Abstract
Subcellular compartmentalization, cell growth, hormone secretion and neurotransmission require rapid, targeted, and regulated membrane fusion. Fusion entails extensive lipid rearrangements by two apposed (docked) membrane vesicles, joining their membrane proteins and lipids and mixing their luminal contents without lysis. Fusion of membranes in the secretory pathway involves Rab GTPases; their bound ‘effector’ proteins, which mediate downstream steps; SNARE proteins, which can ‘snare’ each other, in cis (bound to one membrane) or in trans (anchored to apposed membranes); and SNARE-associated proteins (SM proteins; NSF or Sec18p; SNAP or Sec17p; and others) cooperating with specific lipids to catalyze fusion. In contrast, mitochondrial and cell-cell fusion events are regulated by and use distinct catalysts.
Early studies with bilayer liposomes provided important insights about fusion mechanisms (see review by Chernomordik and Kozlov in this issue1). Liposome fusion can be induced by calcium, polyethylene glycol, diacylglycerol, peptides or high membrane curvature, but is often accompanied by substantial lysis2–4, in which the membrane permeability barrier to polar solutes is lost. Fusion proceeds through a stalk or hemifusion intermediate (Fig. 1) in which the outer bilayer leaflets are merged while the inner leaflets, and aqueous compartments, remain distinct5. Viral fusion proteins can catalyze rapid fusion alone (see review by Harrison in this issue6). While anchored in one bilayer, viral fusion proteins unfold like a flower to reveal a hydrophobic fusion peptide that inserts into an apposed membrane. The fusion protein then oligomerizes and undergoes a conformational change that stresses the apposed bilayers, inducing their fusion7. Viral fusion also entails a hemifusion stalk (see Fig. 1 of ref. 8) and is accompanied by some lysis9,10. Though viral fusion proteins can be indiscriminate in their targeting, the similarity of their structures and actions to SNAREs (see Box 1 for definition) has been a powerful paradigm for studies of intracellular fusion11. Cell-cell fusion events and mitochondrial fusion events rely on quite distinct strategies.
Figure 1.
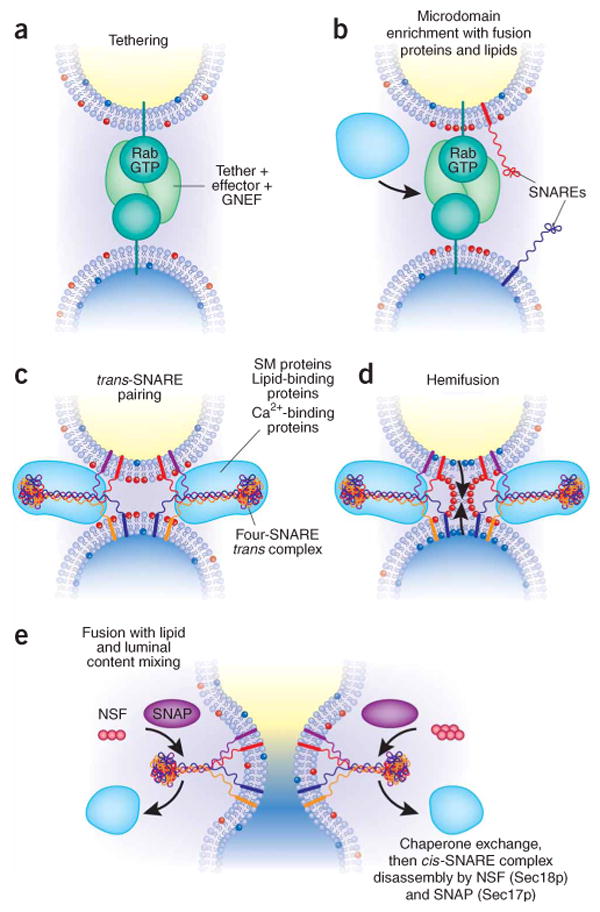
Membrane fusion on the exocytic and endocytic pathways, in five steps. (a) The first association of membranes, termed ‘tethering’, requires a prenyl-anchored Rab-family GTPase and tethering proteins termed ‘effectors’124, which bind to the Rab in its activated, GTP-bound state. Proteins mediating tethering have been studied in the Golgi stacks125,126 and other systems. (b) Rab-regulated enrichment of fusion proteins and lipids in a microdomain. Rabs, their multi-functional effector complexes, and lipids with defined roles in fusion (such as sterols (not shown) or phosphoinositides or diacylglycerol (red polar head groups)) assemble into a microdomain, the site of subsequent steps in the fusion pathway. In some systems, Rab effectors can include guanine nucleotide exchange factors, which activate Rabs; lipid kinases, which synthesize phosphoinositides; and SM proteins, which bind SNAREs. It remains unclear in most instances whether Rab effectors must remain bound to the Rab to be activated for these functions, or whether concentration of these several protein and lipid factors in the fusion microdomain suffices. In some systems, such as the yeast vacuole, one multisubunit complex fulfills tethering, guanine nucleotide exchange, SM protein, and lipid-binding functions. (c) Assembly of trans-SNARE complexes127 with additional regulatory proteins. These include SM proteins48 and can include proteins or domains that bind to Ca2+, to lipids or to SNAREs. These complexes may encircle the site of future fusion44. Lipids with small head groups and negative membrane curvature, which promote hemifusion, are enriched at the cytoplasmic surface of the fusion microdomain (red head groups). (d) Hemifusion, formed by fusion of the halves of the lipid bilayer of each membrane that face the cytoplasm. Arrows indicate the direction of subsequent lipid movement to complete the fusion process. Lipids with positive curvature due to large head groups (shown here as blue head groups) may become enriched at this stage for invasion of the hemifused structure (arrows). (e) Completion of fusion, with mixing of lipid bilayers, membrane proteins and luminal compartments but retention of the barrier between cytoplasm and organellar lumen. This process converts trans-SNARE complexes to post-fusion cis-SNARE complexes; it is unclear whether cis-SNARE complexes can arise by any other route. SNAP (Sec17p) may displace other SNARE-bound proteins and prepare the cis-SNARE complex for ATP-dependent disassembly by NSF (Sec18p).
