Abstract
The stereoselective synthesis of aminooxy-containing proline analogues bearing Fmoc/Boc or Fmoc/Mtt protection that renders them suitable for incorporation into peptides using Fmoc protocols is reported. Acid-catalyzed unmasking at the completion of peptide synthesis yields free aminooxy–functionalities for oxime formation through reaction with libraries of aldehydes. This allows post solid-phase diversification strategies that may facilitate structure activity relationship studies.
The presence of proline residues in peptides and proteins can induce β-turn secondary structures that provide specific bioactivities.1 This is exemplified by “proline rich motifs” (PRMs) that are involved with the formation of protein-protein complexes.2 Libraries of proline analogues could serve as valuable tools for studying these interactions. However, the preparation of peptides bearing 3- and 4-substituted proline congeners has been limited by the need to synthesize each individual proline derivative in protected form prior to peptide synthesis.3
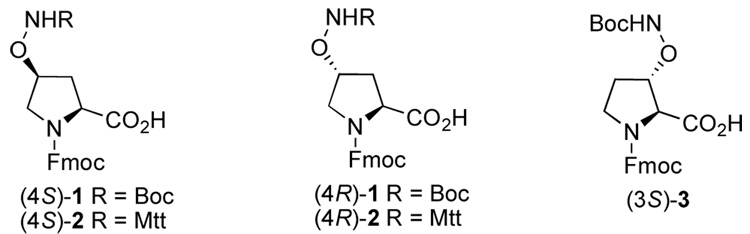
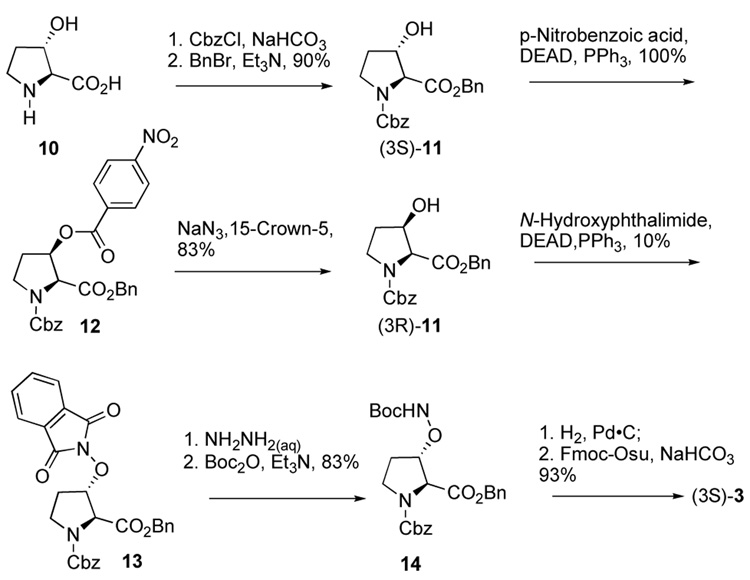
The incorporation of suitably protected aminooxy groups into orthogonally-protected proline residues could result in highly useful biochemical tools amenable to diversification through oxime formation by conjugation with libraries of aldehydes following the completion of peptide synthesis.4–6 The aminooxy-containing proline analogues7 1 and 3, bearing Fmoc and Boc or 2, having Fmoc and Mtt protection, represent examples of proline derivatives that may be suitable for post solid-phase diversification through oxime formation (Figure 1). Reported herein are the synthesis of these new proline analogues and their application in a physiologically-relevant context where proline residues serve as key recognition elements.
Figure 1.
Structures of orthogonally-protected proline analogues.
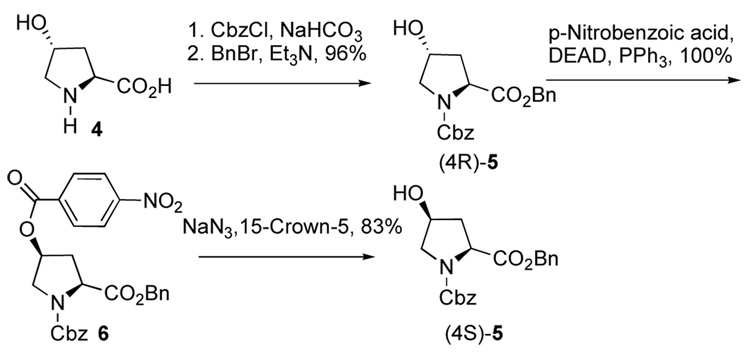
Synthesis
Commercially available (4R)-4-hydroxy-L-proline (4) was protected as its N-Cbz, O-Bn derivative (4R)-5 (Scheme 1).8,9 Inversion of the (4R)-stereocenter was achieved in two steps involving the 4-nitrobenzoate ester (6)10 under Mitsunobu conditions.11 The high cost of (4S)-4 makes its preparation from the relatively inexpensive (4R)-4 attractive.12
Scheme 1.
