 2004; Ogas et al., 1997; Rider et al., 2004). pkl primary roots that express these traits are referred to as "pickle roots" based on their swollen green appearance.
PKL encodes a SWI/SNF class chromatin remodeling factor that belongs to the CHD3 group (Eshed et al., 1999; Ogas et al., 1999). Consistent with the presence of an ATPase domain that is the hallmark of SWI/SNF proteins, animal CHD3 proteins have been demonstrated to exhibit ATP-dependent chromatin remodeling activity in vitro (Brehm et al., 2000; Guschin et al., 2000; Wang and Zhang, 2001). In addition, CHD3 proteins from animal systems have been shown to associate in multisubunit complexes (Mi-2 or NURD) that contain a histone deacetylase, a result that suggests that CHD3 proteins function as negative regulators of transcription (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998). Characterization of mutants from a variety of model systems in which CHD3 activity has been compromised also indicates that CHD3 proteins generally act as repressors of transcription, often of genes that are developmentally regulated (Ahringer, 2000; Kehle et al., 1998; Unhavaithaya et al., 2002; von Zelewsky et al., 2000).
Previous characterization of pkl plants suggested that PKL acts to repress genes that promote embryonic identity (Ogas et al., 1997; Ogas et al., 1999; Rider et al., 2003). The LEAFY COTYLEDON class of genes (LEC1, LEC2, and FUS3) encode transcription factors that are preferentially expressed in developing seeds and act as positive regulators that play a critical role in promoting seed development (Harada, 2001). In particular, LEC1 and LEC2 have the ability to promote somatic embryogenesis when ectopically expressed during germination (Lotan et al., 1998; Stone et al., 2001). In pkl plants, expression of LEC1 and LEC2 is substantially derepressed during germination (Ogas et al., 1999; Rider et al., 2003). Furthermore, expression of all three LEC genes is elevated more than 100-fold in pickle roots, suggesting that the elevated expression of the LEC genes contributes substantially to the manifestation of this unique developmental state (Rider et al., 2003).
Previous data have suggested that PKLand gibberellin (GA) act in concert during germination to establish repression of genes that promote embryonic identity. Inhibition of GA biosynthesis in pkl seedlings results in increased penetrance of the pickle root phenotype, but only if the inhibition occurs during germination (Ogas et al., 1997). Expression analysis of 10 genes that exhibit PKL-dependent expression revealed that the transcript level of all 10 genes were coordinately upregulated during germination of pkl seeds (Rider et al., 2003). Furthermore, transcript levels of PKL increase during germination (Henderson et al., 2004). These observations provide tantalizing yet indirect evidence that PKL acts specifically during germination to repress expression of embryonic traits.
Prior characterization of CHD3 proteins in animal systems makes a strong biochemical prediction that PKL interacts with the promoter of target genes as a component of a multisubunit complex. Yet these data also reveal that CHD3 proteins are directed to a multitude of targets and can function in multiple complexes. In Arabidopsis, for example, characterization of the gymnos mutant (which is allelic to PKL) revealed that PKL also acts to repress ectopic formation of placental meristems during carpel development (Eshed et al., 1999). Determination of when PKL acts to repress embryonic traits would greatly facilitate biochemical characterization of how PKL acts by enabling us to focus on that specific developmental window to evaluate potential targets and co-factors of PKL. We therefore undertook an analysis of PKL expression in an effort to investigate the timing of PKL action. In particular, we fused PKL to the glucocorticoid receptor to generate a conditional version of PKL (PKL:GR) that is dependent on the presence of dexamethasone (Dex) for activity. We then used this fusion to examine when PKL acts to repress embryonic traits as well as to investigate when PKL acts to repress traits that arise during post-germinative development.
RESULTS
PKL protein preferentially accumulates in differentiating tissue
Polyclonal antibodies to PKL recognize a protein of an apparent molecular weight of about 200 kD in crude extracts from 3-day-old seedlings of wild-type plants (Figure 1a, lane 1). The observed molecular weight of PKL is substantially different from the predicted molecular weight of 158 kD; however, western analysis of PKL protein produced in S. cerevisiae and in E. coli indicates that it migrates with the same apparent molecular weight as observed in extracts from Arabidopsis (data not shown), suggesting that the increased apparent molecular weight of PKL is unlikely to be due to some modification of PKL protein that occurs in plant cells. PKL protein is absent in plants carrying a fast neutron-derived allele of PKL, pkl-7 (Ogas et al., 1999) (Figure 1a, lane 3). PKL is substantially reduced in abundance in extracts of carrying an EMS-derived allele of PKL, pkl-1 (Ogas et al., 1997) (Figure 1a, lane 2). Sequence analysis of the pkl-1 allele revealed a G to A transition that alters the consensus sequence for the 3′ splice site upstream of exon #15 leading to the use of an alternate splice site that results in a 9 nucleotide in frame deletion of the PKL transcript. This deletion corresponds to amino acids 633-635 from the PKL protein, which lie in the conserved ATPase domain necessary for the remodeling activity of SWI/SNF proteins.
Figure 1.
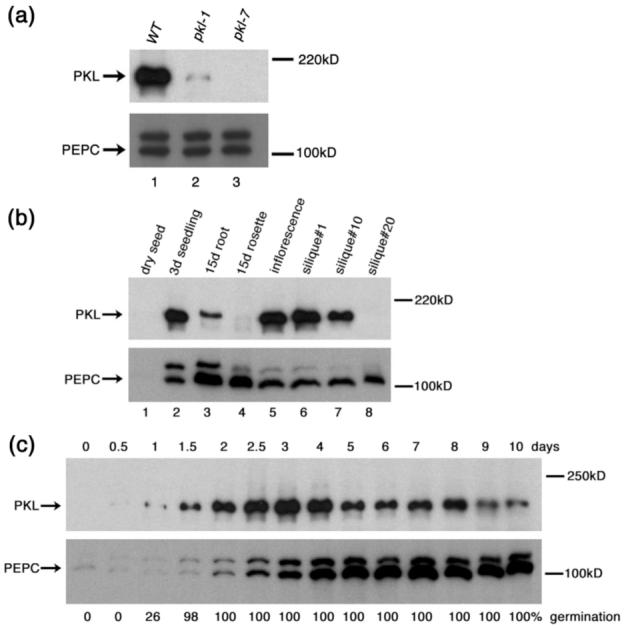
PKL protein preferentially accumulates in differentiating tissue. Western blot analysis of total protein extracts with a polyclonal antibody to the N-terminus of PKL was used to examine the amount of PKL protein levels in a variety of tissues. Levels of PEP carboxylase were similarly determined as a loading control. The size (in kD) of protein molecular weight standards is indicated to the right of each gel.
(a) Levels of PKL protein found in 3-day-old WT (lane 1), pkl-1 (lane 2), and pkl-7 (lane 3) seedlings.
(b) Levels of PKL protein found in dry seed (lane1), 3-day-old seedlings (lane2), 15-day-old roots (lane3), 15-day-old rosettes (lane4), the inflorescence (lane5), silique #1 (lane6), silique #10 (lane7), and silique #20 (lane8). Please see Experimental Procedures for a description of the numbering scheme used for siliques.
(c) Levels of PKL protein found in seedlings that had been imbibed for up to 10 days. The percentage of seedlings that had germinated at each time point is indicated below each lane. 25μg of protein were added per lane in all panels. During this analysis, we observed that PEP carboxylase was only effective as a loading control at 4 days after imbibition and thereafter. Prior to this time, expression of PEP carboxylase is significantly upregulated relative to a desiccated seed as the young seedling develops.