BOX 1. Membrane fusion: a glossary of basic terms.
SNAREs: Soluble NSF/alpha SNAP receptors: proteins with conserved heptad repeats that bind to (snare) each other in four-helix bundles (complexes), in cis (with each transmembrane domain anchored to the same bilayer) or in trans (with transmembrane domains anchored in apposed bilayers, before fusion).
SNAP and Sec17p: Proteins that bind SNAREs and permit NSF or Sec18p to perform ATP-driven SNARE complex disassembly.
NSF and Sec18p: Hexameric AAA ATPases that actively disassemble SNARE complexes into SNARE monomers.
Rab/Ypt: A subfamily of Ras GTPases, with unique members on each organelle and defining roles in tethering, fusion microdomain assembly, and trans-SNARE complex formation.
Rab-effector: A protein that binds to a GTP-activated Rab, then performs a downstream function in fusion.
SM protein: Sec1/Munc18-1 family proteins that bind to SNAREs and are required for their physiological promotion of fusion.
Tethering: An initial, reversible stage of membrane association, requiring Rab and effector(s) but not trans-SNARE associations.
Docking: All stages of membrane association that lead to fusion.
Fzo1/Mfn1: dynamin-like GTPase responsible in part for mitochondrial tethering and outer membrane fusion.
Ugo1: A mitochondrial outer membrane protein that links Fzo1/Mfn1 to the inner membrane and helps coordinate fusion.
Ugo1: A mitochondrial outer membrane protein that links Fzo1/Mfn1 to the inner membrane and helps coordinate fusion.
Mgm1/OPA1: A mitochondrial inner membrane dynamin-like GTPase that is required for inner membrane fusion.
EFF1/AFF1: C. elegans epithelial and anchor cell plasma membrane proteins required for cell fusion.
Fus1: Yeast plasma membrane protein that promotes the bridging of mating cells in preparation for cell fusion.
Fus2: Yeast peripheral membrane protein that acts in conjunction with Fus1 to promote cell wall merger in mating pairs.
Prm1: Polytopic plasma membrane protein that promotes plasma membrane fusion in yeast mating pairs.
Our understanding of intracellular membrane fusion has largely rested on three approaches: genetic screens for the relevant genes; the enumeration of the proteins and their binding associations in the neuronal synapse, which is highly specialized for rapid fusion; and the development of in vitro reactions reconstituting organelle fusion and the biochemical identification of their essential components. The consilience between the proteins identified through these approaches, and the substantial conservation of components from yeast to man and among the intracellular organelles, shows that biological membrane fusion occurs by conserved mechanisms. However, work in the last decade has shown that mitochondrial fusion is catalyzed by a distinct repertoire of proteins.
Molecular insights from a genetic approach
Secretion mutants defective in mucocyst discharge in Paramecium and Tetrahymena were described and characterized morphologically in the mid 1970s (ref. 12). However, the difficulty of molecular and genetic analysis in protozoa limited the potential impact of these early, fascinating studies. Studies on a genetically tractable system began with the isolation of Saccharomyces cerevisiae temperature-sensitive sec mutants, each blocked at a stage of the secretory pathway13,14. Some of the SEC genes encode general fusion chaperones such as Sec17p and Sec18p, whereas others specify proteins that catalyze fusion at one organelle. Genes involved in the budding or cargo-sorting stages of trafficking were resolved from those required for the later fusion step by epistasis analysis15 and by biochemical complementation experiments as in vitro fusion reactions became available16,17. In other screens, defects in endomembrane trafficking yielded vps (vacuole protein sorting) mutants18,19, in which inefficient protein sorting from the Golgi to the vacuole (lysosome of yeast) results in the secretion of normally vacuolar proteins. A screen for abnormal vacuole morphology (vam) mutants with highly fragmented vacuoles had notable success in identifying the genes required for the fusion of this organelle20. The discovery that the SEC18 gene encodes a yeast homolog of mammalian NSF was an early indicator that the proteins of membrane fusion are highly conserved21.