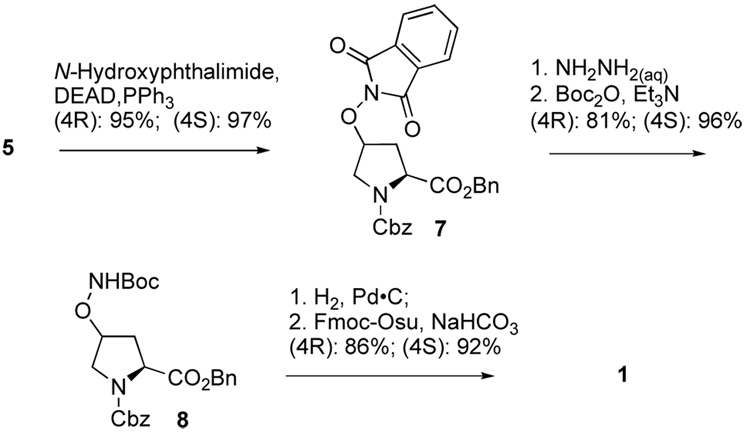
Mitsunobu transformation of the protected isomeric (4R)- and (4S)-4-hydroxyprolines (5)13 was achieved smoothly with inversion of C4 stereochemistry to give the corresponding (4S)- and (4R)- N-phthalimidoaminooxy compounds (7) in nearly quantitative yield (Scheme 2).14,15
Scheme 2.
The phthalimido groups of 7 were cleaved at room temperature using aqueous hydrazine and the free aminooxy primary amines were directly protected by treatment with Boc anhydride and triethylamine. Hydrogenation of the resulting derivatives (8)16,17 over Pd•C removed both the O-Bn and N-Cbz protecting groups to yield the free amino acids, which were converted in situ to their N-Fmoc forms (1).18,19 In this fashion starting from the commercially available free amino acid 4, (4S)-1 was obtained in 82% overall yield (7 steps) and (4R)-1 was obtained in 53% overall yield (9 steps).
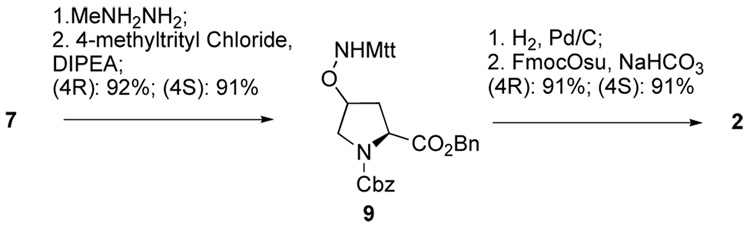
N-Boc-protection of aminooxy functionality is suitable for Fmoc-based solid-phase protocols used in combination with moderately labile acid-sensitive resins, where removal of the Boc groups is concomitant with cleavage of the peptide from the resin. However, when deprotection of the aminooxy group is desired without cleavage of the peptide from the solid-phase resin, more highly acid-labile methyltrityl (Mtt) protection (2) is appropriate. Toward this objective, the isomeric phthalimido-protected aminooxy intermediates (4R)-7 and (4S)-7 were treated with methylhydrazine, then directly reacted with 4-methyltrityl chloride to give the Mtt protected compounds (4R)-9 and (4S)-9 (Scheme 3).20,21 Simultaneous hydrogenolytic cleavage of both the amino and carboxylic protecting groups (Pd•C/H2 in EtOAc : MeOH) and subsequent treatment with Fmoc-OSu provided the final products (4S)-222 and (4R)-223 in 77% overall yield (7 steps) and 63% overall yield (9 steps) respectively, starting from commercially available 4.
Scheme 3.
Preparation of the N-Boc derivatized isomeric 3-substituted products (3) began by protection of commercially available (3S)-3-hydroxy-L-proline (10) using the conditions described above for the conversion of 4 to (4R)-5. The attempted Mitsunobu transformation of the resulting (3S)-1124 using N-hydroxyphthalimide under a variety of reaction conditions predominately gave the corresponding α,β-elimination material with no desired product. With the epimeric (3R)-11, prepared in near quantitative yield from (3S)-11 by a two-step p-nitrobenzoyl ester stereo-inversion sequence,25,26 a low yield (10%) of the desired 3-phthalimidooxy analogue (3S)-13 could be obtained.27,28 This allowed subsequent conversion to the final product (3S)-3 (Scheme 4).29
Scheme 4.
