Abstract
All three single-stranded RNAs of the brome mosaic virus (BMV) genome contain a highly conserved, 193-base 3' noncoding region. To study the recombination between individual BMV RNA components, barley plants were infected with a mixture of in vitro-transcribed wild-type BMV RNAs 1 and 2 and an RNA3 mutant that carried a deletion near the 3' end. This generated a population of both homologous and nonhomologous 3' recombinant BMV RNA3 variants. Sequencing revealed that these recombinants were derived by either single or double crossovers with BMV RNA1 or RNA2. The primary sequences at recombinant junctions did not show any similarity. However, they could be aligned to form double-stranded heteroduplexes. This suggested that local hybridizations among BMV RNAs may support intermolecular exchanges.
Full text
PDF
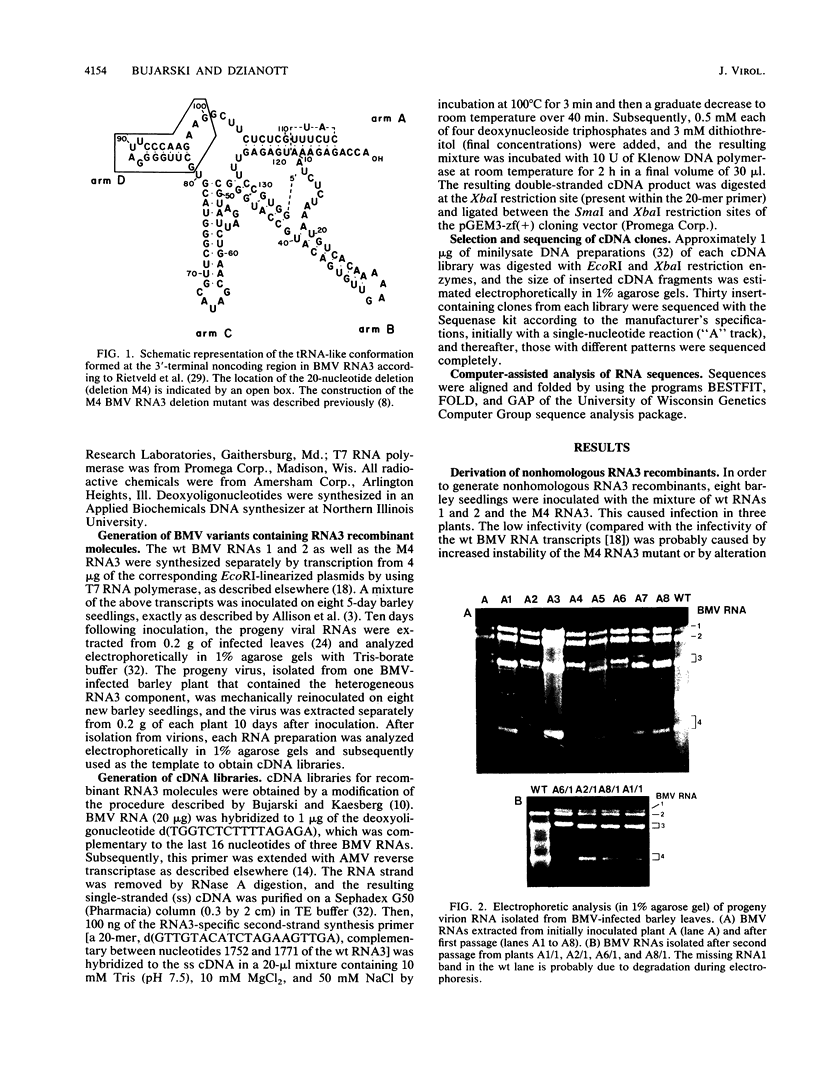

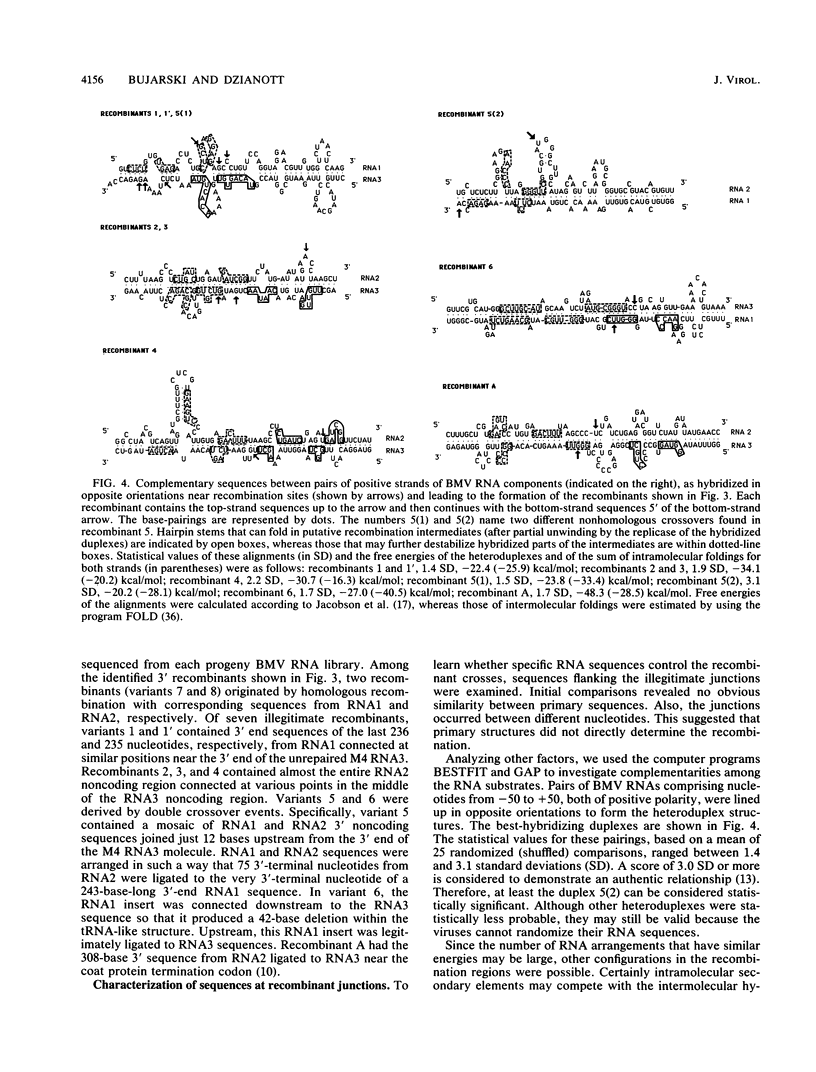
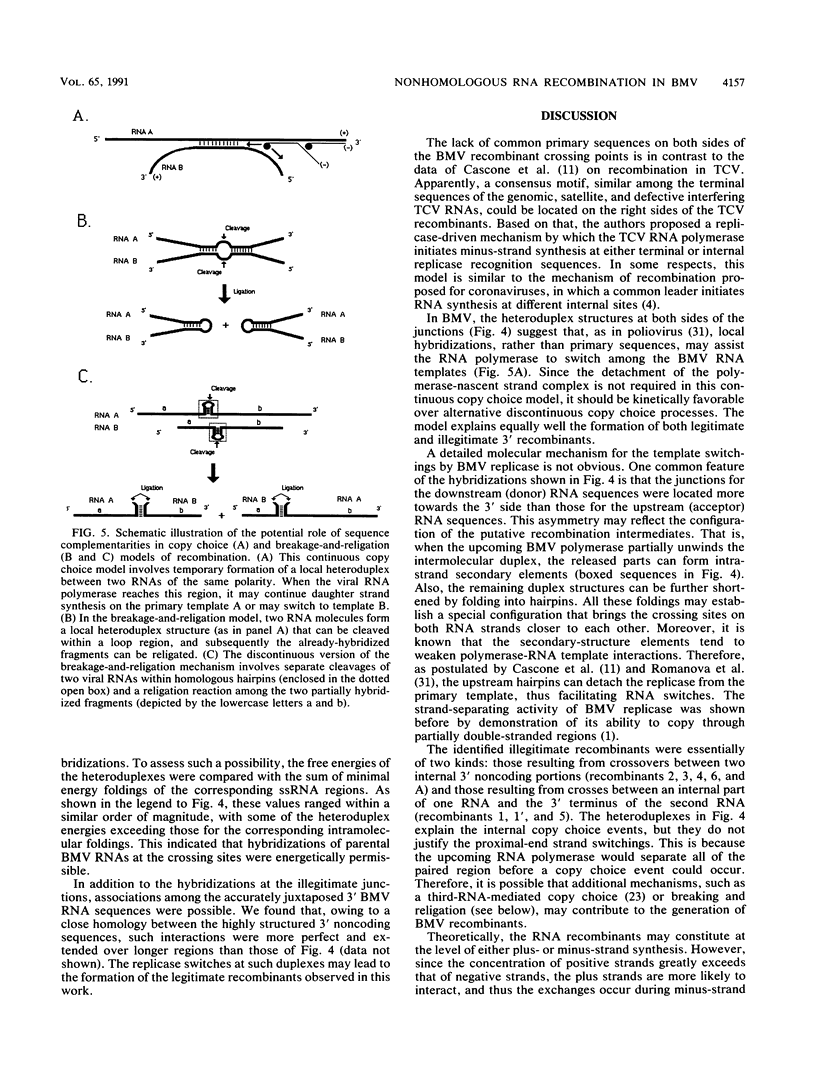


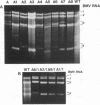
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Allison R., Thompson C., Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Lai M. M. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990 Dec;9(12):4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner L. R., Keck J. G., Lai M. M. A clustering of RNA recombination sites adjacent to a hypervariable region of the peplomer gene of murine coronavirus. Virology. 1990 Apr;175(2):548–555. doi: 10.1016/0042-6822(90)90439-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin M., Dasgupta R., Hall T. C., Kaesberg P. Similarity in structure and function of the 3'-terminal region of the four brome mosaic viral RNAs. J Mol Biol. 1976 Jun 5;103(4):737–745. doi: 10.1016/0022-2836(76)90206-0. [DOI] [PubMed] [Google Scholar]
- Bouzoubaa S., Niesbach-Klösgen U., Jupin I., Guilley H., Richards K., Jonard G. Shortened forms of beet necrotic yellow vein virus RNA-3 and -4: internal deletions and a subgenomic RNA. J Gen Virol. 1991 Feb;72(Pt 2):259–266. doi: 10.1099/0022-1317-72-2-259. [DOI] [PubMed] [Google Scholar]