The neuronal synapse
Synaptic vesicles are highly enriched in proteins that mediate vesicle fusion at the active zone22. Neuronal SNARE proteins were initially discovered by their abundance in synaptic vesicle preparations and their associations with one another23–25, then shown to be the targets of specific endoproteolytic neurotoxins26, establishing their direct role in membrane fusion. SNAREs associate with other proteins, notably Sec1/Munc18 proteins, synaptotagmin and complexin, that regulate their association and function and integrate it with the triggering calcium flux27–29. Two SNARE-bound fusion factors, NSF and SNAP, couple the energy of ATP binding and hydrolysis to the disassembly of SNARE complexes30,31. SNAREs are found in all species and all organelles32. Electrophysiological characterization of SNARE gene knockout mice has established their roles in normal neurotransmission. Synaptotagmin has direct affinity for the SNAREs33. As calcium enters the synapse, the calcium binds to the two C2 domains of synaptotagmin, activating binding of synaptotagmin to lipids and SNAREs, displacing complexin34 and triggering fusion. The inherent interest in understanding the human brain, as well as the leading role of neuronal SNAREs in studies of the SNARE complex structure35 and function in liposome model reactions36–38, has made neuronal fusion catalysis a leading system for molecular analysis.
In vitro fusion reactions
The luminal compartment mixing that occurs upon membrane fusion provides an assay of the fusion event. Golgi fractions, isolated from vesicular stomatitis virus–infected CHO cells lacking an N-acetyl glucosamine transferase, have viral G protein (VSV-G) with abnormal glycosylation. Upon incubation with Golgi from wild-type, uninfected cells, the fusion step of trafficking delivers the VSV-G protein from the infected cell's mutant Golgi to the mannosyl transferase within wild-type Golgi, and the mannose addition can be assayed as a marker of fusion16. This assay is blocked by N-ethylmaleimide (NEM), and complementation of the NEM-blocked reactions by fresh cytosol allowed isolation of the NEM-sensitive factor, or NSF, a soluble peripheral membrane protein39. NSF binding to the Golgi requires a second peripheral membrane protein, the soluble NSF attachment protein, or SNAP40,41. Affinity chromatography of brain detergent extracts on a matrix of immobilized SNAP and NSF yielded a complex of syntaxin, synaptobrevin and SNAP-25 (ref. 31); these proteins had been identified in earlier studies and proposed to be central in synaptic transmission, but their specific association with NSF and SNAP established this role. Syntaxin, synaptobrevin and SNAP-23 were then termed SNAREs, for soluble NEM-sensitive factor attachment protein receptors.
SNAREs are found in all eukaryotic organisms and are required for each step of the exocytic and endocytic trafficking pathways. They have a common heptad-repeat SNARE motif, which forms four-helix coiled-coil structures35 termed SNARE complexes. Though most aminoacyl residues that are buried within the four-helix SNARE complex structure are apolar, the ‘0-layer’ at the center is almost invariably comprised of three glutaminyl residues and one arginyl. This has formed the basis for the classification of SNAREs as Q- or R-SNAREs32, with further refinement of Q-SNAREs according to sequence conservation into subfamilies Qa, Qb and Qc. Virtually all characterized natural SNARE complexes are of the composition QaQbQcR. SNAREs also have variable N-terminal domains that precede the heptad repeat. Most SNAREs have a single transmembrane anchor near their C terminus, though some are anchored by prenyl groups or by a phosphoinositide-binding domain42. SNARE complexes are termed cis if all their membrane anchors are in one bilayer or trans if they have anchors in each of two apposed, docked membranes. SNARE complex disassembly, assembly and function are regulated by SNARE-associated factors. SNARE complexes are disassembled by NSF or Sec18p, which are AAA-family ATPase chaperones, along with SNAP or Sec17p (ref. 30). SNAREs become enriched in fusion-competent microdomains43,44 and pair in trans36,45,46, with oligomerization of trans-SNARE complexes47. SNARE pairs are often associated with other proteins, most generally of the Sec1/Munc18-1 (SM) family.
SM proteins are required for each intracellular fusion event48. Different SM proteins associate with SNARE complexes49,50, with the N-terminal peptide region of certain syntaxin (Qa) family members51,52 or with the folded N-terminal domain of other syntaxins53. Despite structural studies of SNARE–SM cocrystals54, it has been unclear how SM proteins promote trans-SNARE pairing and fusion. Recent studies38 suggest that the neuronal SM protein Munc18-1 catalyzes the formation of ternary complexes of the cognate neuronal SNAREs, activating them for lipid mixing (as discussed below) while suppressing this capacity for other SNARE combinations55. Since each SM protein is physically and functionally specific to a limited number of organelles, the SM proteins may provide a vital layer of specificity to trans-SNARE complex assembly and function.