We next used the α-PKL antibodies to examine developmental regulation of PKL protein (Figure 1b). The greatest levels of PKL protein were observed in tissue undergoing significant differentiation such as young seedlings, inflorescent tissue, and young siliques. This pattern of expression is consistent with previous transcript analysis (Eshed et al., 1999) and with the hypothesis that one of the primary roles of PKL is to act as a determination factor during differentiation. Decreasing amounts of PKL were observed as organs aged. Markedly less PKL protein is found in older siliques than in younger siliques (lane 8 versus lanes 6 and 7) and in 15-day-old rosette tissue versus 3-day-old seedlings (lane 4 versus lane 2). Previous data had suggested that PKL acted during germination to repress expression of genes that promote embryonic identity (Rider et al., 2003), so we used western analysis to examine the level of PKL protein in greater detail after seed imbibition (Figure 1c). As in our previous analyses (Ogas et al., 1997; Rider et al., 2003), we consider germination to begin with seed imbibition (defined as when the seeds are sterilized) and to end with emergence of the radicle from the seed coat. We observed that PKL protein was expressed during germination (0.5d versus 1.5d), consistent with the hypothesis that PKL acts during germination to repress transcription of target genes. The peak of PKL protein accumulation, however, occurred after seedlings had completed germination (2.5d - 4d after seed imbibition). This observation indicated that PKL is also likely to play a significant role in gene expression in young seedlings after germination.
PKL protein accumulates in the nucleus
PKL codes for a CHD3 chromatin remodeling factor that is predicted to function in the nucleus. To examine the localization of the PKL protein, we fused eGFP to the 3′ end of the PKL ORF and expressed the resulting PKL:eGFP fusion under the control of the endogenous PKL promoter and terminator. This fusion construct is capable of rescuing all of the associated mutant phenotypes when transformed into a pkl-1 plant (Figure 2a-b). Thus localization of the PKL:eGFP translational fusion is likely to reflect that of the endogenous PKL protein. We observed that GFP fluorescence was nuclear localized in transgenic plants carrying the PKL:eGFP translational fusion (Figure 2c, e). Identification of the nuclear compartment was confirmed by staining the roots with DAPI (Figure 2f). In contrast, a PKL:eGFP transcriptional fusion in which expression was directed by the same genomic sequences is detected in both the cytoplasm and nucleus of cells (Figure 2g), indicating that the nuclear localization exhibited by the translational fusion is conferred by the PKL amino acid sequence. The transcriptional and translational fusions otherwise exhibit a similar pattern of expression, with the exception that transgenic lines carrying the transcriptional fusion exhibit much greater fluorescence than lines carrying the translation fusion (data not shown). The presence of the PKL:eGFP translational fusion in trichomes (Figure 2c) is consistent with the observation that the trichomes of pkl plants exhibit a reduced branching phenotype. This observation also demonstrates that PKL is expressed even in highly differentiated cells. Although eGFP reporter constructs are ubiquitously expressed (Figure 2c, d, h), the greatest expression was observed in tissue undergoing differentiation (Figure 2h-i). These data are consistent with the western analysis (Figure 1b).
Figure 2.
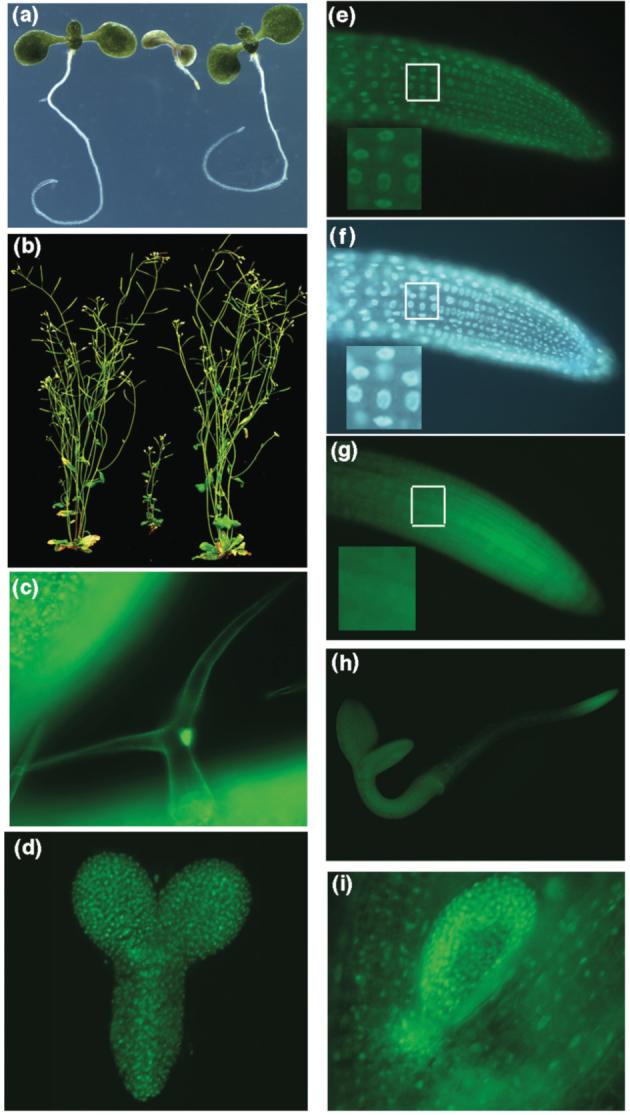
The PKL:eGFP translational fusion rescues pkl-1 and is nuclear localized. Complementation of pkl-1 seedlings (a) and mature pkl-1 plant (b). For both panels, the plant on the left is wild-type, the plant in the center is pkl-1, and the plant on the right is pkl-1 transformed with the PKL:eGFP translational fusion. The plants in panel (a) were grown in the presence of 10-8 M uniconazole-P in continuous light for 14 days, whereas the plants in panel (b) were grown in the presence of 10-8 M uniconazole-P in continuous light for 23 days then moved to soil and grown under continuous light for an additional 21 days.
(c-i) Transgenic plants expressing PKL:eGFPM transcriptional (g,h) or translational (c-f,i) fusions were observed using fluorescence microscopy. (c) GFP fluorescence in a trichome from the first leaf of 9-day-old plant carrying a translational fusion. (d) GFP fluorescence in torpedo stage embryo in a plant expressing a translational fusion. (e) GFP fluorescence in the root tip of a 3-day-old plant expressing a translational fusion. (f) DAPI fluorescence in the same root tip depicted in panel (e). (g) GFP fluorescence in the root tip of a 3-day-old plant expressing a transcriptional fusion. The white box in panels e-g highlights the area magnified 2.5X in the lower left corner of each panel. (h) GFP fluorescence of an entire 3-day-old plant expressing a transcriptional fusion. (i) GFP fluorescence in the first leaf of a 5-day-old plant expressing a translational fusion.
Recent work has highlighted the contribution of the endosperm of the mature seed to storage reserve mobilization and post-germinative growth of dark-grown seedlings (Penfield et al., 2004). The implication of this work is that gene expression occurs in two distinct developmental compartments during germination: the embryo and the endosperm. To address the possibility that PKL functions in the endosperm as well as the seedling during germination, we examined expression of the PKL:eGFP translational fusion in germinating seeds. We were unable to detect GFP fluorescence in the endosperm/seed coat of germinating seeds despite robust detection of GFP fluorescence in the seedling (data not shown). These data suggest that PKL is unlikely to play a role in regulation of gene expression in the endosperm during germination and that PKL-dependent expression that occurs during this time is most likely due to the action of PKL in the developing embryo.
Activation of PKL during germination is sufficient to suppress expression of embryonic traits
The discovery that PKL protein accumulation is constitutively nuclear localized suggested that we might be able to use the glucocorticoid receptor (Lloyd et al., 1994; Picard et al., 1988) to generate an inducible version of PKL protein. We fused the ligand-binding domain of the glucocorticoid receptor to full length PKL ORF to generate a PKL:GR translational fusion. This construct was then placed under the control of the endogenous PKL promoter and terminator as described above for the PKL:eGFP fusions.