Application of Aminooxy-Proline Analogues to Peptide Library Synthesis
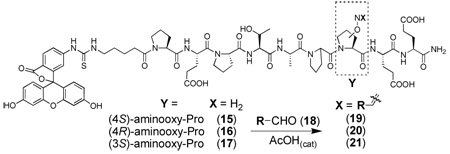
In order to demonstrate the utility of post solid-phase diversification using aminooxy-proline analogues, we employed a Tsg101-binding peptide system. The UEV (ubiquitin E2 variant) domain of the human protein Tsg101 (tumor susceptibility gene 101) is recruited by major structural proteins of HIV-1 to facilitate viral budding. This involves the direct interaction of the Tsg101 UEV domain with a Pro-Thr-Ala-Pro (“PTAP”) sequence in the viral Gagp6 protein.30,31 Using a p6 – derived sequence, we had previously replaced critical Pro residues with hydrazone- and hydrazide-containing N-substituted glycines as peptoid surrogates that allowed the expedited synthesis of libraries of Tsg101-directed binding antagonists.32 In our current study, we utilized the wild-type p6-derived “PEPTAPPEE” sequence to examine libraries of peptides having proline-oximes situated in place of the Pro7 residue (underlined).33,34
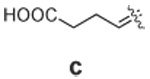
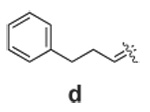
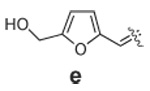
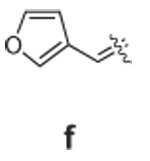
Libraries were generated from parent aminooxy-proline containing peptides prepared using the orthogonally protected residues described above. TFA-mediated cleavage of peptides from the resin gave the globally-deprotected aminooxy-proline containing peptides 15 – 17 (Table 1) which were purified by HPLC. Condensation of 15 – 17 with a series of aldehydes (18a – t) was conducted overnight in DMSO with a catalytic amount of AcOH and a molar ratio of peptide to aldehyde of 1:1. As indicated by HPLC, the reactions went to completion, yielding proline-oxime containing peptides 19 – 21 in sufficient purity (>90%) for direct biological evaluation (Table 1).35 Certain of these peptides (for example, 21o; IC50 = 11 µM) exhibited up to five-fold higher Tsg101-binding affinity than wild-type peptide (Kd = 54 µM),36 indicating the potential utility of the approach.
Table 1.
Tsg101-Binding Affinities of Proline-Oxime Containing Peptides Determined as in Reference 32.
 | |||
|---|---|---|---|
| IC50 (µM) | |||
| X | Peptide: 19 | 20 | 21 |
| H2 | 55 [15] | 43 [16] | 21 [17] |
 |
nd* | 54 | 41 |
| *nd=Not determined | |||
 |
42 | 41 | 23 |
 |
75 | 27 | 16 |
 |
83 | 26 | 20 |
 |
nd | 26 | 25 |
 |
48 | 30 | 20 |
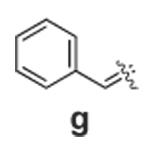 |
64 | 26 | 15 |
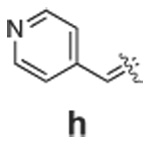 |
nd | 29 | 22 |
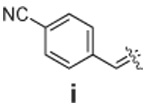 |
nd | 20 | 22 |
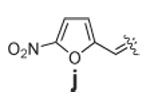 |
nd | 12 | 18 |
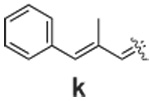 |
58 | 13 | 12 |
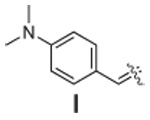 |
42 | 17 | 12 |
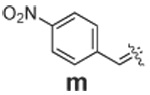 |
91 | 21 | 16 |
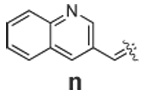 |
nd | 25 | 19 |
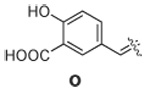 |
93 | 16 | 11 |
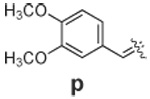 |
114 | 16 | 22 |
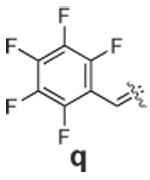 |
139 | 32 | 19 |
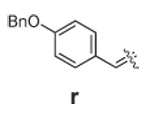 |
54 | 30 | 18 |
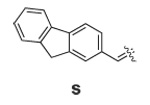 |
nd | 27 | 17 |
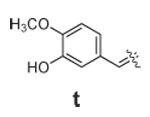 |
nd | 23 | 13 |
Conclusions
