Abstract
Pax proteins, characterized by the presence of a paired domain, play key regulatory roles during development. The paired domain is a bipartite DNA-binding domain that contains two helix–turn–helix domains joined by a linker region. Each of the subdomains, the PAI and RED domains, has been shown to be a distinct DNA-binding domain. The PAI domain is the most critical, but in specific circumstances, the RED domain is involved in DNA recognition. We describe a Pax protein, originally called Lune, that is the product of the Drosophila eye gone gene (eyg). It is unique among Pax proteins, because it contains only the RED domain. eyg seems to play a role both in the organogenesis of the salivary gland during embryogenesis and in the development of the eye. A high-affinity binding site for the Eyg RED domain was identified by using systematic evolution of ligands by exponential enrichment techniques. This binding site is related to a binding site previously identified for the RED domain of the Pax-6 5a isoform. Eyg also contains another DNA-binding domain, a Prd-class homeodomain (HD), whose palindromic binding site is similar to other Prd-class HDs. The ability of Pax proteins to use the PAI, RED, and HD, or combinations thereof, may be one mechanism that allows them to be used at different stages of development to regulate various developmental processes through the activation of specific target genes.
Pax genes play key roles in regulating developmental processes and are characterized by a conserved motif, the paired box, which encodes the paired domain (PD; ref. 1). The PD is a DNA-binding domain (2) that is composed of two separable and independent subdomains, the N-terminal PAI and the C-terminal RED domains (3, 4), each containing a helix–turn–helix (HTH) motif (5). The PAI domain seems to be the most critical part of the PD (2, 5), because it is necessary and sufficient for DNA binding in vitro (2–4, 6). In fact, the Drosophila Prd protein does not need the RED domain to confer full function to the protein in vivo (7, 8). There are, however, situations in which the RED domain may play a role (3). The bipartite organization of the PD allows for the recognition of composite sites by both PAI and RED domain. Furthermore, there is an isoform of the Pax-6 protein (Pax-6 5a) that contains, in the middle of the recognition helix of the PAI domain, an 11-residue insertion that inactivates DNA binding to sequences normally bound by the Pax-6 PD (6). Pax-6 5a, however, is able to bind to a recently described sequence called 5aCON (6). Other Pax proteins also contain insertions in the PAI domain that inactivate DNA binding and uncover RED-domain DNA-binding activity (9). However, to date, there has been no report of a Pax protein containing only a PAI or a RED domain.
About half of the known Pax genes (1, 10–16) also encode a second DNA-binding domain, a Prd-class homeodomain (HD). The Prd-class HDs recognize DNA through cooperative dimerization mediated by the HD (17, 18). There are several Prd HD subclasses that are defined by the identity of the residue at position 50 of their HD (19). This residue is of critical importance for DNA recognition, and variations of this residue lead to dramatic changes in the specificity of DNA binding (18, 20, 21). It also affects the nature of the dimer formed between Prd-class HDs (19, 22). All members of the Prd-class HD that contain a serine at position 50 also encode a PD as a second DNA-binding domain (1).
Here, we report the identification of a Drosophila Pax gene that encodes a protein with only a RED domain and no PAI domain. This protein, initially named Lune, is the product of the gene eye gone (eyg; ref. 23) and herein will be referred to as Eyg. Eyg also contains a typical Prd-class HD with a serine at position 50. We show that Eyg is able to bind DNA either through the HD or through the RED domain. The RED-domain binding site is highly related to part of a site (5aCON) identified for the Pax-6 5a and Pax-5S isoforms (6, 9). eyg is expressed strongly in the embryo in the salivary gland placode (SGP) and in a striped pattern in the trunk of the embryo. As its name indicates, eyg plays a critical role in eye development (23) as well as in salivary-duct development (24).
MATERIALS AND METHODS
In Situ Hybridization.
Embryos were dechorionated in 100% bleach (Clorox) and were fixed in a mixture of 1× PBS/10% formaldehyde/heptane, 1:1:2 (vol/vol). The embryos were devitellinized in a methanol/heptane, 1:1 (vol/vol), mixture and were fixed again. RNA probes were made with digoxigenin-labeled uracil by using the Genius kit (Boehringer Ingelheim). Embryos were hybridized with RNA probes to the full-length eyg and to lacZ. The probes were detected by antibodies to digoxigenin conjugated to alkaline phosphatase. Antibodies to engrailed (En) were a gift from Steve Dinardo (The Rockefeller University, New York) and were detected with secondary antibodies that were coupled to horseradish peroxidase. Mutant alleles were placed in trans to lacZ-marked balancer chromosomes TM3β or SM6β. Homozygous mutant embryos were identified by the absence of staining when hybridized with a lacZ probe.
