Abstract
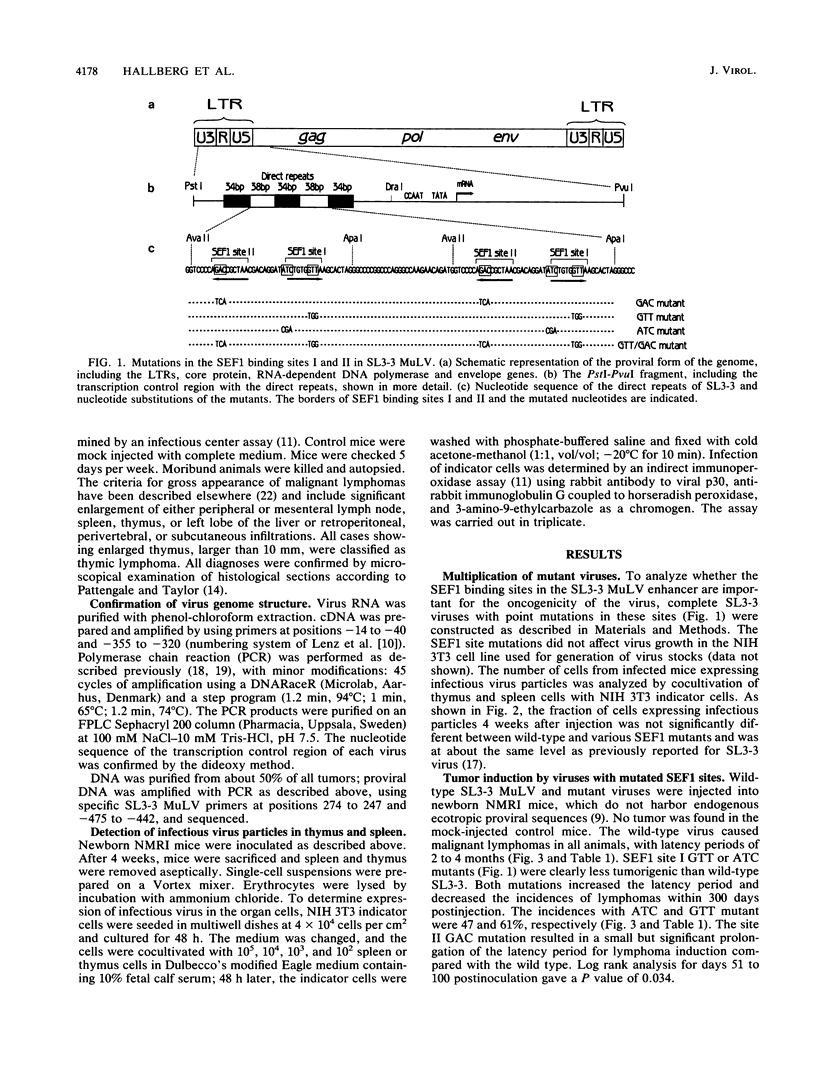
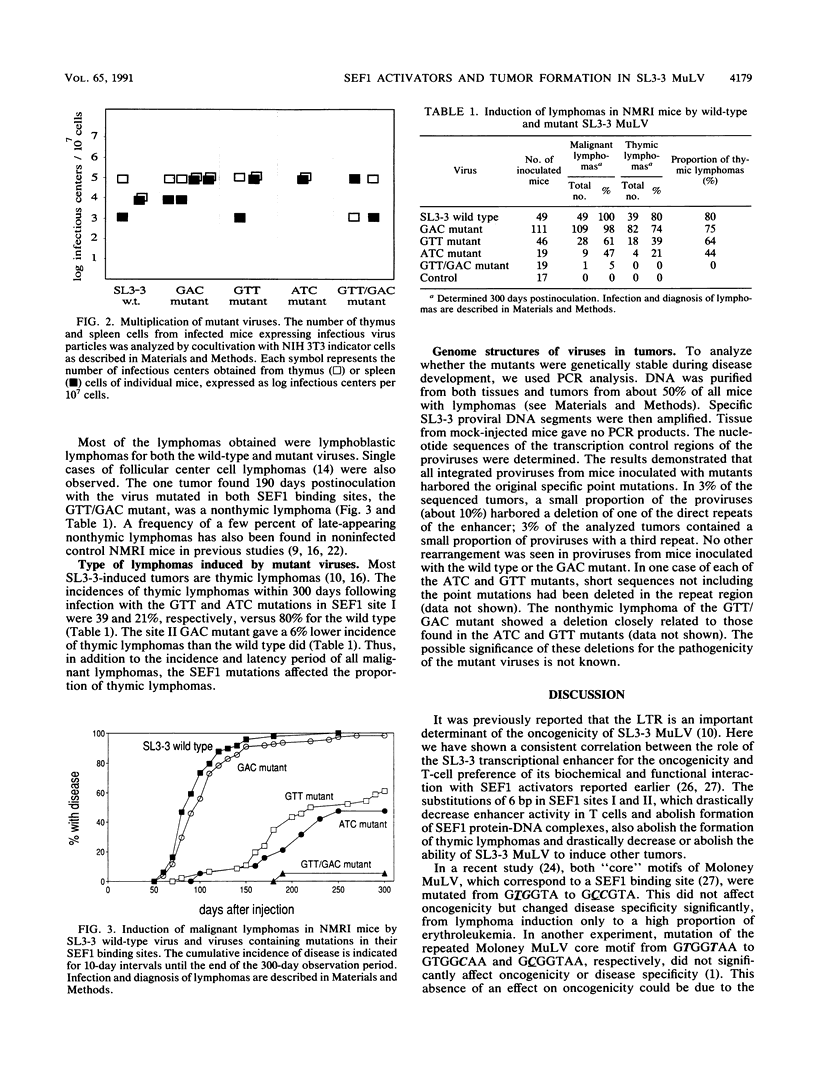
The transcriptional enhancers of retroviruses that lack an oncogene are important determinants of their oncogenicity. However, no specific cellular transcriptional activator has yet been found to determine the oncogenicity for any of these viruses. The SL3-3 enhancer factor 1 (SEF1) cellular transcriptional activators are expressed preferentially in T lymphocytes. In the SL3-3 murine leukemia virus enhancer, two different sequences can bind SEF1 activators. We show that mutation of the SEF1 binding sites disrupts the disease potential of SL3-3 murine leukemia virus, implying that SEF1 transcriptional activators are required for tumor induction by SL3-3. The SEF1 site mutations did not appear to affect the pathogenicity of SL3-3 by impairment of virus multiplication, but rather by a specific defect in the ability of neoplastic transformation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Case R. S., Khang Y. H., Kumar A., Yuen P. H. A single mutation in one of the CORE elements of Moloney murine leukemia virus reduced binding of a 42-kDa T lymphoma cell nuclear factor but did not affect lymphomagenesis. Mol Carcinog. 1990;3(2):93–102. doi: 10.1002/mc.2940030207. [DOI] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Gallagher R. E., Gallo R. C. Type C RNA tumor virus isolated from cultured human acute myelogenous leukemia cells. Science. 1975 Jan 31;187(4174):350–353. doi: 10.1126/science.46123. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hallberg B., Grundström T. Tissue specific sequence motifs in the enhancer of the leukaemogenic mouse retrovirus SL3-3. Nucleic Acids Res. 1988 Jul 11;16(13):5927–5944. doi: 10.1093/nar/16.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., Ogawa E., Asano M., Ishida S., Murakami Y., Satake M., Ito Y., Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990 Oct;64(10):4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib-Mösch C., Schmidt J., Etzerodt M., Pedersen F. S., Hehlmann R., Erfle V. Oncogenic retrovirus from spontaneous murine osteomas. II. Molecular cloning and genomic characterization. Virology. 1986 Apr 15;150(1):96–105. doi: 10.1016/0042-6822(86)90269-2. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Nexo B. A. A plaque assay for murine leukemia virus using enzyme-coupled antibodies. Virology. 1977 Apr;77(2):849–852. doi: 10.1016/0042-6822(77)90504-9. [DOI] [PubMed] [Google Scholar]