GTPases of the Rab family are also essential for fusion. Sec4p, required for docking secretory vesicles at the yeast plasma membrane, was found to be a Ras-superfamily GTPase56 whose cycling between its GDP and GTP-bound forms57 is controlled by GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GNEFs)58. In its GTP-bound form, Sec4p binds to a complex of proteins termed the exocyst, linking the secretory vesicle to the plasma membrane59–61. As at the exocyst, docking and fusion at each organelle requires a unique large tethering complex—for example, the TRAPP complex at the cis-Golgi62 and the HOPS complex at the vacuole63—that interacts with the Rab GTPase and other factors for tethering and to catalyze the exchange of GTP for GDP and thereby activates the Rab64–66. GTPases of the same family as Sec4p, termed Rab or Ypt proteins, were found for every organelle and throughout the various taxa studied. For example, the Rab5 GTPase is required for mammalian endosomal homotypic fusion67. When detergent extracts of endosomes were passed over a column of immobilized Rab5p–GTP, a broad selection of specifically bound proteins was found, each subsequently shown to be active in cell-free assays of endosome fusion68–71. Some of these Rab5–GTP ‘effectors’ catalyze phosphoinositide synthesis71, bind directly to phosphoinositides72, or both73, providing an early link between the proteins and lipids of membrane fusion. These lipids contribute to the formation of fusion-active microdomains (Fig. 1b), serve as a platform for binding fusion proteins to the membrane, and themselves undergo non-bilayer rearrangements during hemifusion and fusion (Fig. 1d,e).
Membrane microdomains can form specialized platforms for fusion43,74. This has been especially clear in studies of the fusion of mammalian endosomes and of isolated yeast vacuoles. The endosomal Rab5 is activated by the GNEF Rabex5 and, in its active form, binds the effector Rabaptin5 (ref. 68) to form a ternary complex69. Rabaptin5 in turn activates Rabex5 (ref. 75), insuring that Rab5 remains in its GTP-bound form and is not extracted by the guanine-nucleotide dissociation inhibitor. Activated Rab5 also binds phosphatidylinositol-3-OH kinases71 and phosphoinositide 4- and 5-phosphatases, which create more phosphatidylinositol substrate for the 3-kinase to act upon76. The local synthesis of PI(3)P allows high-affinity localization of the effector protein EEA1, which has affinity for both activated Rab5 and, through its FYVE domain, for PI(3)P (ref. 73). These microdomains, organized by Rab5, provide favored sites of endosomal membrane fusion70,77. Yeast vacuole homotypic fusion is another example of Rab-directed microdomain assembly. Vacuoles have diameters of a micron or more, allowing fluorescence light microscopic visualization of docking, of the ensuing spatial distribution of fusion proteins and lipids on docked vacuoles, and of the fusion event itself. Docked vacuoles are drawn against each other to form pairs of apposed disc-like microdomains, the ‘boundary membrane’. The region surrounding the boundary membrane, termed the ‘vertex ring’, becomes highly enriched in the Rab (Ypt7p), Rab-effector complex (HOPS), SNAREs, and ‘regulatory lipids’ (ergosterol, diacylglycerol and phosphoinositides) that are required for fusion44,78,79. Fusion itself then occurs around the vertex ring, joining the two boundary membranes and yielding a luminal vesicle within the larger, fused organelle. The regulatory lipids (diacylglycerol, ergosterol and phosphoinositides) depend on one another for vertex ring enrichment, and the Rab, Rab-effector and SNAREs are interdependent for vertex accumulation. Unexpectedly, regulatory lipid enrichment also depends on these proteins, and these proteins require the lipids as well44,79. As shown for endosomal fusion components, the relevant vacuolar proteins and lipids undergo a complex and highly interdependent process to establish a fusion-competent microdomain79. Each of the proteins and lipids needed for the establishment of a vertex ring microdomain is needed until fusion, presumably for microdomain maintenance80.
Fusion reconstitution
Reconstitution of fusion with all-pure components, and in a manner that faithfully reflects the biology seen in in vivo and in in vitro studies with the intact organelle, is an essential, if challenging, step toward understanding the chemistry of the protein-catalyzed lipid rearrangements of fusion. Viral fusion proteins provide an important model, though they are not likely to reveal how essential catalysts such as Rabs and Rab-effectors work.
The fusion of proteoliposomes bearing pure, recombinant SNAREs has been studied extensively. In this system, one set of ‘donor’ liposomes is prepared with two self-quenching fluorescent lipids, rhodamine-phosphatidylethanolamine and NBD-phosphatidylethanolamine, while the ‘recipient’ liposomes are not fluorescent. Upon fusion, dilution of the fluorescent lipids causes dequenching81. Complementary sets of recombinant SNARE proteins are reconstituted into either the donor or acceptor liposomes36, representing the SNAREs believed to function on the target organelle (t-SNARE) or vesicle (v-SNARE) for particular fusion events. After overnight pre-incubation at 4 °C, presumably allowing trans-SNARE pairing, samples are warmed and the kinetics of fluorescence increase is recorded. This system, despite technical limitations82,83, has shown that, for those cognate pairs of SNAREs that normally function together in the cell, SNAREs alone can drive fusion with some specificity84,85. SNARE-driven fusion proceeds through a hemifusion intermediate86 and is sensitive to the distance between the SNARE domain and transmembrane anchor87. Studies with neuronal SNARE liposomes have shown that far lower amounts of SNAREs suffice for fusion when calcium and synaptotagmin are included88; these are key to neuronal fusion, but are not required for other intracellular fusion events. Recent studies have shown that SNARE proteoliposomes undergo extensive lysis as well as fusion, suggesting that much, but by no means all89, of the SNARE-dependent lipid mixing reflects lysis followed by reannealing rather than true fusion90,91. The balance between lysis and fusion has also been confirmed with yeast vacuole fusion92: vacuole fusion with endogenous levels of fusion proteins is not accompanied by lysis, but the fusion of vacuoles from strains with elevated amounts of each vacuolar SNARE becomes Rab-independent and is accompanied by massive organellar lysis. It is not known which factors, in response to SNARE-mediated bilayer destabilization, guide docked organelles to fusion instead of lysis.