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) Assay.
Fusion proteins of glutathione S-transferase (GST) or N-terminal histidine with the RED DNA-binding domain or the HD DNA-binding domain were made by PCR with primers to either the RED domain or the HD. The RED domain included the regions defined by the Prd PD crystal structure as the linker region (from residue 36, GSLRPP) and the entire C-terminal RED subdomain (to residue 94, ILRNRA). The HD includes the 60 residues from residue 231 (FRRNRT) to the end of the HD (residue 291, WRRHQ). The fusion proteins were bound to beads.
Random nucleotide libraries consisting of oligonucleotides with variable regions of 30 nucleotides (RED-domain experiments) or 12 nucleotides (HD experiments; ref. 19) were used. SELEX experiments were performed as described (19). Briefly, the oligonucleotide library was incubated with fusion proteins bound to either GST-coated bead-bound fusion proteins (GST-tag) or nickel/nitrilotriacetic acid matrix-bound fusion proteins (histidine-tag) in a buffer containing 20 mM Tris (pH 8.0), 50 mM KCl, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, 20 μg/ml BSA, and 2 μg/ml poly(dI)⋅poly(dc). Samples were washed briefly with the same buffer [lacking BSA and poly(dI)⋅poly(dc)] and were boiled. Subsequently, the bound library sequences were amplified by PCR. Sequences for the RED and HD underwent five SELEX cycles before sequence analysis.
30-mer random oligonucleotide:
5′-CTAGGATCCGATACTCT CAGAN30CACAAGTGCAGTGGATCCTCG-3′
3′-TTCACGTCACCTAGGACG-5′
Binding Assays.
SELEX-derived binding sequences were radioactively labeled and incubated with bead/matrix-bound RED or HD fusion proteins. Samples were subsequently washed, resuspended in 1× loading buffer, boiled for 5 min, and loaded on an acrylamide gel. Incubation and wash-buffer compositions were identical to those used in the SELEX assay. Gel-shift assays were performed as described (19).
Oligonucleotides used for these studies contained the corresponding target sequence flanked on each side by 11 bp designed for low homology to the binding site. The exact sequences of the oligonucleotides used are shown in Table 1 (see also Fig. 6C).
Table 1.
Oligonucleotides used in binding assays
| Binding site | Sequence | No. of bp |
|---|---|---|
| 5aCON | GCATC ATGCTCAGTGAATGTTCATTGA CTACG | 32 |
| CGTAG TCAATGAACATTCACTGAGCAT GATGC | ||
| 5aCON1 | GTCAGAGTAAG ATGCTCAGTGA CAGGATC | 33 |
| CTCATTC TACGAGTCACT GTCCTAGCATG | ||
| 5aCON2 | GTCAGAGATAG ATGTTCATTGA CAGGATC | 33 |
| CTCTATC TACAAGTAACT GTCCTAGCATG | ||
| Lune RD1 | GATCCTGAGTC TCACT CTGTACG | 27 |
| GACTCAG AGTGA GACATGCCTAG | ||
| Lune RD2 | GATCCTGAGTC TCACTG CTGTACG | 28 |
| GACTCAG AGTGAC GACATGCCTAG | ||
| Lune RD3 | GTACTGCATAG ATGTTCACTGA AACTAGC | 33 |
| ACGTATC TACAAGTGACT TTGATCGCATG | ||
| Lune RD4 | GCTAGGATCGG TCACTGA GGGTCGG | 29 |
| CCTAGCC AGTGACT CCCAGCCATCG |
Boldface distinguishes the sequences of binding sites from flanking sequences.
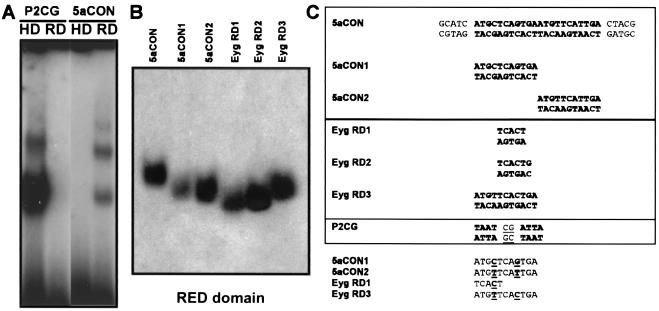
Figure 6.
DNA Binding of Eyg RED and HD. (A) Gel shift with the optimal HD (P2CG) or RED-domain (5aCON) sites bound by the HD or RED domains of Eyg. (B) Precipitation of radioactively labeled oligonucleotides containing various RED-binding sites by an Eyg GST-RED-domain fusion protein attached to glutathione beads. (C) Alignment of the various binding sites containing subsets of the Pax-6 5aCON sites or the consensus obtained by the SELEX experiment.