Reported herein are the design and synthesis of 3- and 4-aminooxy-substituted praline analogues suitably protected for Fmoc-based peptide synthesis. As demonstrated by application in a Tsg101-binding system, these non-natural amino acid analogues may be highly useful for post solid-phase diversification strategies that could facilitate rapid structure-activity relationship studies, potentially leading to new biologically active motifs.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick and the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.(a) Mauger AB. J. Natural Prod. 1996;59:1205. doi: 10.1021/np9603479. [DOI] [PubMed] [Google Scholar]; (b) Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S. FASEB J. 1995;9:736. [PubMed] [Google Scholar]; (c) Ruzza P, Siligardi G, Donella-Deana A, Calderan A, Hussain R, Rubini C, Cesaro L, Osler A, Guiotto A, Pinna LA, Borin G. J. Peptide Sci. 2006;12:462. doi: 10.1002/psc.750. [DOI] [PubMed] [Google Scholar]; (d) Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. J. Mol. Biol. 1998;279:449. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 2.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Angew. Chem. Int. Ed. 2005;44:2852. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 3.(a) Jacquot Y, Broutin I, Miclet E, Nicaise M, Lequin O, Goasdoue N, Joss C, Karoyan P, Desmadril M, Ducruix A, Lavielle S. Bioorg. Med. Chem. 2007;15:1439. doi: 10.1016/j.bmc.2006.11.002. [DOI] [PubMed] [Google Scholar]; (b) Kim W, Hardcastle KI, Conticello VP. Angew. Chem. Int. Ed. 2006;45:8141. doi: 10.1002/anie.200603227. [DOI] [PubMed] [Google Scholar]; (c) Taylor CM, Hardre R, Edwards PJB. J. Org. Chem. 2005;70:1306. doi: 10.1021/jo0490043. [DOI] [PubMed] [Google Scholar]
- 4.(a) Johnson SM, Petrassi HM, Palaninathan SK, Mohamedmohaideen NN, Purkey HE, Nichols C, Chiang KP, Walkup T, Sacchettini JC, Sharpless KB, Kelly JW. J. Med. Chem. 2005;48:1576. doi: 10.1021/jm049274d. [DOI] [PubMed] [Google Scholar]; (b) Lees A, Sen G, LopezAcosta A. Vaccine. 2006;24:716–729. doi: 10.1016/j.vaccine.2005.08.096. [DOI] [PubMed] [Google Scholar]; (c) Nazarpack-Kandlousy N, Chernushevich IV, Meng L, Yang Y, Eliseev AV. J. Am. Chem. Soc. 2000;122:3358. [Google Scholar]; (d) Su S, Acquilano DE, Arumugasamy J, Beeler AB, Eastwood EL, Giguere JR, Lan P, Lei X, Min GK, Yeager AR, Zhou Y, Panek JS, Snyder JK, Schaus SE, Porco JA., Jr Org. Lett. 2005;7:2751. doi: 10.1021/ol051023r. [DOI] [PubMed] [Google Scholar]
- 5.Jencks WP. J. Am. Chem. Soc. 1959;81:475. [Google Scholar]
- 6.Baindur N, Harris SM, Labroo VM. Combinatorial non-peptide libraries. No. 5,646,285. Zymogenetics, Inc., USA; U.S. Patent. 1997 July 8;
- 7.The synthesis of unprotected (2S,4S)-4-aminooxyproline hydrochloride has been reported: Phuket SRN, Trifonov LS, Yu CX, Worthen DR, Crooks PA, Rosenthal GA, Freeman JW. Drug Dev. Res. 1999;47:170.
- 8.(4R)-4-hydroxy-L-proline is available from Sigma-Aldrich; (3S)-3-hydroxy-L-proline is available from Acros Organics.
- 9.(4R)-5: [α]20D −53.2 (c 2.02, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.32 – 7.18 (m, 10 H), 5.23 – 5.11 (m, 2 H), 5.01 – 4.95 (m, 2 H), 4.53 (m, 1 H), 4.40 (m, 1 H), 3.65 – 3.51 (m, 2 H), 2.91 (brs, 1 H), 2.26 (m, 1 H), 2.02 (m, 1 H).