We isolated transgenic pkl-1 lines carrying the PKL:GR fusion, selected lines in which segregation of the resistance marker indicated that a single insertion event had taken place, and scored homozygous T3 progeny of these lines for penetrance of the pickle root phenotype in the presence and absence of dexamethasone (Dex). We observed that penetrance of the pickle root phenotype varied in individual T3 lines, which is consistent with prior characterization of this trait (Ogas et al., 1997 and unpublished observations). Penetrance of the pickle root phenotype depends on the genotype and ecotype of the plants, how the parent plants are grown, the age of the seeds, and the conditions under which the seeds are plated. As a result, penetrance of the pickle root phenotype does vary from experiment to experiment as it is technically impossible to carry out all of the experiments with the same batch of seed at the same time. In any given experiment, all seeds are obtained from plants grown in the same chamber at the same time so as to provide a suitable control, analogous to what must be done for studies of seed germination. In the case of characterization of penetrance of the pickle root phenotype in T3 lines, the position of the T-DNA integration event as well as any extraneous mutations introduced by transformation are also likely to have contributed to the observed variation in pickle root penetrance.
We examined 6 T3 lines in all, of which 5 exhibited at least a 30% Dex-dependent decrease in pickle root penetrance (Figure 3a). The ability of Dex to rescue the pickle root phenotype of transgenic pkl-1 plants carrying the PKL:GR fusion was consistent with the hypothesis that PKL functions in the nucleus. This result furthermore demonstrated that activation of PKL:GR after imbibition is sufficient to rescue the pickle root phenotype, an observation that is consistent with the hypothesis that PKL acts during germination to repress genes that promote embryonic identity.
Figure 3.
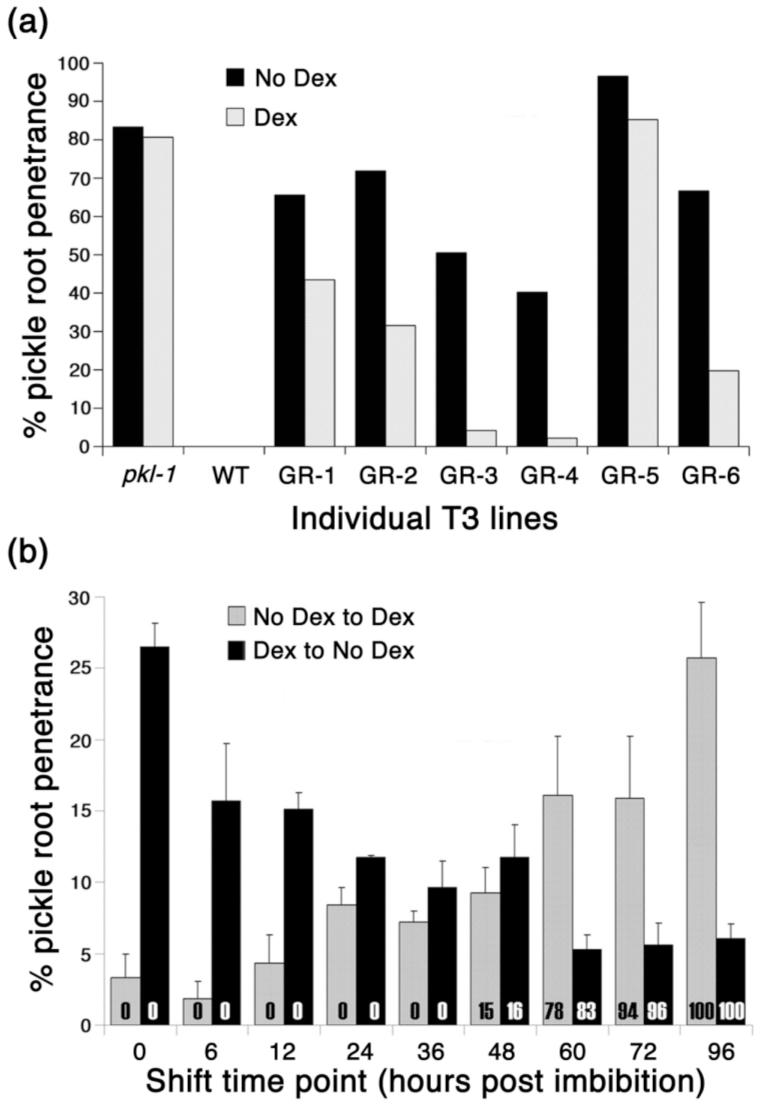
Action of PKL during germination is sufficient to suppress embryonic traits.
(a) 6 independent T3 pkl-1 lines that were homozygous for PKL:GR were scored for pickle root penetrance when grown in the presence or absence of Dex.
(b) Seeds from the GR-3 pkl-1/PKL:GR T3 line were imbibed in water and immediately plated on media not containing Dex and then shifted at the indicated times to media containing Dex (gray columns), or on media containing Dex and then shifted at different times to media without Dex (black columns). The x axis indicates the times at which seeds were shifted. Error bars represent the standard error of the mean. The numbers within the columns represent the percent of germinated seeds at the time the seeds were shifted.
For both panels, pickle root penetrance was scored fifteen days after imbibition. All media used contained 10-7 M uniconazole-P to increase penetrance of the pickle root phenotype.
To determine if there was a particular phase of development during which PKL:GR was capable of rescuing the pickle root phenotype, PKL:GR transformants of pkl-1 plants (henceforth referred to as pkl-1/PKL:GR plants) were transferred to or from Dex-containing media at different times during the first 96 hours after seed imbibition. Specifically, seed from the GR-3 T3 line (Figure 3a) was used in this and all subsequent analyses involving pkl-1/PKL:GR plants. Previous work has demonstrated that transgenic plants expressing a GR fusion can respond to application of Dex prior to germination (Sanchez and Chua, 2001), indicating that the initial presence of an intact seed coat would not prevent Dex from reaching the seedling. In fact, we observed that as time after imbibition increased, the pkl-1/PKL:GR seedlings became less responsive to induction/repression of PKL:GR (Figure 3b). We found that after 60 hours in either the absence or the presence of Dex, the fate of the primary root of pkl-1/PKL:GR seedlings was largely determined even though 100% germination was not obtained until 96 hours. Thus PKL:GR can only suppress expression of the pickle root phenotype if induced prior to the completion of germination. Conversely, transient activation of PKL:GR during germination is sufficient to establish repression of embryonic traits; continuous exposure to Dex is not required for repression of the pickle root fate in pkl-1/PKL:GR seedlings.
Although these data indicate that PKL can act during germination to repress expression of embryonic traits, they do not exclude the possibility that PKL can also act during seed formation to enable subsequent repression of embryonic traits during germination. To test this hypothesis, we examined pickle root penetrance in the progeny of pkl-1/PKL:GR plants that had been grown in the absence or presence of Dex (Figure 4). A GR fusion expressed in the seed has previously been demonstrated to exhibit Dex-dependent activity (Baudry et al., 2004), and PKL is abundantly expressed in young embryos (Figure 2d), suggesting that the PKL:GR fusion was likely to be Dex-responsive during this period of development. Consistent with this supposition, we found that siliques of pkl-1/PKL:GR plants grown in the presence of Dex were restored to nearly wild-type length, indicating that application of Dex can rescue pkl-associated traits in developing pkl-1/PKL:GR siliques (Figure 4a). Penetrance of the pickle root trait in the subsequent generation, however, was insensitive to application of Dex to the parent (Figure 4b). Application of Dex during pkl-1/PKL:GR seed formation did not decrease pickle root penetrance in the resulting progeny (black bars denote absence of Dex during seed formation whereas white bars denote presence of Dex during seed formation). In fact, application of Dex during seed formation resulted in an increase in pickle root penetrance in a manner that was not dependent on the presence of the GR gene as it occurred in both pkl-1 and pkl-1/PKL:GR lines. This effect, albeit surprising at first glance, is consistent with prior characterization of the pickle root penetrance as being responsive to the conditions under which the parent plant is grown. In contrast, application of Dex during germination of pkl-1/PKL:GR seeds suppressed pickle root penetrance regardless of the treatment during seed formation but had no measurable effect on pickle root penetrance of pkl-1 seed. These data reveal that action of PKL during seed formation is not sufficient to repress expression of embryonic traits in the subsequent generation.