- Bujarski J. J., Ahlquist P., Hall T. C., Dreher T. W., Kaesberg P. Modulation of replication, aminoacylation and adenylation in vitro and infectivity in vivo of BMV RNAs containing deletions within the multifunctional 3' end. EMBO J. 1986 Aug;5(8):1769–1774. doi: 10.1002/j.1460-2075.1986.tb04425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J. J., Dreher T. W., Hall T. C. Deletions in the 3'-terminal tRNA-like structure of brome mosaic virus RNA differentially affect aminoacylation and replication in vitro. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5636–5640. doi: 10.1073/pnas.82.17.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J. J., Kaesberg P. Genetic recombination between RNA components of a multipartite plant virus. 1986 May 29-Jun 4Nature. 321(6069):528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascone P. J., Carpenter C. D., Li X. H., Simon A. E. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 1990 Jun;9(6):1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson W. O., Lewandowski D. J., Hilf M. E., Bubrick P., Raffo A. J., Shaw J. J., Grantham G. L., Desjardins P. R. A tobacco mosaic virus-hybrid expresses and loses an added gene. Virology. 1989 Sep;172(1):285–292. doi: 10.1016/0042-6822(89)90130-x. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Fields S., Winter G. Nucleotide sequences of influenza virus segments 1 and 3 reveal mosaic structure of a small viral RNA segment. Cell. 1982 Feb;28(2):303–313. doi: 10.1016/0092-8674(82)90348-8. [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Carrington J. C., Morris T. J. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell. 1987 Nov 6;51(3):427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- Huisman M. J., Cornelissen B. J., Groenendijk C. F., Bol J. F., van Vloten-Doting L. Alfalfa mosaic virus temperature-sensitive mutants. V. The nucleotide sequence of TBTS 7 RNA 3 shows limited nucleotide changes and evidence for heterologous recombination. Virology. 1989 Aug;171(2):409–416. doi: 10.1016/0042-6822(89)90609-0. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M., French R., Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cdna and effects of 5' extensions on transcript infectivity. Virology. 