Current models of intracellular fusion
Membrane fusion is governed by layers of specific interactions, reversible80,93,94 and subject to regulation, such as by phosphorylation95–97. Organelle-specific Rabs and Rab-effectors mediate tethering98,99 (Fig. 1a) and the assembly of fusion-competent microdomains (Fig. 1b), on the basis of a web of mutual affinities of fusion regulatory lipids for one another, of fusion regulatory lipids for proteins with direct capacity to bind to these lipids and regulate their synthesis, and of the proteins for one another. Microdomain assembly may be the key to achieving efficient fusion without lysis. After enrichment in microdomains, SNAREs assemble with each other in trans (Fig. 1c). trans-SNARE complexes include additional bound factors such as SM proteins, complexin and synaptotagmin (at synapses) or large complexes such as HOPS63,100 (at the vacuole or lysosome). trans-SNARE pairs have transmembrane anchors in each bilayer, and the formation of continuous straight α-helices between these transmembrane anchors and their SNARE domains in the four-helix trans complex has been proposed to drive bilayer distortion87, triggering hemifusion (Fig. 1d) and then completion of fusion (Fig. 1e). Although insertion of aminoacyl residues between SNARE and transmembrane domains inhibits fusion, fusion is not blocked by insertion of helix-disrupting prolyl or glycyl resides, suggesting that this may not be the sole means by which SNAREs trigger fusion. Two other mechanisms have been suggested. SNARE transmembrane domains can be inherently bilayer disrupting101–103, perhaps because of their insertion at an angle to the bilayer104; trans-SNARE pairing may localize their bilayer-disrupting property to the fusion microdomain on the two docked membranes. SNAREs are also required for the enrichment of lipids, including DAG, that inherently disrupt bilayer structure3,105 (red lipid head-groups in Fig. 1).
Mitochondrial fusion
An entirely distinct process of membrane fusion and fission regulates and executes the dynamic state of mitochondrial development in yeast and metazoan cells. The balance of fusion and fission permits mitochondrial mixing when yeast cells mate and partition the parental mitochondrial genomes in daughter diploid progeny. Genetic studies in yeast, Drosophila and mammalian cells have identified the main components of fusion and fission, defining processes quite distinct from the events that mediate membrane fusion among secretory organelles.
Because mitochondria possess a two-membrane envelope, the fusion process (Fig. 2a) begins by the apposition of outer membranes in a reaction dependent on the transmembrane protein Fzo1 (fuzzy onion; first identified in Drosophila melanogaster)106–108. In mammals, equivalent molecules termed mitofusins (Mfn1 and Mfn2) have been characterized structurally and seem to provide an outer-membrane tethering function through antiparallel coiled coils109. Fzo1 is proposed to be a dynamin-like GTPase, although it is not clear that it acts analogously to dynamin to promote membrane fusion. The outer-membrane fusion event has been reconstituted in a cell-free reaction that depends on Fzo1 and shows distinct requirements for inner-membrane apposition and fusion110.
Figure 2.
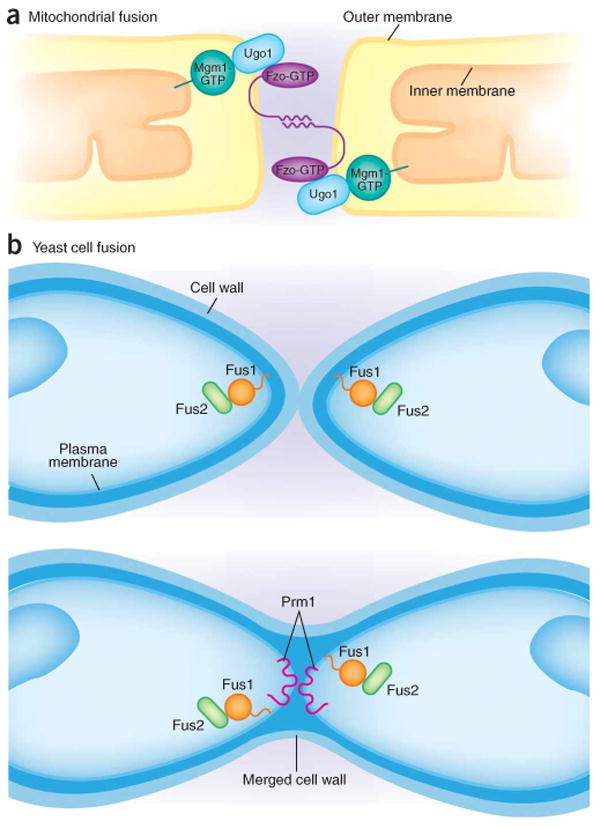
Mitochondrial fusion and cell fusion in mating yeast cells. (a) Mitochondria initiate contact and fusion through the interaction of Fzo1 (or Mfn in mammals), a dynamin-like GTPase located in the outer mitochondrial membrane. The fusion of the outer membrane occurs first, and then inner membrane contact and fusion is regulated by another dynamin-like GTPase, Mgm1 (or OPA1 in mammals). Ugo1 in the outer membrane provides a physical link between Fzo1 and Mgm1. (b) Yeast cells initiate cell fusion by regulated expression of two membrane proteins, Fus1p (a single-pass membrane protein) and Prm1p (a multispanning membrane protein), and a cytoplasmic protein, Fus2p. Fus1p and Fus2p localize to the cell mating tip and are required for the dissolution of the cell wall separating the plasma membranes of the mating pair. Prm1p is required for some as-yet-undefined reaction, possibly the formation of a fusion pore, that occurs when the mating cell plasma membranes come into contact.