- 10.6: [α]20D −63.8 (c 1.36, CHCl3). 1H NMR (400 MHz, CDCl3) δ 8.16 (AB, JAB = 8.8, 2.0 Hz, 2 H), 8.03 – 7.97 (m, 2 H), 7.38 – 7.28 (m, 5 H), 7.22 – 7.15 (m, 5 H), 5.58 (m, 1 H), 5.25 – 5.02 (m, 4 H), 4.73 (dd, J = 9.2, 1.6 Hz, 0.5 H), 4.63 (dd, J = 9.2, 1.6 Hz, 0.5 H), 3.92 – 3.85 (m, 2 H), 2.66 – 2.46 (m, 2 H). ESI-MS (+VE) m/z: 527.1(M+Na)+. HR-ESI/APCI MS cacld for C27H25N2O8 (M+H) +: 505.1611, Found: 505.1606.
- 11.Gomez-Vidal JA, Silverman RB. Org. Lett. 2001;3:2481. doi: 10.1021/ol0161054. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Vidal J, Forrester MT, Silverman RB. Org. Lett. 2001;3:2477. doi: 10.1021/ol016104b. [DOI] [PubMed] [Google Scholar]
- 13.(4S)-5: [α]20D −17.1 (c 2.56, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.35 – 7.24 (m, 10 H), 5.30 – 5.00 (m, 4 H), 4.49 (dd, J = 9.6, 1.6 Hz, 0.4 H), 4.42 (dd, J = 9.6, 1.6 Hz, 0.6 H), 4.34 (m, 1 H), 3.72 – 3.57 (m, 2 H), 3.27 (brs, 1 H), 2.29 (m, 1 H), 2.12 (m, 1 H). 13C NMR (100 MHz, CDCl3) δ 174.12, 174.04, 154.98, 154.31, 136.36, 136.25, 135.23, 135.04, 128.58, 128.48, 128.44, 128.37, 128.27, 128.15, 128.08, 128.03, 127.90, 127.84, 70.97, 70.00, 67.46, 67.42, 67.38, 67.34, 67.25, 58.25, 57.88, 55.91, 55.57, 38.68, 37.80.
- 14.(4S)-7: Mp 130–132 °C. [α]20D −4.20 (c 0.88, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.84 – 7.81 (m, 2 H), 7.77 – 7.74 (m, 2 H), 7.45 – 7.23 (m, 10 H), 5.33 – 4.99 (m, 4 H), 4.94 (m, 1 H), 4.65 (dd, J = 10.0, 2.0 Hz, 0.5 H), 4.58 (dd, J = 9.6, 2.0 Hz, 0.5 H), 4.05 (m, 1 H), 3.92 (m, 1 H), 2.70 (m, 0.5 H), 2.66 (m, 0.5 H), 2.45 (m, 1 H). ESI-MS (+VE) m/z: 523.1(M+Na)+. HR-ESI/APCI MS cacld for C28H24N2NaO7 (M+Na) +: 523.1481, Found: 523.1475.
- 15.(4R)-7: Mp. 124–126°C. [α]20D −44.1 (c 1.09, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.85 – 7.82 (m, 2 H), 7.77 – 7.75 (m, 2 H), 7.40 – 7.18 (m, 10 H), 5.26 – 5.14 (m, 3 H), 4.99 – 4.96 (m, 2 H), 4.78 (m, 1 H), 4.08 (m, 1 H), 3.75 (dd, J = 12.8, 4.0 Hz, 1 H), 2.71 (m, 1 H), 2.15 (m, 1 H). ESI-MS (+VE) m/z: 523.1 (M+Na)+. HR-ESI/APCI MS cacld for C28H24N2NaO7 (M+Na)+: 523.1481, Found: 523.1473.
- 16.(4S)-8: [α]20D −37.6 (c 1.27, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.35 – 7.23 (m, 10 H), 6.82 (s, 0.50 H), 6.74 (s, 0.50 H), 5.23 – 4.99 (m, 4 H), 4.56 - 4.45 (m, 2 H), 3.78 (d, J = 12.8 Hz, 0.5 H), 3.73 (d, J = 12.8 Hz, 0.5 H), 3.59 (dd, J = 12.8, 4.8 Hz, 0.5 H), 3.56 (dd, J = 12.8, 4.8 Hz, 0.5 H), 2.51 (d, J = 4.8 Hz, 0.5 H), 2.48 (d, J = 4.8 Hz, 0.5 H), 2.21 (ddd, J = 18.8, 9.2, 4.8 Hz, 0.5 H), 2.16 (ddd, J = 18.8, 9.2, 4.8 Hz, 0.5 H), 1.41 (s, 4.5 H), 1.40 (s, 4.5 H). ESI-MS (+VE) m/z: 493.1(M+Na)+. HR-ESI/APCI MS cacld for C25H30N2NaO7 (M+Na) +: 493.1951, Found: 493.1955.