1987 May;158(1):259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- Keck J. G., Stohlman S. A., Soe L. H., Makino S., Lai M. M. Multiple recombination sites at the 5'-end of murine coronavirus RNA. Virology. 1987 Feb;156(2):331–341. doi: 10.1016/0042-6822(87)90413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M., McCahon D., Slade W. R., Newman J. W. Recombination in RNA. Cell. 1982 Jul;29(3):921–928. doi: 10.1016/0092-8674(82)90454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986 Nov 7;47(3):433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Kawamura N., Nomoto A. Genetic variation occurring on the genome of an in vitro insertion mutant of poliovirus type 1. J Virol. 1989 Mar;63(3):1069–1075. doi: 10.1128/jvi.63.3.1069-1075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCahon D., King A. M., Roe D. S., Slade W. R., Newman J. W., Cleary A. M. Isolation and biochemical characterization of intertypic recombinants of foot-and-mouth disease virus. Virus Res. 1985 Jul;3(1):87–100. doi: 10.1016/0168-1702(85)90044-9. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Rao A. L., Hall T. C. Requirement for a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J Virol. 1990 May;64(5):2437–2441. doi: 10.1128/jvi.64.5.2437-2441.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A. L., Sullivan B. P., Hall T. C. Use of Chenopodium hybridum facilitates isolation of brome mosaic virus RNA recombinants. J Gen Virol. 1990 Jun;71(Pt 6):1403–1407. doi: 10.1099/0022-1317-71-6-1403. [DOI] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L. I., Blinov V. M., Tolskaya E. A., Viktorova E. G., Kolesnikova M. S., Guseva E. A., Agol V. I. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology. 1986 Nov;155(1):202–213. doi: 10.1016/0042-6822(86)90180-7. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Inglis S. C. The mutation rate and variability of eukaryotic viruses: an analytical review. J Gen Virol. 1987 Nov;68(Pt 11):2729–2740. doi: 10.1099/0022-1317-68-11-2729. [DOI] [PubMed] [Google Scholar]
- Tolskaya E. A., Romanova L. I., Blinov V. M., Viktorova E. G., Sinyakov A. N., Kolesnikova M. S., Agol V. I. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology. 1987 Nov;161(1):54–61. doi: 10.1016/0042-6822(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]