Figure 4.
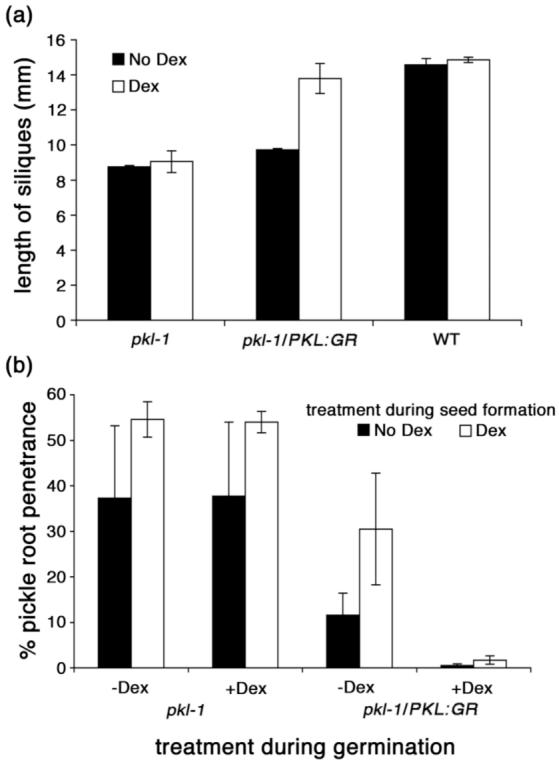
Action of PKL during seed formation does not suppress embryonic traits.
(a) The length of siliques was determined of pkl-1, pkl-1/PKL:GR T3, and WT plants that were grown in the absence or presence of Dex. The values plotted are the mean ± SD of three biological replicates, in which 10 siliques were measured for each replicate.
(b) Seeds from pkl-1 and pkl-1/PKL:GR T3 plants that were grown in the absence or presence of Dex were scored for pickle root penetrance when imbibed in the presence or absence of Dex. All seeds were imbibed on media that contained 10-8 M uniconazole-P to increase penetrance of the pickle root phenotype. Expression of the pickle root trait was scored fifteen days after imbibition. The values plotted are the mean ± SD of three biological replicates, in which 144 seedlings were scored for each replicate.
Taken together, our data strongly suggest that PKL acts specifically during germination to repress the potential of the young seedling to express embryonic traits and thus enable the developmental switch to post-germinative growth.
PKL can also act after germination to regulate development
In addition to the pickle root trait, pkl plants exhibit pleiotropic defects in shoot development including dark green leaves, reduced stature, and delayed flowering (Henderson et al., 2004; Ogas et al., 1997). We examined the shoot phenotype of pkl-1/PKL:GR plants that were grown in the presence or absence of Dex. We found that Dex-treated pkl-1/PKL:GR plants exhibited partial to complete suppression of various pkl-associated traits assayed, including rescue of the rosette size, height, and flowering time (Figure 5a, b, data not shown). These data suggest that the PKL:GR fusion protein can also act in the shoot to regulate gene expression.
Figure 5.
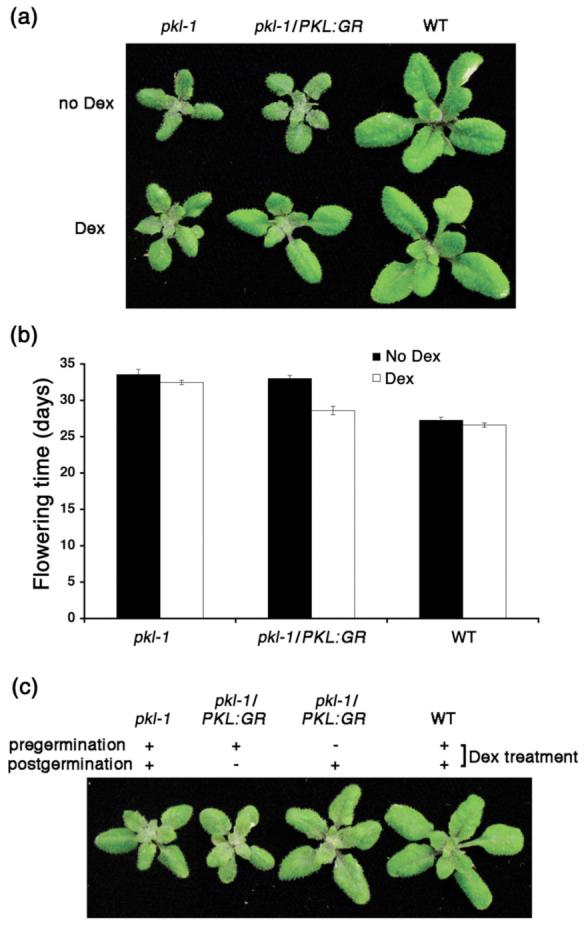
PKL:GR can act after germination to rescue the shoot phenotype of pkl plants.
(a) pkl, pkl-1/PKL:GR, and wild-type plants were grown for 19 days in the absence or presence of Dex.
(b) Flowering time was determined for pkl, pkl-1/PKL:GR, and wild-type plants grown in the presence or absence of Dex. The values plotted are the mean +/- SE of 15 plants measured.
(c) Comparison of shoots of 19-day-old pkl-1/PKL:GR plants that were imbibed in the presence of Dex until completion of germination (2 days) and then not treated with Dex subsequently (-/+) or that were imbibed in the absence of Dex until completion of germination (2 days) and then sprayed with Dex subsequently (+/-). 19-day-old pkl-1 and wild-type plants that were treated continuously with Dex (+/+) are shown for comparison.
Our analysis of PKL protein expression (Figures 1 and 2) in combination with previous analyses of PKL transcript accumulation (Eshed et al., 1999; Ogas et al., 1999) indicated that PKL was likely to act at other points in the plant life cycle in addition to germination to regulate gene expression. Our characterization of the time of PKL action (Figure 3b), however, was consistent with an alternative hypothesis that the pkl shoot phenotypes that arise after germination are primarily due to the establishment of improper patterns of gene regulation during germination of pkl seedlings. In support of such a hypothesis, it has previously been observed that pkl leaves accumulate elevated levels of the FUS3 transcript (Rider et al., 2003) and that transgenic plants exhibiting ectopic expression of FUS3 in the L1 layer of the shoot exhibit GA-deficient phenotypes (Gazzarrini et al., 2004) reminiscent of those exhibited by pkl plants. To examine the contribution of PKL action during germination to post-germinative development, we examined the shoot phenotype of pkl-1/PKL:GR plants that were only treated with Dex after seed germination. We found that these Dex-treated plants exhibited a substantially wild-type shoot phenotype whereas plants that were treated with Dex only during germination but not afterwards did not rescue the pickle shoot phenotype as effectively (Figure 5c). Thus these data suggest that the shoot phenotype of pkl plants is primarily due to the absence of PKL activity after germination.