Two other proteins, Ugo1 and Mgm1 (OPA1 in mammalian cells), coordinate outer-membrane fusion and inner-membrane contact. Ugo1 is an outer-membrane integral protein that seems to link Fzo1 to Mgm1 in the inner membrane111. Mgm1 is a GTPase and is more clearly related to mammalian dynamin than is Fzo1 (ref. 112). The in vitro fusion reaction allowed a demonstration that Mgm1, though present in a functional complex with Fzo1 and Ugo1, serves a distinct role after inner-membrane contact. Mutant studies in yeast have also shown that Mgm1/OPA1 also influences the morphology of normal inner-membrane cristae, but the strongest evidence favors a direct role of this dynamin in inner-membrane fusion113. Intra-allelic complementation studies using the in vitro fusion assay suggest that Mgm1 function may require self-oligomerization, which may promote a SNARE-like close juxtaposition of inner-membrane partners. Another possibility, by analogy to endocytic dynamin, is that Mgm1 may promote membrane tubules or buds that project fusogenic, highly curved ends. Clearly, a more refined analysis will require reconstitution of fusion events with separated outer and inner membrane fractions.
Cell fusion
Fusion at the cell surface employs fusogenic proteins independent of the regulatory molecules available to intracellular fusion events. Classic studies on viral fusion have identified mechanisms mediated by viral envelope glycoproteins, but thus far little similarity has been seen in the potentially related event of cell-cell fusion. Genetic analysis of Caenorhabditis elegans development and yeast cell mating have yielded important information on cell fusion. Cells in a number of body tissues in C. elegans harbor multiple nuclei resulting from cell-cell fusion. Two related molecules, EFF-1 and AFF-1, are required for epithelial and anchor cell fusion, respectively114,115. These single membrane–spanning proteins oligomerize, and they may also associate in trans, as EF-1 function seems to be required in both fusion partner cells116. Although other membrane protein partners may be required for cell fusion, the expression of EFF-1 in a distant surrogate, insect Sf-9 cells, is sufficient to promote the formation of multinucleate syncytia116,117. EFF-1 is likely to promote fusion directly, as it becomes enriched at the point of contact between cells in a prelude to fusion118.
Genetic, molecular cloning and localization studies have identified several cell-surface proteins that mediate yeast cell fusion during mating (Fig. 2b). Yeast cells secrete one of two peptides that interact with G protein–coupled receptors specific for the a or α mating type. Signal transduction mediated by binding of pheromone to the G protein–coupled receptor produces a cell-cycle-arrest program of transcription, protein processing and polarized cell-tip growth that results in the deposition of membrane and secretory proteins responsible for local cell fusion. An initial contact through the cell wall, promoted by cell type–specific agglutinins, leads to the dissolution of cell wall glycans and permits the generation of a fusion intermediate with two cells encased within a continuous wall. Two proteins required for fusion, Fus1p and Fus2p, were identified in a genetic screen wherein both mating partners must contain the mutation to display the defect. Fus1p is a single-spanning membrane protein and Fus2p is a cytosolic protein bound peripherally on the inner surface of the mating tip119,120. Mutations in the FUS1 and FUS2 genes arrest mating pairs at a step before the dissolution of the mating-tip cell wall, and before the direct apposition of the plasma membranes of the mating cells. Thus, these proteins may not have a direct role in membrane fusion121.
PRM1 encodes a multispanning membrane protein that seems to play a more direct role in mating cell membrane fusion. Prm1p is expressed only during mating and is localized to the mating-tip membrane. As with Fus1p and Fus2p, a mating defect is observed only when both partners lack Prm1p (ref. 122). When mating partners arrest in a prm1 mutant, the defect is only partial, with 40% proceeding to fusion. Nonetheless, the remaining pairs arrest with close contact between the surface membranes, clearly subsequent to the step defined by Fus1p and Fus2p. Interestingly, another consequence of the prm1 mating arrest is cell lysis123, possibly similar to the membrane lysis that accompanies aborted SNARE-mediated fusion in the vacuole fusion reaction. The exact role of Prm1p is not known, nor is it clear whether it constitutes the core of the cell-fusion machinery. An experiment such as that performed with C. elegans EFF-1 and AFF-1, wherein Prm-1 is expressed in a surrogate cell, could determine whether this protein has a central role in cell fusion.
What's next?