- Nilsson P., Hallberg B., Thornell A., Grundström T. Mutant analysis of protein interactions with a nuclear factor I binding site in the SL3-3 virus enhancer. Nucleic Acids Res. 1989 Jun 12;17(11):4061–4075. doi: 10.1093/nar/17.11.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan K., Dai H. Y., Duch M., Jørgensen P., Kjeldgaard N. O., Pedersen F. S. Different relative expression from two murine leukemia virus long terminal repeats in unintegrated transfected DNA and in integrated retroviral vector proviruses. J Virol. 1989 Dec;63(12):5201–5207. doi: 10.1128/jvi.63.12.5201-5207.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale P. K., Taylor C. R. Experimental models of lymphoproliferative disease. The mouse as a model for human non-Hodgkin's lymphomas and related leukemias. Am J Pathol. 1983 Nov;113(2):237–265. [PMC free article] [PubMed] [Google Scholar]
- Pedersen F. S., Crowther R. L., Tenney D. Y., Reimold A. M., Haseltine W. A. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumour. Nature. 1981 Jul 9;292(5819):167–170. doi: 10.1038/292167a0. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Satake M., Ibaraki T., Yamaguchi Y., Ito Y. Loss of responsiveness of an AP1-related factor, PEBP1, to 12-O-tetradecanoylphorbol-13-acetate after transformation of NIH 3T3 cells by the Ha-ras oncogene. J Virol. 1989 Sep;63(9):3669–3677. doi: 10.1128/jvi.63.9.3669-3677.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Erfle V., Pedersen F. S., Rohmer H., Schetters H., Marquart K. H., Luz A. Oncogenic retrovirus from spontaneous murine osteomas. I. Isolation and biological characterization. J Gen Virol. 1984 Dec;65(Pt 12):2237–2248. doi: 10.1099/0022-1317-65-12-2237. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985 Jul;4(7):1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991 Jan;65(1):42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Imler J. L., Chatton B., Schatz C., Wasylyk C. Negative and positive factors determine the activity of the polyoma virus enhancer alpha domain in undifferentiated and differentiated cell types. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7952–7956. doi: 10.1073/pnas.85.21.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]