PKL acts continuously after germination to repress PHE1 expression in the shoot
In order to examine when PKL acted after germination to regulate development, we needed a distinct marker for PKL action in the shoot similar to that of the pickle root phenotype. Ongoing analysis of candidate genes for PKL-dependent expression in young seedlings (selected on the basis of their predicted/demonstrated role in embryo development) uncovered 2 genes that exhibited robust PKL-dependent expression after germination (Table 1, data not shown). PHERES1 (PHE1) and PHERES2 (PHE2) code for type I MADS box proteins that are transiently expressed in young embryos (Kohler et al., 2003). We extracted RNA from plants that were staged at 50% germination (i.e. 50% of the seedlings had completed germination based on emergence of the radicle from the seed coat), 4-day-old seedlings, and from 21-day-old rosette tissue from wild-type and pkl plants and analyzed relative transcript levels by qRT-PCR (Table 1). We observed that PHE1 and PHE2 exhibit strong PKL-dependent expression, but only after germination. This observation is in striking contrast to previously characterized PKL-dependent genes such as LEC1 and LEC2 that exhibit strong PKL-dependent expression during germination but do not exhibit PKL-dependent expression in the mature shoot (Rider et al., 2003).
Table 1.
Relative transcript levels for PHERES1 and PHERES2 in pkl mutants. Transcript levels were determined for pkl-1 and wild-type plants at 50% radicle emergence (Germination), in young seedings (4 day) and in 21-day-old leaves (21 day). 18s rRNA was used as a standardization control. levels are normalized relative to wild-type plants.
| Fold Change (pkl/wt) | ||||
|---|---|---|---|---|
| AGI Code | Gene | Germination | 4 day | 21 day |
| At1g65330 | PHERES1 | 0.9 | 161 | 151 |
| At1g65300 | PHERES2 | 3 | 40 | 27 |
The discovery that two type I MADS box genes exhibited PKL-dependent expression after germination prompted us to examine whether other type I MADS box genes exhibited similar regulation. The expression of 45 type I MADS box genes (Alvarez-Buylla et al., 2000)(De Bodt et al., 2003) was examined by qRT-PCR at the same developmental stages assayed for PHE1 and PHE2. We identified 4 other predicted type I MADS box genes for which the level of the corresponding transcript was altered at least 5-fold in the absence of PKL. Unlike PHE1 and PHE2, however, none of these genes exhibited PKL-dependent expression at more than one of the developmental stages that were assayed. At5gt26630 and At3g66656 transcripts were elevated 5- and 7-fold respectively in germinating seedlings whereas At2g28700 transcripts were up 17-fold in 4-day-old seedlings. In contrast, At1g18750 was 12-fold decreased at 50% germination, suggesting that PKL is a positive regulator of this locus. Our analysis failed to reveal a large effect of the pkl mutation on expression of the other type I MADS box genes (Supplementary Data, Table S1), indicating that PKL does not play a general regulatory role for this class of transcription factor.
The identification of PHE1 and PHE2 as genes that exhibit robust PKL-dependent expression in the shoot provided us with the opportunity to determine if PKL could also act after germination to regulate gene expression. We examined PHE1 transcript levels in pkl-1/PKL:GR plants grown continuously in the presence or absence of Dex. These experiments demonstrated that application of Dex to pkl-1/PKL:GR plants results in 3-fold repression of PHE1 transcript levels relative to growth in the absence of Dex (Figure 6a). Thus the PKL:GR fusion is capable of partial rescue of PHE1 expression in pkl plants, an observation that is consistent with the observation that application of Dex to pkl-1/PKL:GR plants can completely rescue some pkl-associated traits (Figures 4a and 5b) but only partially rescue others (Figure 5a). Similarly, application of Dex to pkl-1/PKL:GR plants results in less than 2-fold repression of PHE2 transcript levels (data not shown).
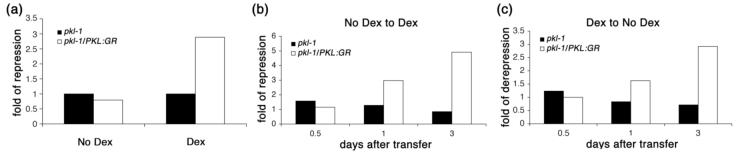
Figure 6.
PKL can act after germination to determine expression of PHE1. Quantitative RTPCR was used to determine the relative transcript levels of PHE1 in plants in response to a variety of Dex treatments. 18S rRNA was used as a standardization control.
(a) Relative PHE1 transcript levels were determined in 21-day-old pkl-1/PKL:GR plants and pkl plants that were grown in the presence and absence of 10-5 M Dex. Expression levels are normalized relative to pkl plants.
(b) Relative PHE1 transcript levels were determined in pkl-1 and pkl-1/PKL:GR seedlings that were grown on media that did not contain Dex for 10 days and then transferred to media containing 10-5 M Dex and allowed to grow for the number of days indicated on the x axis. Expression levels (fold repression) are normalized relative to plants of the same genotype that underwent a mock treatment regime (transfer to no Dex media).
(c) Relative PHE1 transcript levels were determined in pkl-1 and pkl-1/PKL:GR seedlings that were grown on media that contained 10-5 M Dex for 10 days and then transferred to media that did not contain Dex and allowed to grow for the number of days indicated on the x axis. Expression levels (fold derepression) are normalized relative to plants of the same genotype that underwent a mock treatment regime (transfer to media that contained 10-5 M Dex).
We were able to use the more robust Dex-dependent PHE1 expression to examine when PKL is capable of acting to regulate gene expression in the shoot. pkl-1/PKL:GR seedlings were grown in the absence or presence of Dex for 10 days and then transferred to or from Dex-containing media. Transcript levels of PHE1 were followed by qRT-PCR at 0.5, 1, and 3 days after transferring the seedlings as well as in plants that were subjected to a mock transfer (Dex to Dex or no Dex to no Dex). We found that after 10 days of growth in the absence of Dex, treatment of pkl-1/PKL:GR seedlings with Dex resulted in repression of PHE1 transcript levels within 1 day (Figure 6b). Conversely, we found that transfer of 10-day-old pkl-1/PKL:GR seedlings from media containing Dex to Dex-free media resulted in elevation of transcript levels within 1 day (Figure 6c). Thus in contrast to what is observed during germination, PKL appears to be continuously needed to repress expression of at least one gene in the shoot after germination. In both cases, 3 days of the new treatment was largely sufficient to switch PHE1 transcript levels to reflect the new regime. Similar results were observed if pkl-1/PKL:GR plants are transferred after 21 days of growth (data not shown). Thus these data reveal that PKL continuously acts after germination to regulate expression of some genes in the shoot.
PKL acts during germination to repress expression of LEC1
Our success at examining the effect of activation of PKL:GR on PHE1 expression in the shoot prompted us to examine the effect of activation of PKL:GR on gene expression during germination. Although the previous analysis indicated that PKL acted during germination to repress expression of embryonic traits in the primary root (Figure 3b), it did not address when PKL was capable of acting to repress gene expression. We have previously observed that transcript levels of LEC1 are elevated during germination of pkl seeds (Ogas et al., 1999; Rider et al., 2003). To test the hypothesis that PKL acted early during germination to repress expression of LEC1, we used qRT-PCR to examine LEC1 transcript levels in germinating pkl and pkl-1/PKL:GR seedlings that were treated with Dex either during early germination or late germination. We also examined PHE1 transcript levels as a control for developmental specificity.