RESULTS
Cloning of a Pax-Like Gene in Drosophila.
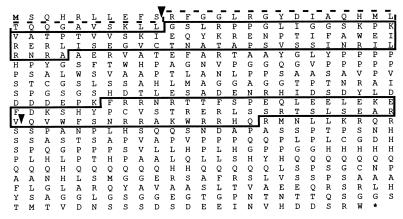
To identify Pax genes encoding both PD and HD, we searched for proteins that contain a Prd-class HD with a specific serine at position 50. All current members of this class possess a complete PD (i.e., vertebrate Pax-3 and Pax-7, Pax-4, Pax-6, Drosophila Prd, and the Gsb proteins; ref. 1). One such clone (BK27) had been identified in a southwestern screen by using oligomerized nonpalindromic HD binding sites (25) as a probe (26). The BK27 partial cDNA was used to isolate a full-length cDNA from a Drosophila embryonic library (a gift from N. Brown, University of Cambridge, Cambridge, U.K.). It encodes a protein with a partial PD that contains a RED but not a PAI domain (Fig. 1). This partial paired domain suggested that the RED domain might be able to act as an independent DNA-binding domain in Pax proteins and that there may be other Pax proteins that contain only the RED domain that have not been identified.
Figure 1.
Deduced protein sequence of Eyg. The solid lines frame the RED domain and the HD. The dashed line frames the weak region of homology to the PAI domain. Arrowheads indicate the locations of the two introns.
Molecular Characterization of Eyg.
The full-length protein was recently shown to be encoded by eyg and to have a critical role in salivary gland and eye development (23, 24). Indeed, two alleles of eyg exhibit fragment-size changes in their genomic DNA, and the eyg cDNA under the control of the heat-shock promoter is able to rescue the eyg phenotype (23). Eyg encodes a 523-aa protein (Fig. 1) with a RED domain and a HD (Fig. 2 A and B). Most PDs are located in close proximity to the N terminus of Pax proteins, and the RED domain of Eyg starts at position 36 of the protein. It exhibits very limited homology with the end of the PAI domain until residue 47 of the full-length PD. At position 48 (Fig. 2A), the homology becomes higher and extends 7 aa to position 54. The sequence is then interrupted by a 6-aa gap, at the position of a splice site that is not found in other PDs. The homology then continues from position 61 in the PD and remains uninterrupted to include the entire RED domain. There is a proline residue inserted after position 65 that is not found in other PDs.
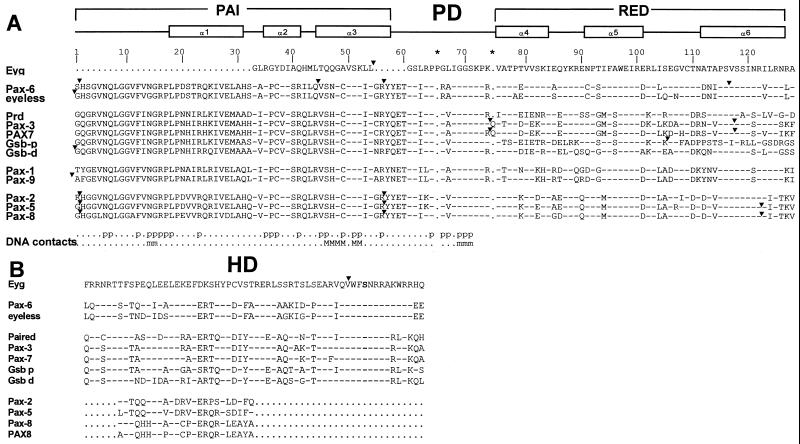
Figure 2.
Comparison of Eyg RED and HD sequences with other Pax proteins. Dashes indicate identity. (A) Comparison of Eyg RED domain with PD from each of the major subgroups. Above the comparison is a schematic that illustrates the position of the α-helices for the PAI and RED domains. Below the comparison, the residues that contact the DNA-binding site through phosphate (p), major groove (M) or minor groove (m) contacts have been indicated (5). The asterisks indicate additional residues within the PD that are found only in Eyg, Pax-3, or Pax-7. (B) Comparison of Eyg HD with that of other Pax genes. Arrowheads indicate the positions of introns.
Alignment of the Eyg RED domain with that of other complete PDs indicates that it is distinct from other known Pax classes (Fig. 2A). The linker region between PAI and RED domains is most related to that of Pax-2, Pax-5, and Pax-8, whereas the HTH region of its RED domain is most related to Pax-6. The most closely related PDs are those of Pax-6 (63 identical residues) and Pax-8 (60 identical residues).