Membrane fusion has progressed from the genetic and biochemical identification of the relevant proteins and lipids and determination of their associations and structures to the current era of mechanistic studies. In several systems, the pure proteins and lipids of fusogenic microdomains are being reconstituted into proteoliposomes. Understanding the factors that guide fusion without lysis and reconstituting efficient fusion that depends on the physiological mixture of Rab, Rab effector and lipids, as well as SNAREs, will set the stage for a coming era of quantitative and chemical studies. Clearly, our understanding of cell fusion is at a more primitive stage, but genetic and limited molecular studies make it clear that this process is quite distinct and worthy of a vigorous biochemical approach.
Acknowledgments
We thank J. Shaw, A. Engel and P. Walter for discussions. Work in the authors' laboratories was supported by grants from the US National Institutes of Health and funds from the Howard Hughes Medical Institute (R.S.).
References
- 1.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malinin VS, Frederik P, Lentz BR. Osmotic and curvature stress affect PEG-induced fusion of lipid vesicles but not mixing of their lipids. Biophys J. 2002;82:2090–2100. doi: 10.1016/S0006-3495(02)75556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel DP, et al. Physiological levels of diacylglycerols in phospholipid membranes induce membrane fusion and stabilize inverted phases. Biochemistry. 1989;38:3703–3709. doi: 10.1021/bi00435a012. [DOI] [PubMed] [Google Scholar]
- 4.Burgess SW, McIntosh TJ, Lentz BR. Modulation of poly(ethylene glycol)-induced fusion by membrane hydration: importance of interbilayer separation. Biochemistry. 1992;31:2653–2661. doi: 10.1021/bi00125a004. [DOI] [PubMed] [Google Scholar]
- 5.Lee J, Lentz BR. Evolution of lipidic structures during model membrane fusion and the relation of this process to cell membrane fusion. Biochemistry. 1997;36:6251–6259. doi: 10.1021/bi970404c. [DOI] [PubMed] [Google Scholar]
- 6.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melikyan GB, Brener SA, Cok D, Cohen FS. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wharton SA, Martin SR, Ruigrok RWH, Skehel JJ, Wiley DC. Membrane fusion by peptide analogues of influenza haemagglutinin. J Gen Virol. 1988;69:1847–1857. doi: 10.1099/0022-1317-69-8-1847. [DOI] [PubMed] [Google Scholar]
- 10.Frolov VA, Dunina-Barkovskaya AY, Samsonov AV, Zimmerberg J. Membrane permeability changes at early stages of Influenza hemagglutinin-mediated fusion. Biophys J. 2003;85:1725–1733. doi: 10.1016/S0006-3495(03)74602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skehel JJ, Wiley DC. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 12.Beisson J, Lefort-Tran M, Pouphile M, Rossignol M, Satir B. Genetic analysis of membrane differentiation in Paramecium. Freeze-fracture study of the trichocyst cycle in wild-type and mutant strains. J Cell Biol. 1976;69:126–143. doi: 10.1083/jcb.69.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 16.Fries E, Rothman JE. Transport of vesicular stomatitis virus glycoprotein in a cell-free extract. Proc Natl Acad Sci USA. 1980;77:3870–3874. doi: 10.1073/pnas.77.7.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- 18.Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 19.Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuole morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- 21.Wilson DW, et al. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- 22.Südhof T. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 23.Trimble WS, Cowan DM, Scheller RH. VAMP-1: A synaptic vesicle-associated integral membrane protein. Proc Natl Acad Sci USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumert M, Maycox PR, Navone F, De Camilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18 000 daltons present in small synaptic vesicles of rat brain. EMBO J. 1989;8:379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett MK, Calakos N, Scheller RH. Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 26.Schiavo G, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 27.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Tokumaru H, et al. SNARE complex oligomerization by Spaphin/Complexin is essential for synaptic vesicle exocytosis. Cell. 2001;104:421–432. doi: 10.1016/s0092-8674(01)00229-x. [DOI] [PubMed] [Google Scholar]
- 29.Roggero CM, et al. Complexin/synaptotagmin interplay controls acrosomal exo-cytosis. J Biol Chem. 2007;282:26335–26343. doi: 10.1074/jbc.M700854200. [DOI] [PubMed] [Google Scholar]
- 30.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 31.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 32.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickman C, Davletov B. Mechanism of calcium-independent synaptotagmin binding to target SNAREs. J Biol Chem. 2003;278:5501–5504. doi: 10.1074/jbc.C200692200. [DOI] [PubMed] [Google Scholar]
- 34.Tang J, et al. A complexin/synaptotagmin1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 35.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 36.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 37.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 38.Shen J, Tareste DC, Paumet F, Rothman JE, Mella TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 39.Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci USA. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clary DO, Rothman JE. Purification of three related peripheral membrane proteins needed for vesicular transport. J Biol Chem. 1990;265:10109–10117. [PubMed] [Google Scholar]