- 17.(4R)-8: [α]20D −30.4 (c 1.40, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.85 (s, 0.56 H), 7.67 (s, 0.44 H), 7.30 – 7.10 (m, 10 H), 5.19 – 5.09 (m, 2 H), 5.02 – 4.94 (m, 2 H), 4.55 – 4.45 (m, 2 H), 3.93 (d, J = 12.4 Hz, 0.56 H), 3.82 (d, J = 12.4 Hz, 0.44 H), 3.56 (m, 1 H), 2.55 (m, 1 H), 1.99 (m, 1 H), 1.41 (s, 9H). ESI-MS (+VE) m/z: 493.1(M+Na)+. HR-ESI/APCI MS cacld for C25H30N2NaO7 (M+Na)+: 493.1951, Found: 493.1968.
- 18.(4S)-1: [α]20D −26.4 (c 0.65, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.86 – 7.55 (m, 6 H), 7.42 – 7.35 (m, 2 H), 7.33 – 7.29 (m, 2 H), 4.61 – 4.33 (m, 4 H), 4.22 (m, 1 H), 3.92 (d, J = 12.8 Hz, 0.4 H), 3.78 (d, J = 12.4 Hz, 0.6 H), 3.50 (m, 1 H), 2.79 (d, J = 14.8 Hz, 0.6 H), 2.50 (d, J = 14.4 Hz, 0.4 H), 2.36 (m, 0.4 H), 2.16 (m, 0.6 H), 1.46 (s, 9 H). ESI-MS (+VE) m/z: 491.1(M+Na)+. HR-ESI/APCI MS cacld for C25H28N2NaO7 (M+Na) +: 491.1794, Found: 491.1788.
- 19.(4R)-1: [α]20D −41.2 (c 0.97, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.76 (d, J = 7.6 Hz, 1 H), 7.69 (d, J = 7.6 Hz, 1 H), 7.59 – 7.51 (m, 2 H), 7.41 – 7.25 (m, 4.35 H), 7.18 ( 0.65 H), 4.53 – 4.33 (m, 4 H), 4.24 (t, J = 6.8 Hz, 0.65 H), 4.13 (t, J = 6.8 Hz, 0.35 H), 3.96 (d, J = 12.6 Hz, 0.35 H), 3.85 (d, J = 12.6 Hz, 0.65 H), 3.53 (m, 1 H), 2.23 (m, 0.65 H), 2.10 (m, 0.35 H), 1.49 (s, 5.5 H), 1.46 (s, 3.5 H). ESI-MS (+VE) m/z: 491.1 (M+Na)+. HR-ESI/APCI MS cacld for C25H28N2NaO7 (M+Na)+: 491.1794, Found: 491.1800.
- 20.(4R)-9: [α]20D −30.5 (c 0.94, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.31 – 7.19 (m, 19 H), 7.16 (m, 1 H), 7.10 (d, J = 9.0 Hz, 2 H), 7.04 (d, J = 9.0 Hz, 2 H), 6.44 (s, 0.5 H), 6.41 (s, 0.5 H), 5.19 – 4.93 (m, 4 H), 4.35 (t, J = 8.0 Hz, 0.5 H), 4.24 (t, J = 8.0 Hz, 0.5 H), 4.10 (m, 1 H), 3.85 (d, J = 12.0 Hz, 0.5 H), 3.67 (d, J = 12.0 Hz, 0.5 H), 3.47 (dd, J = 8.0, 4.4 Hz, 0.5 H), 3.44 (dd, J = 8.0, 4.4 Hz, 0.5 H), 2.90 (s, 3 H), 2.25 (m, 1 H), 1.87 (dd, J = 8.2, 5.0 Hz, 0.5 H), 1.83 (dd, J = 8.2, 5.0 Hz, 0.5 H). ESI-MS (+VE) m/z: 649.3 (M+Na)+.HR-ESI/APCI MS cacld for C40H38N2NaO5 (M+Na)+: 649.2678, Found: 649.2674.