We found that imbibition of pkl-1/PKL:GR seedlings in the presence of Dex until they reached 50% germination resulted in a >5-fold decrease in transcript levels of LEC1 relative to pkl-1/PKL:GR seedlings imbibed in the absence of Dex (Figure 7a). In contrast, if pkl-1/PKL:GR seeds were imbibed in the absence of Dex until they had reached 75% germination and then treated with Dex for 48 hours, we observed a < 2-fold decrease in transcript levels of LEC1 relative to pkl-1/PKL:GR seedlings that were not exposed to Dex (Figure 7b). Thus these data reveal that the ability of PKL:GR to repress expression of LEC1 (Figure 7) and to repress expression of the pickle root phenotype (Figure 3b) is largely restricted to early germination, consistent with the hypothesis that expression of the pickle root trait is a reflection of the ability of PKL to act on LEC1 and related genes during early germination.
Figure 7.
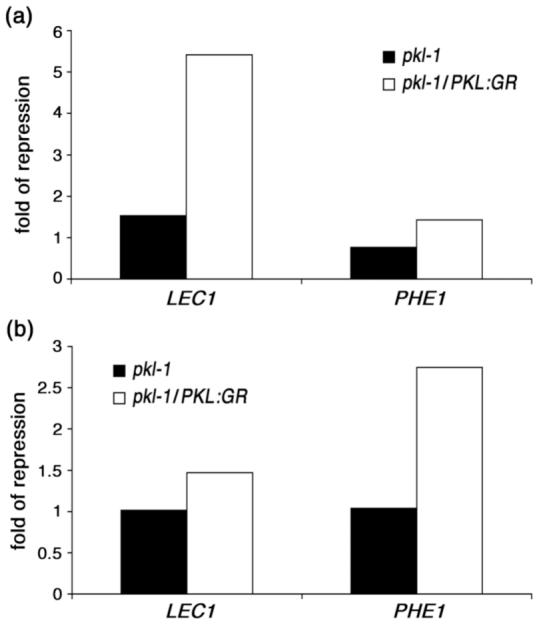
PKL acts early during germination to determine expression of LEC1. Quantitative RT-PCR was used to determine the relative transcript levels of LEC1 and PHE1 in pkl and pkl-1/PKL:GR seedlings in response to a variety of Dex treatments. 18S rRNA was used as a standardization control.
(a) Relative LEC1 and PHE1 transcript levels were determined in pkl and pkl-1/PKL:GR seedlings that had been imbibed in the absence or presence of 10-5 M Dex until they reached 50% germination. Expression levels in Dex-treated plants are normalized relative to plants grown in the absence of Dex.
(b) Relative LEC1 and PHE1 transcript levels were determined in pkl and pkl-1/PKL:GR seedlings that had been imbibed in the absence of Dex until they reached 75% germination and then exposed to 10-5 M Dex for 48 hours or grown for 48 hours in the continued absence of Dex. Expression levels in Dex-treated plants are normalized relative to plants grown in the absence of Dex.
PHE1 transcript levels exhibited a pattern of expression that was reciprocal to that of LEC1. PHE1 transcript levels were relatively unchanged when pkl-1/PKL:GR seedlings were imbibed in Dex until 50% germination (Figure 7a) whereas they were repressed nearly 3-fold when pkl-1/PKL:GR seedlings were treated with Dex after they had reached 75% germination (Figure 7b). These data are consistent with our observation that PKL is necessary after germination for repression of PHE1 expression and further demonstrate that PKL:GR is capable of acting to regulate gene expression in response to Dex after 75% germination.
As expected, LEC1 and PHE1 transcript levels were not responsive to either regimen of Dex application in pkl seedlings.
DISCUSSION
Activation of PKL during germination is sufficient to repress expression of embryonic traits
We have shown that PKL is a nuclear localized protein that is preferentially expressed in differentiating tissue and specifically acts during germination to repress genes that promote embryonic identity, in strong agreement with our previous observation that genes that exhibit PKL-dependent expression exhibit elevated transcript levels during germination of pkl seedlings (Ogas et al., 1999; Rider et al., 2003). We have previously demonstrated that GA also acts during germination to repress expression of the pickle root trait (Ogas et al., 1997). Thus our data reveal that PKL and GA act during the same period of development to repress expression of embryonic traits. The implication of our data is that a GA-dependent PKL-independent repression pathway exists that operates concurrently with the PKL-dependent pathway and that can in some way ameliorate the effect of loss of PKL on repression of embryonic traits. Given that PKL is a chromatin remodeling factor, it seems reasonable to anticipate that the mechanism by which GA suppresses the consequences of loss of PKL is likely to be chromatin-based. Determination of whether or not this GA-dependent PKL-dependent repression pathway plays a similar role in the presence of PKL has yet to be determined. Suppression of GA biosynthesis in the presence of a wild-type copy of PKL is not sufficient to generate a pickle root phenotype or to elevate expression of any of the LEC genes during germination (JO and JH unpublished observations). Nonetheless, these observations do not preclude the possibility that reduced levels of GA lead to an alteration of chromatin that is not sufficient to alter expression of affected genes except in the absence of PKL.
Our data are consistent with a model in which the seed is developmentally reprogrammed during germination in a PKL-and GA-dependent manner to repress expression of embryonic traits and thereby ensure the switch to the developmental status of a seedling. Thus the ability to express embryonic potential is not silenced during seed maturation but rather during seed germination. This type of regulatory control would provide an explanation of why treatments that promote somatic embryogenesis during germination are not effective during subsequent shoot development. For example, ectopic expression of LEC1 during germination is sufficient to promote somatic embryogenesis (Lotan et al., 1998; Zuo et al., 2002), whereas it is not sufficient after germination (Zuo et al., 2002). By the time germination is complete, PKL and GA have acted to render the differentiation state of an embryo less accessible, most likely through alteration of the chromatin structure of genes that promote embryonic identity.
PKL acts continuously during shoot development to repress expression of PHE1
Our analysis of pkl-1/PKL:GR plants clearly demonstrates that PKL also acts after germination. Although activation of PKL:GR during germination is sufficient to suppress pickle root penetrance (Figure 3b), it is not sufficient to robustly rescue pkl-associated shoot traits (Figure 5c). In contrast, we observe that activation of PKL:GR after germination is sufficient to substantially rescue the shoot phenotype of pkl plants, suggesting that the shoot phenotype of pkl plants is largely due to the loss of PKL-dependent regulation after germination. An interesting implication of this observation is that subsequent shoot development appears rather insensitive to a substantial prior perturbation of the expression of developmentally regulated genes. Based on our expression analysis and phenotypic characterization of pkl-1/PKL:GR plants, it is likely that many of the genes that exhibit elevated transcript levels in pkl plants exhibit similarly elevated transcript levels in pkl-1/PKL:GR plants imbibed in the absence of Dex. Yet despite this extensive alteration in the pattern of gene expression during germination, pkl-1/PKL:GR plants are substantially restored to wild-type appearance when treated with Dex after germination.
Our analysis of PKL-dependent regulation of PHE1 and PHE2 clearly illustrates that PKL can act after germination to regulate gene expression. PHE1 and PHE2 code for closely related type I MADS box proteins that are transiently expressed during early embryogenesis (Kohler et al., 2003). Previous analysis of embryo-related genes for PKL-dependent expression, such as LEC1 and LEC2, identified genes that exhibited PKL-dependent expression during germination (Rider et al., 2003). PHE1 and PHE2, in contrast, do not exhibit PKL-dependent expression during germination but instead exhibit strong PKL-dependent expression after germination (Table 1). Furthermore, we observed that PKL acts continuously during shoot development to regulate expression of PHE1. Transcript levels of PHE1 in 10-day-old pkl-1/PKL:GR plants responded within one day of addition or removal of Dex (Figure 6).