Eyg also contains a Prd-class HD (26) with a serine at position 50 (Fig. 2B). Overall, the HD is most closely related to that of Pax-3 and Pax-7, although it is significantly distinct. The numbers of identical residues between Eyg and other HDs are 40 for Pax-7, 38 for Pax-6, and 36 for Gsb-p. The rest of the Eyg protein has few distinguishing features other than a glutamine-rich region found frequently in Drosophila proteins.
Therefore, although it is clearly a distinct Pax gene, the closest relative of Eyg seems to be Pax-6. This feature is interesting, because these two genes seem to act at the same level in eye development (23, 27). However, the RED domain of Eyg is most closely related to those of Pax-2, Pax-5, and Pax-8. Interestingly, the Pax-2, Pax-5, and Pax-8 proteins have only remnants of a HD, whereas Eyg and Pax-6 have a complete and functional HD.
Genomic Structure of Eyg.
A λ-EMBL3 library was screened with a DNA probe derived from the RED domain. The eyg coding region contains two introns and three exons. The first 680-bp intron interrupts the PD in the region preceding the linker region (Figs. 1 and 2) at a gap in the sequence where there seems to be a remnant of the recognition helix of the PAI subdomain (residue 54 of the PD). Although several of the splice sites are conserved among Pax genes belonging to the same class, this splice site is not (Fig. 2). The second 200-bp intron found within the homeobox is also found at this position in eve as well as other Prd-class homeobox sequences (e.g., gcs and otx; ref. 28).
Embryonic Expression Pattern of eyg.
eyg has an unique spatial and temporal expression pattern that is unlike other Pax genes in Drosophila. eyg seems to play a role in later stages of embryonic development, more specifically in salivary gland organogenesis (24) and eye development (23). In situ hybridization showed a dynamic pattern of expression (Fig. 3). eyg is first expressed ventrally in the labial lobe, in a cluster of cells that correspond to the region of the salivary gland progenitor cells that form the SGP (24, 29). At first, eyg is expressed nonuniformly in the placode (Fig. 3a), but expression then becomes more uniform; eventually, eyg is expressed strongly in all of the SGP cells. In embryos that have been double-stained with En antibody, eyg overlaps the anterior but not the posterior En stripe flanking the SGP (Fig. 3b). Thus, eyg is expressed in PS2. This expression is transient and fades away before the cells invaginate to form the salivary gland (24). Fading begins in the dorsoposterior regions of the SGP and progresses in a ventroanterior direction, forming a crescent shape before disappearing, hence the original name, lune (Fig. 3 g–i). eyg expression in the subantennal region of the head begins at the same time as in the SGP (Fig. 3c, arrowhead) and is persistent throughout embryonic development. It is reminiscent of the expression of the eyeless gene in the primordium of the eye, and it is described in more detail elsewhere (30). In the trunk, eyg is expressed in a segmental pattern, initially in single cells (Fig. 3c) within each segment, but then the expression expands to a patch of cells that are restricted ventrally, forming a striped pattern (Fig. 3d). Double-stained embryos show that this expression alternates with the En stripes (Fig. 3f). This expression fades to a narrow stripe during germ-band retraction and disappears (Fig. 3 d–f). It was determined, by using markers for the tracheal placodes, that eyg is expressed near, but not in, the tracheal placodes (N. Hacohen and M. Krasnow, personal communication). Finally, eyg expression is also found in the region around the posterior spiracle. This signal seems coincident with segmental expression and persists late in embryonic development after germ-band retraction (Fig. 3e, arrow).
Figure 3.
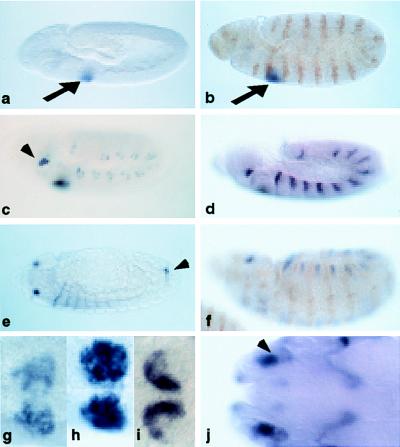
eyg expression. Embryos were hybridized with a digoxigenin-labeled RNA probe generated from a full-length eyg cDNA. (a) Expression begins in early stage 11 in the SGP. (b–f) eyg expression appears in the head and torso slightly later and persists through germ-band retraction. (e, f, and j) During later stages of development, eyg expression remains in the head. (g–i) These ventral views of the embryo focus on the SGP. Expression begins in nonuniform patches (g) and soon forms a uniform patch outlining the SGP (h). It is transient and begins to fade during stage 12, leaving a crescent-shaped patch of expression before fading (i). Embryos double-stained with anti-En antibody (b and d, in brown) show how eyg is expressed in PS2. Expression in the torso alternates and does not overlap with the En stripes (f). During later stages of development, eyg is expressed in several regions of the head (j). These cells may be neurons or may become sensory organs in the larva.