- 21.(4S)-9: [α]20D −25.5 (c 1.00, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.33 – 7.30 (m, 2 H), 7.25 – 7.18 (m, 16 H), 7.15 – 7.02 (m, 6 H), 6.40 (s, 0.5 H), 6.38 (s, 0.5 H), 5.18 – 5.05 (m, 2 H), 4.96 (d, J = 11.6 Hz, 1 H), 4.87 (d, J = 12.8 Hz, 0.5 H), 4.75 (d, J = 12.4 Hz, 0.5 H), 4.51 (dd, J = 8.0, 1.2 Hz, 0.5 H), 4.40 (dd, J = 8.0, 1.2 Hz, 0.5 H), 4.07 (m, 0.5 H), 4.03 (m, 0.5 H), 3.70 (d, J = 12.4 Hz, 0.5 H), 3.62 (d, J = 12.0 Hz, 0.5 H), 3.45 (dd, J = 12.4, 4.8 Hz, 0.5 H), 3.40 (dd, J = 12.4, 4.8 Hz, 0.5 H), 2.39 (t, J = 12.0 Hz, 1 H), 2.28 (s, 3 H), 2.00 (m, 1 H). ESI-MS (+VE) m/z: 649.2 (M+Na)+.HR-ESI/APCI MS cacld for C40H38N2NaO5 (M+Na)+: 649.2678, Found: 649.2665.
- 22.(4S)-2: [α]20D −17.9 (c 0.79, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.72 – 7.62 (m, 2 H), 7.50 – 7.36 (m, 2 H), 7.36 – 7.28 (m, 2 H), 7.28 – 7.12 (m, 14 H), 7.10 – 6.6 (m, 5 H), 4.40 – 4.00 (m, 4 H), 3.90 (m, 1 H), 3.40 (m, 1 H), 3.26 (m, 1 H), 2.60 (m, 1 H), 2.27 (s, 3 H) 1.90 (m, 1 H). ESI-MS (+VE) m/z: 647.2 (M + Na)+. HR-ESI/APCI MS cacld for C40H36N2NaO5 (M+Na)+: 647.2522, Found: 647.2518.
- 23.(4R)-2: [α]20D −35.6 (c 0.64, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.71 (d, J = 7.8 Hz, 1 H), 7.66 (d, J = 7.8 Hz, 1 H), 7.54 (d, J = 7.2 Hz, 0.5 H), 7.48 (d, J = 7.2 Hz, 0.5 H), 7.42 (dd, J = 12.6, 7.4 Hz, 1 H),7.35 (t, J = 7.6 Hz, 1 H), 7.30 (dd, J = 11.8, 2.2 Hz, 1 H), 7.25 – 7.16 (m, 13 H), 7.13 (d, J = 8.4 Hz, 2 H), 7.08 – 7.02 (m, 3 H), 6.92 (d, J = , 0.5 H), 6.82 (d, J = , 0.5 H), 4.31 – 3.99 (m, 5 H), 3.80 (d, J = 12.0 Hz, 0.3 H), 3.60 (d, J = 12.0 Hz, 0.7 H), 3.32 (dd, J = 11.8, 4.2 Hz, 0.7 H), 3.24 (dd, J = 12.2, 4.2 Hz, 0.3 H), 2.30 (s, 3 H), 2.20 (m, 1 H), 1.87 (m, 0.7 H), 1.73 (m, 0.3 H). ESI-MS (+VE) m/z: 647.2 (M+Na)+. HR-ESI/APCI MS cacld for C40H36N2NaO5 (M+Na)+: 647.2522, Found: 647.2511.
- 24.(3S)-11: [α]20D −24.7 (c 1.64, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.38 – 7.20 (m, 10 H), 5.22 – 5.13 (m, 2 H), 5.09 – 5.00 (m, 2 H), 4.45 (m, 1 H), 4.42 (m, 0.5 H), 4.33 (m, 0.5 H), 3.75 – 3.62 (m, 2 H), 2.24 (brs, 1 H), 2.08 (m, 1 H), 1.91 (m, 1 H). ESI-MS (+VE) m/z: 356.1 (M+H)+. HR-ESI/APCI MS cacld for C20H21NNaO5 (M+Na)+: 378.1317, Found: 378.1316.
- 25.12: [α]20D −91.4 (c 0.80, CHCl3). 1H NMR (400 MHz, CDCl3) δ 8.14 (t, J = 9.2 Hz, 2 H), 7.93 (t, J = 9.2 Hz, 2 H), 7.41 – 7.26 (m, 5 H), 7.15 – 7.07 (m, 5 H), 5.75 (m, 1 H), 5.24 – 5.05 (m, 3 H), 4.98 – 4.78 (m, 2 H), 3.85 – 3.69 (m, 2 H), 2.38 – 2.25 (m, 2 H). ESI-MS (+VE) m/z: 527.1 (M+Na)+. HR-ESI/APCI MS cacld for C27H24N2NaO8 (M+Na)+: 527.1430, Found: 527.1428.