One interesting implication of our data is the existence of multiple regulatory pathways for expression of embryonic identity. Both GA and PKL act during germination to repress expression of the pickle root trait, whereas PHE1 and PHE2 transcript levels are elevated after germination. PHE1 and PHE2 were initially identified in an effort to identify targets of MEDEA (MEA), a PcG protein that plays an essential role in developing embryos (Kohler et al., 2003). The characterization of PHE1 and PHE2 expression in wild-type and mea embryos suggested that transient elevated expression of PHE1 and PHE2 was likely to play a significant role in early embryogenesis (Kohler et al., 2003). The observation that expression of PHE1 and PHE2 is not elevated during germination of pkl seedlings reveals that altered expression of these genes does not contribute to the establishment of embryonic identity in this context.
Our data also suggest that the response of the plant to altered PHE1 expression is likely to be dependent on the stage of development. Suppression of elevated PHE1 levels in mea seeds is sufficient to rescue mea-associated seed abortion, suggesting that elevated PHE1 expression is lethal in this context (Kohler et al., 2003). In contrast, we observe that elevated PHE1 levels (Table 1) are not sufficient to lead to termination of development in the pkl shoot. Furthermore, we also find that Dex can substantially rescue the shoot phenotype of pkl-1/PKL:GR plants despite the fact that transcript levels of PHE1 are elevated approximately 50-fold in Dex-treated pkl-1/PKL:GR plants relative to wild-type plants (Table1, Figure 6b, and data not shown). This observation reveals that elevated PHE1 levels are not sufficient to generate the pkl shoot phenotype. Determination of whether or not elevated PHE1 expression is necessary for development of the pkl shoot phenotype will require reducing PHE1 transcript levels in pkl plants by mutational analysis or RNAi.
Although these data do not reveal if PKL directly regulates expression of LEC1 and PHE1, it is intriguing to note that our data are consistent with the possibility that PKL acts in a similar fashion to regulate expression of both genes. The level of PHE1 transcript was determined in whole plants. Possible expression patterns range from PKL-dependent expression occurring in every cell throughout development to PKL-dependent expression occurring in a specific subset of cells during a particular stage of leaf development. The latter type of regulation would be very similar to the manner in which PKL acts during germination to repress expression of embryonic traits in the developing root. This model would also explain why PKL is only required after germination to repress expression of PHE1; PKL acts after germination during organogenesis to shut down transient expression of PHE1. More detailed characterization of developmental regulation of PHE1 expression in pkl-1/PKL:GR plants with the assistance of reporter genes and/or in situ hybridization will be necessary to distinguish between these possible patterns of expression.
CHD3 chromatin remodeling factors are known to play a role in promoting various developmental transitions in eukaryotes (Ahringer, 2000). In the absence of a conditional version of a CHD3 protein, however, it has not been possible to address whether or not CHD3 action during a specific developmental window is sufficient to prevent inappropriate expression of prior differentiation traits during subsequent developmental stages. Our studies demonstrate that the CHD3 protein PKL can act during a specific developmental window to determine fate in plants: PKL is required during germination to repress embryonic identity. Determination of the timing of PKL action greatly facilitates elaboration of other aspects of the mechanism of PKL action. It is now known when to undertake biochemical characterization of the PKL protein (e.g. identification of interacting proteins and direct targets) to understand how PKL promotes the developmental transition that occurs during germination.
The identity of differentiated plant tissue is commonly perceived as being significantly more malleable than that of differentiated animal tissue. It is less often recognized that limitations to this developmental plasticity occur quite frequently. Many agronomically important plants are resistant to in vitro manipulation (e.g. many species of forest trees) (Merkle and Dean, 2000). Although it is relatively easy in Arabidopsis to transform roots into shoots and vice versa by manipulating hormone concentrations, reacquisition of embryonic identity after germination is much more difficult to achieve (Zuo et al., 2002). PKL plays a critical role in repression of embryo-associated differentiation traits. It seems reasonable to anticipate that PKL may play a general role in restricting access to alternate differentiation states in plants. Characterization of the role of CHD3 proteins in development and differentiation in animal systems is consistent with such a hypothesis (Ahringer, 2000; Fujita et al., 2004; Unhavaithaya et al., 2002). Further characterization of the mechanism by which PKL regulates gene expression during germination may thus provide general insights into how plants - like animals - restrict developmental potential.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Wild type, pkl-1 (Ogas et al., 1997), and pkl-7 (Ogas et al., 1999) are in the Col background. Plants were grown on synthetic media or in pots as previously described, with exceptions noted below (Ogas et al., 1997). Plants were transformed by using an in planta transformation protocol with the Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998).
For the analysis of PKL protein expression during development (Figure 1b), the seedling tissue and the 15-day-old shoot and root tissue was collected from plants grown on synthetic media. The inflorescences and siliques were collected from plants that were grown for 3 weeks on plates and then transferred to pots and grown for another 5 weeks. Siliques were numbered sequentially where the first open flower corresponded to silique number one, the second open flower to silique number two, and etc. Siliques #10 and #20 contained embryos at the globular stage and the mature embryo stage respectively (Bowman, 1993).
For the analysis of Dex-dependent shoot traits, plants were imbibed 2 days with an aqueous solution containing 10-5M Dex or with a mock solution containing 0.2% methanol. Seedlings were rinsed with water for 5 times, sown on soil, grown in a Percival growth chamber with continuous illumination (150-180 μE m-2 s-1) at 22°C. Plants were sprayed every other day with an aqueous solution containing 10-5 M Dex and 0.05% (w/v) Tween 20 or with a mock solution containing 0.02% methanol and 0.05% (w/v) Tween 20. The rosette size of pkl-1, PKL:GR, and WT were compared after 3 weeks. The length of siliques was measured when seed maturation was complete. Eight to ten siliques were measured for each treatment. Plants for Dex-dependent flowering time analysis were grown in a Percival growth chamber with 16 hours of illumination. Flowering was scored as the number of days until the first flower opened. For analysis of the effect of application of Dex during seed formation on subsequent penetrance of the pickle root phenotype, seeds were plated on MS media supplemented with 10-8M uniconazole or 10-8M uniconazole and 10-5M Dex, and the pickle root trait was determined for 144 seedlings 10 days after imbibition.
For analysis of PHE1 expression in pkl-1/PKL:GR transgenic lines (Figure 6), seeds were plated on MS media for 10 days, then transferred to MS media supplemented with 10-5M Dex, and harvested 0.5, 1, or 3 days after transfer.
Plasmid construction
A complete description of the construction of all recombinant DNA molecules generated for this study can be found in Supplementary Material.
Antibody preparation
Polyclonal anti-PKL antiserum was obtained from rabbits immunized with the N-terminal (1-291aa) or C-terminal (740-1385aa) region of PKL. E. coli Tuner DE3 cells (Novagen, Madison, WI) were used for the production of recombinant antigen. Growth of the expression host strain and induction of protein expression were performed according to the manufacturer's recommendations. Whole cell lysates from E. coli expressing recombinant PKL peptide were separated on 10% SDS-PAGE (Laemmli, 1970). Gels were stained with copper chloride (Lee et al., 1987) and the band corresponding to the PKL N- or C-terminal fragment was excised and equilibrated in 40 volumes of 0.5 M EDTA (pH 8.0) for 30 minutes, followed by 3 washes (30 minutes each) in 40 volumes of Tris-Glycine gel running buffer (25 mM Tris, 250 mM glycine, 0.1% SDS) to remove the copper stain. The recombinant PKL peptides were electroeluted from the gel slices using an S&S Elutrap apparatus (Schleicher and Schuell, Inc., Keene, NH) according to the manufacturers instructions. Eluted peptides were concentrated using Microcon concentrators (Millipore Corp., Billerica, MA), quantified, and adjusted to a final concentration of approximately 1 mg/ml. The partially purified PKL fragments (∼5 mg) were used for the production of polyclonal antibodies (Alpha Diagnostic, San Antonio, TX).