Regulation of eyg Expression.
Based on its pattern of expression, eyg may play a role in salivary gland organogenesis (30). SGP development responds to positional information. On the anteroposterior axis, Sex combs reduced (Scr) specifies PS2. In Scr− embryos, no salivary glands are formed (31) and eyg expression is lost (Fig. 4a), except for a small patch of cells present at the PS1/PS2 border. In a teashirt− mutation (32), Scr is expanded to both PS2 and PS3 and results in enlarged SGPs. The SGP expression of eyg is duplicated in PS3, although its appearance and fading are delayed slightly (Fig. 4b). Along the dorsoventrad axis, the SGP is restricted dorsally by decapentaplegic (dpp), and the spitz group of genes restricts it ventrally (31, 33). In dpp− embryos, eyg expression expands dorsally to form a ring that is interrupted ventrally (Fig. 4c). In several spitz-group mutant embryos, such as single minded (sim; Fig. 4d), the SGPs from each side move ventrally, and eyg expression expands ventrally. Expression in the trunk is also disordered, which may be a secondary effect of the disruption of the mesoderm.
Figure 4.
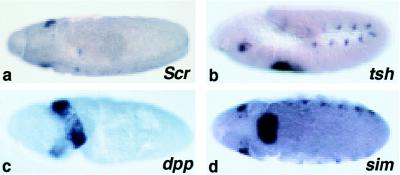
Regulation of eyg expression in mutants. Embryos were hybridized with RNA probes for eyg and lacZ. Scr determines the identity of PS2 and the anteroposterior position of the SGP in the embryo. In an Scr mutant embryo, no salivary placodes are formed, and eyg expression in the SGP is lost, except for a small patch of cells that correspond to the PS1/PS2 border (a) Expression in other segments and in the head is unaffected. In a teashirt− mutant embryo, Scr is ectopically expressed in PS3, and eyg expression is also expanded in PS3 (b) In a dpp− mutant embryo, eyg expression expands dorsally to form an incomplete ring that is only interrupted ventrally (c) dpp− embryos fail to gastrulate properly and the embryos appear twisted. (d) In a sim− embryo, the SGPs on each side fuse, and eyg expression expands ventrally and crosses the midline to form a band.
The RED Domain and HD of Eyg Recognize Specific DNA Sequences.
eyg encodes a PD without a PAI domain. Although the RED domain possesses a three-helix bundle, including an HTH structure similar to the PAI domain, it has been shown to play little or no role in the DNA binding of the PD of several Pax proteins, both in vitro and in vivo. However, it seems to provide additional specificity to binding by the PAI domain of Pax-5 and to stabilize DNA binding (3, 4).
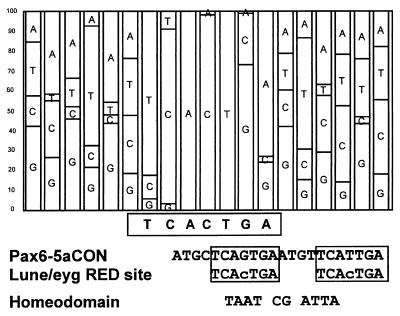
To address whether the RED domain of Eyg could act as an independent DNA-binding domain, we employed the SELEX assay to determine an in vitro DNA-binding site by using GST-RED or histidine-RED fusion proteins and a 30-nucleotide random oligonucleotide library (22). The 7-bp consensus sequence that can be derived (Fig. 5) contains five highly conserved residues forming a TCACT core and strong preferences for a G and an A at the sixth and seventh positions, respectively. The TCACT minimum site is sufficient for the RED domain to recognize and bind specifically (Fig. 6B). The TCA C TGA consensus forms a palindromic sequence that is very similar to either half of the Pax-6 5aCON binding site. Pax-6 5aCON is made of two direct repeats of an 11-bp sequence (Fig. 5; ref. 6). Therefore, we tested whether the RED domain of Eyg could also bind to the Pax-6 5aCON site or to either of the halves of the site, designated 5aCON1 and 5aCON2. Indeed, Eyg can bind with high affinity to the 5aCON site (Fig. 6A). Based on electrophoretic mobility-shift assay and methylation interference data for the Pax-6 5a protein, Epstein et al. (6) proposed that binding occurs not only as monomers and dimers but also as trimers and tetramers on the 22-nucleotide direct-repeat sequence. Gel retardation assays with the Eyg RED domain showed multiple bands, although two major bands were detected, and these results were consistent with binding of Eyg RED-domain multimers (Fig. 6A).