- 26.(3R)-11: [α]20D −33.0 (c 1.10, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.34 – 7.22 (m, 10 H), 5.25 – 5.00 (m, 4 H), 4.56 (m, 1 H), 4.48 (d, J = 6.8 Hz, 0.4 H), 4.44 (d, J = 6.8 Hz, 0.6 H), 3.68 (m, 1 H), 3.51 (m, 1 H), 2.85 (d, J = 5.0 Hz, 0.6 H), 2.79 (d, J = 5.0 Hz, 0.4 H), 2.04 (m, 2 H). ESI-MS (+VE) m/z: 378.1 (M+Na)+. HR-ESI/APCI MS cacld for C20H21NNaO5 (M+Na)+: 378.1317, Found: 378.1312.
- 27.13: [α]20D + 1.25 (c 1.65, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.84 – 7.81 (m, 2 H), 7.78 – 7.75 (m, 2 H), 7.41 – 7.25 (m, 9 H), 7.16 (m, 1 H), 5.21 – 4.83 (m, 6 H), 3.87 (m, 1 H), 3.78 (m, 1 H), 2.32 (m, 1 H), 2.17 (m, 1 H). ESI-MS (+VE) m/z: 523.1 (M+Na)+. HR-ESI/APCI MS cacld for C28H24N2NaO7 (M+Na)+: 523.1481, Found:523.1481.
- 28.14: [α]20D −17.2 (c 0.74, CHCl3). 1H NMR (400 MHz, CDCl3) δ 7.32 – 7.19 (m, 11 H), 5.20 – 5.10 (m, 2H), 5.06 – 4.98 (m, 2 H), 4.71 (s, 0.45 H), 462 (s, 0.55 H), 4.48 (m, 1 H), 3.70 – 3.56 (m, 2 H), 2.13 (m, 1 H), 2.02 (m, 1 H), 1.41 (s, H). ESI-MS (+VE) m/z: 493.1 (M+Na)+.HR-ESI/APCI MS cacld for C25H30N2NaO7 (M+Na)+: 493.1951, Found: 493.1968.
- 29.(3S)-3: [α]20D −33.0 (c 0.60, CHCl3). 1H NMR (400 MHz, CDCl3) 8.85 (brs, 1 H), δ 7.77 – 7.70 (m, 2.6 H), 7.56 – 7.50 (m, 2.4 H), 7.37 – 7.30 (m, 2 H), 7.29 – 7.23 (m, 2 H), 4.67 – 4.52 (m, 2 H), 4.44 (m, 1 H), 4.35 (m, 1 H), 4.21 (t, J = 7.2 Hz, 0.6 H), 4.12 (t, J = 7.2 Hz, 0.4 Hz), 3.69 – 3.58 (m, 2 H), 2.23 – 2.00 (m, 2 H), 1.44 (s, 9 H). ESI-MS (+VE) m/z: 491.1 (M+Na)+. HR-ESI/APCI MS cacld for C25H28N2NaO7 (M+Na)+: 491.1794, Found: 491.1800.
- 30.Demirov DG, Freed EO. Virus Res. 2004;106:87. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Mazze FM, Degreve L. Acta Virologica (English Edition) 2006;50:75. [PubMed] [Google Scholar]
- 32.Liu F, Stephen AG, Adamson CS, Gousset K, Aman MJ, Freed EO, Fisher RJ, Burke TR., Jr Org. Lett. 2006;8:5165. doi: 10.1021/ol0622211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Substitution of the Pro7 residue by alanine results in significant reduction of Tsg101-binding affinity, indicating the importance of the 7-position.
- 34.N-Terminal labeling with fluorosceine isothiocyanate linked via an aminovaleric acid spacer (FITC-Ava-) was employed.
- 35.A mixture of HPLC-purified aminooxy-proline containing peptide (15 – 17) (15 mM in DMSO, 10 µL), aldehdye (18a – 18t) (15 mM in DMSO, 10 µL) and acetic acid (70 mM in DMSO, 10 µL) was gently agitated at room temperature (overnight). Examination by HPLC showed the reactions had gone to completion to produce oxime products in ≥90% purity. Crude reaction mixtures were used directly for biological evaluation.
- 36.Based on a fluorescence anisotropy binding assay requirement of 20 mL of 5 mM peptide inhibitor solution, 0.20 mmol of TGR solid-phase resin is theoretically sufficient to prepare a 740 member oxime library.