Protein extraction
For analysis of PKL protein in E. coli, crude whole cell extracts were prepared from cells that were resuspended in 0.5 ml PBS with 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 mM PMSF. Equal volumes of hot (80°C) 1x PBS and hot 2x sample (2% SDS, 80mM Tris pH6.8, 0.6% bromophenol blue, 15% glycerol and 0.1M DTT) buffer were added, and samples were heated at 80°C for 15 min.
For analysis of PKL protein in S. cerevisiae, crude whole cell extracts were prepared from cells grown to saturation in minimal media that were lysed with an equal volume of 2x sample buffer (0.2 M Tris pH6.8, 20% β-mercaptoethanol, 4% SDS, 10% glycerol, 0.002% bromophenol blue) supplemented with 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 mM PMSF. Cells were beat twice for 30 sec with 0.5 mm zirconia beads (Biospec Products, Inc., Bartlesville, OK), boiled for 3 min, and supernatants were transferred to a new tube.
Crude whole cell protein extracts of A. thaliana were prepared as previously described (Rider et al., 2004).
Protein quantification and western analysis
Total proteins were quantified with Bio-Rad RCDC protein assay (Bio-Rad Lab., Hercules, CA) as per manufacturer's recommendation. Equal amounts of total proteins were separated on 6% SDS-polyacrylamide gels and transferred to immobilon-P membrane (Millipore Corp., Billerica, MA). Membranes were blocked with 1x BSA (0.01 M Tris pH8, 0.1 M NaCl, 0.1% Tween-20, 1% BSA), and probed with anti-PKLN or anti-PKLC terminal antiserum in 1:5000x dilution. Anti-PEP carboxylase antibody at a dilution of 1:5000 (Rockland, Gilbertsville, PA) was used as a control. Secondary antibody (goat anti-rabbit IgG-HRP conjugate; Pierce Biotech. Inc., Rockford, IL) was used at a dilution of 1:10000.
PKL:eGFP characterization
Arabidopsis seed, seedlings and leaves were mounted in water and observed with a fluorescence microscope (model DMR HC, Leica Inc., Deerfield, IL). eGFP was visualized with EN GFP filter from Chroma Technology (part # 41017). Nuclei were visualized with DAPI as described by Copenhaver (http://www.biology.wustl.edu/pikaard/protocols/chromcount.html).
pkl-1/PKL:GR inducible lines
Seedlings were grown on synthetic media supplemented with 10-7M uniconazole-P and with or without 10-5 M Dex (Sigma) dissolved in methanol. Seedlings were shifted from plates containing Dex to plates that did not contain Dex or vice versa using fine point tweezers in a sterile hood. At each time point 144 seeds of pkl-1/PKL:GR (transgenic line GR-3), 72 seeds of pkl-1, and 72 wild-type seeds were transplanted from Dex to no Dex plates or vice versa. Seedlings were scored for the pickle root trait at 15 days post seed imbibition. The experiment was carried out at 22°C in continuous light (60 μE m-2 s-1). Three biological replicates of this analysis were performed.
RNA isolation and quantitative RT-PCR
Total RNA was isolated as described previously by Verwoerd et al. (Verwoerd et al., 1989). Quantitative PCR was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems), as previously described (Rider et al., 2003). 18S ribosomal RNA was used as a normalization control for the relative quantification of transcript levels. Data presented are derived from three unpaired technical replicates as recommended by ABI. All qRT-PCR experiments were repeated with a biological replicate with similar results (data not shown). All oligonucleotide primer sequences and primer concentrations used, the critical threshold values for the figures presented, and the complete analysis of type I MADS box genes can be found in the Supplementary Materials.
Supplementary Material
Table 2.
Relative transcript levels for Type I MADS-box transcription factors in pkl mutants. Transcript levels were determined for pkl-1 and wild-type plants at 50% radicle emergence (Germination), in young seedlings (4 day) and in 21-day-old leaves (21 day). 18s rRNA was used as a standardization Expression levels are normalized relative to wild-type plants. Only those loci that showed greater a 5-fold change are presented. ND indicates transcript not detected.
| Fold Change (pkl/wt) | ||||
|---|---|---|---|---|
| AGI Code | Gene | Germination | 4 day | 21 day |
| At2g28700 | ND | 17 | 3.1 | |
| At3g66656 | 7.1 | 1.5 | 0.83 | |
| At5g26630 | AGL 35 | 5.1 | 0.57 | 0.74 |
| At1g18750 | AGL 65 | 0.08 | 0.96 | 0.71 |
ACKNOWLEDGEMENTS
We thank Sean Cutler for sharing the pEGAD vector and Alan Lloyd for sharing the pBI-ΔGR vector. We thank Clint Chapple for thoughtful discussion. We thank Vasundhara Kandachar for assistance with some of the GR-related work. We thank Jody Banks for use of her microscope. We thank the undergraduates in the laboratory for numerous contributions. This work was supported by the National Institutes of Health (grant no. R01GM059770 to J.O.) and by the National Science Foundation (IGERT Fellowships to K.C. and to J.T.H.). This is journal paper no. 17751 of the Purdue University Agricultural Experiment Station.
REFERENCES
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–6. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martinez-Castilla L, Yanofsky MF. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci U S A. 2000;97:5328–33. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiology and Biochemistry. 2002;40:151–160. [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–80. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. Plenum Press; New York, USA: 1994. [Google Scholar]
- Bowman JL. Arabidopsis: an atlas of morphology and development. Springer-Verlag New York, Inc.: 1993. [Google Scholar]
- Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. Embo J. 2000;19:4332–41. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Florquin K, Rombauts S, Rouze P, Theissen G, Van de Peer Y. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J Mol Evol. 2003;56:573–86. doi: 10.1007/s00239-002-2426-x. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD Complex Regulate Cell Fate during B Lymphocyte Differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 2001;126:835–48. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 2002;129:823–37. doi: 10.1104/pp.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The Transcription Factor FUSCA3 Controls Developmental Timing in Arabidopsis through the Hormones Gibberellin and Abscisic Acid. Dev Cell. 2004;7:373–85. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–81. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Guschin D, Wade PA, Kikyo N, Wolffe AP. ATP-Dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry. 2000;39:5238–45. doi: 10.1021/bi000421t. [DOI] [PubMed] [Google Scholar]
- Harada JJ. Role of Arabidopsis LEAFY COTYLEDON genes in seed development. Journal of Plant Physiology. 2001;158:405–409. [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, De Vries SC, Ogas J. PICKLE Acts throughout the Plant to Repress Expression of Embryonic Traits and May Play a Role in Gibberellin-Dependent Responses. Plant Physiol. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci. 1999;4:275–80. [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003;17:1540–53. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of bacteriophage T. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C, Levin A, Branton D. Copper staining: a five-minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1987;166:308–12. doi: 10.1016/0003-2697(87)90579-3. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–9. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Merkle SA, Dean JF. Forest tree biotechnology. Curr Opin Biotechnol. 2000;11:298–302. doi: 10.1016/s0958-1669(00)00099-9. [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–4. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–44. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell. 2004;16:2705–18. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–80. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- Rider SD, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313x.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Jr., Hemm MR, Hostetler HA, Li HC, Chapple C, Ogas J. Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta. 2004;219:489–99. doi: 10.1007/s00425-004-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JP, Chua NH. Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell. 2001;13:1143–54. doi: 10.1105/tpc.13.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl Acad. Sci. USA. 2001;98:11806–11. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a Homolog of the NURD Complex Component Mi-2 Act Together to Maintain Germline-Soma Distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Muller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–84. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 1998;8:843–6. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wang HB, Zhang Y. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 2001;29:2517–21. doi: 10.1093/nar/29.12.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30:349–59. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.