Figure 5.
(Upper) In vitro binding-site assays. SELEX-derived sequences for the RED domain of Eyg. The panels display base preferences in the form of a consensogram. The height of each bar represents the total percentage of instances that a given base occurs at a position. To construct the consensogram, 40 sequences were assessed and aligned. (Lower) Comparison of the Pax-6 5a binding site (5aCON) with the Eyg RED-domain binding site. Boxed regions emphasize regions of similarity.
There is only one difference between the Eyg 7-bp consensus sequence (RED domain, RD) and either 5aCON1 or 5aCON2 (Fig. 6C). We compared the affinity of Eyg to these three sites, which are different only at the central base pair. 5aCON1 and 2 also are different at the positions that precede the palindrome. Fig. 6B shows that the Eyg GST-RED fusion prefers RD3 and 5aCON2 over 5aCON1 and RD1. However, this difference is not significant and may only reflect minor requirements for sequences flanking the consensus. In particular, the presence of ATGT on the left of 5aCON1 may influence its recognition. Therefore, we conclude that the Eyg RED domain recognizes a sequence very similar to that of Pax-6. In the case of Pax-6, the binding can be performed either with a complete PD whose PAI domain is interrupted by an insertion (Pax-6 5a) or by a construction of the RED domain very similar to that used in this work (6, 9). This conclusion is consistent with recent crystallographic data of the Pax-6 PD bound to a composite site for PAI and RED domains that show that the important contact residues are identical between Eyg and Pax-6 (E. Xu, M. Rould, J. Epstein, R. Maas, and C. Pabo, personal communication).
The HD of Eyg is a Prd-class HD with a serine at position 50 and was expected to bind as a cooperative dimer to a palindromic site (22) made of two inverted core HD TAAT sites separated by 2 bp. This site is called P2; a similar palindromic site with 3 bp between the TAATs is recognized by other Prd–class HDs and is called P3 (19, 22). By using the SELEX (19) and gel-mobility shift assays, we showed that the Eyg HD prefers to bind to the palindromic sequence TAAT CG ATTA (Fig. 6A). After five rounds of selection, greater than 90% of the sequenced oligonucleotides contained this sequence, indicating that the Eyg HD has a strong preference for CG between the two TAAT motifs. This finding is unusual for Prd-class HDs with serine at position 50, which usually prefer to bind P2 or P3 sequences with CG or TG at the center of the consensus (19, 22).
DISCUSSION
Eyg Is an Additional Member of the Pax Gene Family.
Eyg contains a Prd-class HD with a serine at residue 50, which is always associated with a PD. The PD of Eyg contains only a RED and not a PAI domain. The Eyg RED domain most closely resembles the Pax-6 class, and its HD most closely resembles the Pax-3 and Pax-7 classes. However, the Eyg protein is clearly distinct from any Pax protein and therefore represents a distinct class of Pax proteins (Fig. 2A). Based on the crystal structure of the Prd PD, we infer that the RED domain of Eyg contains a portion of the α3 recognition helix of the HTH in the PAI domain, the linker region, and the entire HTH RED domain. The remnant of a PAI domain within the Eyg PD is reminiscent of the 26-aa N-terminal HD remnant found in Pax genes such as Pax-2, Pax-5, and Pax-8 (Fig. 2B). In these proteins, the remnant HD is highly unlikely to be involved in DNA binding. The crystal structure of the Prd HD (18) does not indicate a clear role for it. Similarly, the remnant PAI-domain α3-recognition helix found in Eyg is unlikely to contribute to DNA binding.
Regulatory Role of eyg in Embryogenesis.
The most prominent region of eyg expression is in the SGP. The salivary gland in the larva develops through differentiation of cells that eventually invaginate to form the organ. Based on its expression pattern, eyg may play a role in the early determination and differentiation of the cells as they begin to invaginate. It may play a similar regulatory role in the other regions where it is expressed. However, eyg does not seem to be involved in the development of tracheal placodes, the development of which is similar to that of SGPs.
The phenotypic analysis of an eyg mutant (23, 30) confirms that eyg is required for salivary gland development, but eyg also plays a crucial role in eye development. Furthermore, ectopic eyg expression induces ectopic eyes in the ventral margin of the eye (23).
DNA-Binding Specificity of Eyg.
The Eyg HD belongs to the Prd-class and binds to dimeric HD sites (19). However, its preferred sequence is clearly distinct from that of Prd, which prefers P2 or P3 sites (TAAT RY ATTA, in which R represents purine and Y, pyrimidine; ref. 22), or Pax-6, which prefers the P3 site (TAAT TGA ATTA; refs. 34 and 35). The significance of these preferences is not clear and is not explained by the structure of the Prd HD.
The PD is a bipartite DNA-binding domain (3, 5) made of two three-helix bundles. The PAI domain seems to be critical for the function of the Pax proteins (2–4, 6, 7), whereas the RED domain seems to have a role accessory to the PAI domain for Pax-5 and Pax-6 (3, 4). However, both PAI and RED domains are able to bind to DNA in some circumstances (6, 9). The PAI domain is always able to function independently, whereas the RED domain can bind DNA only when its activity is uncovered by disruption of the PAI domain (e.g., Pax-6 5a and Pax-5S isoforms) where insertions in the recognition helix disrupt DNA binding (6, 9). However, the DNA-binding role of the RED domain remains unclear.
Crystallographic studies of the Prd PD bound to DNA have shown that the recognition helix within the PAI domain contacts the major groove, whereas an N-terminal β-turn and a long linker region between the PAI and RED domains enter deep into the minor groove and establish extensive contacts with the DNA (5). The RED domain folds very similarly to the PAI domain and was predicted to contact DNA through its recognition helix. Eyg contains the entire linker region that is found between the PAI and RED domains, which in the case of the Prd protein, fits deeply into the minor groove and establishes numerous essential contacts with the DNA. Eyg contains the three-helix bundle that defines the RED domain and was predicted to recognize DNA in the major groove (5). Recent data from the structure of the Pax-6 PD bound to a composite PAI/RED-domain binding site confirmed this prediction and showed that the linker region may contribute extensively to the binding by the RED domain (E. Xu, M. Rould, J. Epstein, R. Maas, and C. Pabo, personal communication). The Pax-6 PD structure indicates that the residues in the RED-domain recognition helix contacting the major groove are highly conserved among all PDs (except Prd) and, in particular, are identical in Eyg and in Pax-6 (Fig. 2A). Therefore, the region present in the Eyg protein should be sufficient to bind DNA with high affinity.
The DNA-binding specificity of the Eyg RED domain most closely resembles that of the Pax-6 PD subclass (5aCON site). However, the 5aCON site contains a direct repeat of a palindromic sequence, whereas the Eyg RED-domain site seems to prefer a single palindromic sequence. It was argued (9) that two copies of the RED domain of Pax-6 and Pax-8 bind on both faces of the helix on each repeat of the 5aCON site. The palindromic nature of each repeat allows for each molecule to recognize the short half of the palindrome. Although Pax-6 prefers the direct repeat structure, Eyg does not seem to require it.
There is limited potential for discrimination between the DNA-binding activities of the Pax-6 and Eyg RED domains, and they are likely to bind to the same sequences in vivo. This close relation of DNA-binding sites is a common occurrence in many DNA-binding proteins and is particularly striking in the case of homeoproteins (25, 36). Therefore, it is unlikely that the presence of both a RED domain and an HD in Pax-6 and Eyg allows for a different set of target genes. Instead, these two genes may compete for the same target sites in the promoter of genes involved in eye development.
A Role for Independent HTH Motifs Within Complex DNA-Binding Proteins.
Although previous studies showed that the RED domain may act as an independent DNA-binding domain, Eyg does not contain both a PAI and RED domain. There may be other Pax genes that encode only a PAI or a RED domain. Studies with Pax-5 clearly show that some Pax proteins require both PAI and RED domains to recognize and bind to specific DNA sequences. Studies with Prd show that its PAI domain alone is sufficient for full function in Drosophila (7). Our studies with Eyg show that the RED domain may act as an independent DNA-binding domain and does not require an interaction with the PAI domain to recognize and bind to specific DNA sequences. We propose that there may be a family of Pax genes that contain only the PAI or RED domain. Even within Pax proteins that do contain both PAI and RED domains, the RED domain may act independently of or interdependently with the PAI domain to increase the functional diversity and range of binding sites that the whole protein can recognize. Similarly, studies with Prd and other Pax proteins have shown that the PAI domain may interact with the HD, again, to increase the range of functional diversity (4).
It seems that a series of Pax proteins have evolved with different combinations of at least partially independent DNA-binding domains. Some proteins have lost one of the domains—Pax-2, Pax-5, and Pax-8 have lost most of the HD; Eyg has lost its PAI domain—or have lost the functional requirement for a domain—Prd does not require the RED domain for its in vivo function (7).
This ability to use the PAI domain, RED domain, HD, or any combination thereof would give Pax genes a large regulatory potential that could be reused at different stages of development. For instance, Pax-6 is required for early definition of the eye progenitor region, then in the developing eye disc, and finally during terminal differentiation of the photoreceptors (35). In this way, these genes become “master” regulatory genes that play very specific roles during organogenesis (37).
Acknowledgments
We thank Kathy Neal, Rachel Stott, David Wilson, Ernst Wimmer, Pat O’Farrell, Yien Ming Kuo, Steve Beckendorf, Laurent Fasano, and Henry Sun. This work was supported by a William O’Baker Fellowship (to S.J.) and a National Health and Medical Research Council (Australia) C. J. Martin Fellowship (to B.K.).
ABBREVIATIONS
- En
engrailed
- GST
glutathione S-transferase
- HD
homeodomain
- HTH
helix–turn–helix
- PD
paired domain
- RD
RED domain
- SELEX
systematic evolution of ligands by exponential enrichment
- SGP
salivary gland placode
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF091739).
References
- 1.Noll M. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 2.Treisman J, Harris E, Desplan C. Genes Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- 3.Czerny T, Schaffner G, Busslinger M. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 4.Jun S, Desplan C. Development (Cambridge, UK) 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Rould M, Jun S, Desplan C, Pabo C. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 6.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 7.Bertuccioli C, Fasano L, Jun S, Sheng G, Desplan C. Development (Cambridge, UK) 1996;122:2673–2685. doi: 10.1242/dev.122.9.2673. [DOI] [PubMed] [Google Scholar]
- 8.Cai J, Lan Y, Appel L F, Weir M. Mech Dev. 1994;47:139–150. doi: 10.1016/0925-4773(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 9.Kozmik Z, Czerny T, Busslinger M. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- 11.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Cell. 1986;47:1033–1049. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 12.Bopp D, Jamet E, Baumgartner S, Burri M, Noll M. EMBO J. 1989;8:3447–3457. doi: 10.1002/j.1460-2075.1989.tb08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther C, Guenet J L, Simon D, Deutsch U, Jostes B, Goulding M D, Plachov D, Balling R, Gruss P. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 14.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 15.Wallin J, Mizutani Y, Imai K, Miyashita N, Moriwaki K, Taniguchi M, Koseki H, Balling R. Mamm Genome. 1993;4:354–358. doi: 10.1007/BF00360584. [DOI] [PubMed] [Google Scholar]
- 16.Stapleton P, Weith A, Urbanek P, Kozmik Z, Busslinger M. Nat Genet. 1993;3:292–298. doi: 10.1038/ng0493-292. [DOI] [PubMed] [Google Scholar]
- 17.Wilson V, Rashbass P, Beddington R S. Development (Cambridge, UK) 1993;117:1321–1331. doi: 10.1242/dev.117.4.1321. [DOI] [PubMed] [Google Scholar]
- 18.Wilson D S, Guenther B, Desplan C, Kuriyan J. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 19.Wilson D S, Sheng G, Jun S, Desplan C. Proc Natl Acad Sci USA. 1996;93:6886–6891. doi: 10.1073/pnas.93.14.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 21.Hanes S D, Brent R. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 22.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 23.Jang, C. C., Chao, J. L., Jones, N., Bessarab, D., Kuo, Y., Jun, S., Desplan, C., Beckendorf, S. & Sun, H. Y. (1998) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 24.Kuo Y M, Jones N, Zhou B, Panzer S, Larson V, Beckendorf S K. Development (Cambridge, UK) 1996;122:1909–1917. doi: 10.1242/dev.122.6.1909. [DOI] [PubMed] [Google Scholar]
- 25.Desplan C, Theis J, O’Farrell P H. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalionis B, O’Farrell P H. Mech Dev. 1993;43:57–70. doi: 10.1016/0925-4773(93)90023-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazelett, D., Bourouis, M., Walldorf, U. & Tresisman, J. E. (1998) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 28.Duboule D. Guidebook to the Homeobox Genes. New York: Oxford Univ. Press; 1994. [Google Scholar]
- 29.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer; 1985. [Google Scholar]
- 30.Jones, N. A., Kuo, Y. M., Sun, Y. S. & Beckendorf, S. K. (1998) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 31.Panzer S, Weigel D, Beckendorf S K. Development (Cambridge, UK) 1992;114:49–57. doi: 10.1242/dev.114.1.49. [DOI] [PubMed] [Google Scholar]
- 32.Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, Kerridge S. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- 33.Hacker U, Grossniklaus U, Gehring W J, Jackle H. Proc Natl Acad Sci USA. 1992;89:8754–8758. doi: 10.1073/pnas.89.18.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng G, Thouvenot E, Schmucker D, Wilson D, Desplan C. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- 36.Biggin M D, McGinnis W. Development (Cambridge, UK) 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 37.Callaerts P, Halder G, Gehring W J. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]