- 41.Rice LM, Brunger AT. Crystal structure of the vesicular transport protein Sec17: Implications for SNAP function in SNARE complex disassembly. Mol Cell. 1999;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- 42.Cheever ML, et al. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 43.Lang T, et al. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Seeley S, Wickner W, Merz A. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 45.Collins KM, Wickner WT. trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc Natl Acad Sci USA. 2007;104:8755–8760. doi: 10.1073/pnas.0702290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichols BJ, Ungermann C, Pelham HRB, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- 47.Cho SJ, et al. SNAREs in opposing bilayers interact in a circular array to form conducting pores. Biophys J. 2002;83:2522–2527. doi: 10.1016/s0006-3495(02)75263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizo J, Südhof TC. SNAREs and Munc18 in synaptic vesicle fusion. Nat Rev Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 49.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott BL, et al. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dulubova I, et al. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi T, et al. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 53.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Südhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: Role of Munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 55.Peng R, Gallwitz D. Sly1 protein bound to Golgi syntaxin Sec5 allows assembly and contributes to specificity of SNARE fusion complexes. J Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 57.Walworth NC, Goud B, Kabcenell AK, Novick PJ. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins RN, Brennwald P, Garrett M, Lauring A, Novick P. Interactions of nucleotide release factor Dss4p with Sec4p in the post-Golgi secretory pathway of yeast. J Biol Chem. 1997;272:18281–18289. doi: 10.1074/jbc.272.29.18281. [DOI] [PubMed] [Google Scholar]
- 59.TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 60.Finger FP, Novick P. Spatial regulation of exocytosis; lessons from yeast. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo W, Roth D, Walch-Solumena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sacher M, et al. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C–Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–295. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medkova M, France YE, Coleman J, Novick P. The rab exchange factor Sec1p reversibly associates with the exocyst. Mol Biol Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stenmark H, et al. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 69.Horiuchi H, et al. A novel Rab5 GDP/GTP exchange factor complexed to rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 70.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999a;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 71.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 72.De Renzis S, Sönnichsen B, Zerial M. Divalent Rab effectors regulate the subcompartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 73.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 74.Miaczynska M, Zerial M. Mosaic organization of the endocytic pathway. Exp Cell Res. 2002;272:8–14. doi: 10.1006/excr.2001.5401. [DOI] [PubMed] [Google Scholar]
- 75.Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector rabaptin-5 and exchange factor rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shin HW, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McBride HM, et al. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 78.Wang L, Merz A, Collins K, Wickner W. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J Cell Biol. 2003;160:365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fratti R, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific “regulatory” lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jun Y, et al. Reversible, cooperative reactions of yeast vacuole docking. EMBO J. 2006;25:5260–5269. doi: 10.1038/sj.emboj.7601413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 82.Düzgünes N, Allen TM, Fedor J, Papahadjopoulos D. Lipid mixing during membrane aggregation and fusion: Why fusion assays disagree. Biochemistry. 1987;26:8435–8442. doi: 10.1021/bi00399a061. [DOI] [PubMed] [Google Scholar]
- 83.Meers P, Ali S, Erukulla R, Janoff AS. Novel inner monolayer fusion assays reveal differential monolayer mixing associated with cation-dependent membrane fusion. Biochim Biophys Acta. 2000;1467:227–243. doi: 10.1016/s0005-2736(00)00224-8. [DOI] [PubMed] [Google Scholar]
- 84.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 85.Zwilling D, et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 87.McNew JA, Weber T, Engelman DM, Söllner TH, Rothman JE. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 88.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 89.Nickel W, et al. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc Natl Acad Sci USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen X, et al. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Starai V, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jun Y, Xu H, Thorngren N, Wickner W. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 2007;26:4935–4945. doi: 10.1038/sj.emboj.7601915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Melia TJ, You D, Tareste DC, Rothman JE. Lipidic antagonists to SNARE-mediated fusion. J Biol Chem. 2006;281:29597–29605. doi: 10.1074/jbc.M601778200. [DOI] [PubMed] [Google Scholar]
- 95.Hirling H, Scheller R. Phosphorylation of synaptic vesicle proteins: Modulation of the aSNAP interaction with the core complex. Proc Natl Acad Sci USA. 1996;93:11945–11949. doi: 10.1073/pnas.93.21.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fujita Y, et al. Phosphorylation of Munc-18/nSec1/rbSec1 by protein kinase C. J Biol Chem. 1996;271:7265–7268. doi: 10.1074/jbc.271.13.7265. [DOI] [PubMed] [Google Scholar]
- 97.LaGrassa TJ, Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakamura N, Hirata A, Ohsumi Y, Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membrane and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- 101.Langosch D, et al. Conformation of the synaptobrevin transmembrane domain. J Mol Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 102.Hofmann MW, et al. Self-interaction of a SNARE transmembrane domain promotes the hemifusion to fusion transition. J Mol Biol. 2006;364:1048–1060. doi: 10.1016/j.jmb.2006.09.077. [DOI] [PubMed] [Google Scholar]
- 103.Siegel DP, et al. Transmembrane peptides stabilize inverted cubic phases in a biphasic length-dependent manner: implications for protein-induced membrane fusion. Biophys J. 2006;90:200–211. doi: 10.1529/biophysj.105.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das S, Rand RP. Diacylglycerol causes major structural transitions in phospholipid bilayer membranes. Biochem Biophys Res Commun. 1984;124:491–496. doi: 10.1016/0006-291x(84)91580-8. [DOI] [PubMed] [Google Scholar]
- 106.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 107.Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 109.Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 110.Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- 111.Sesaki H, Jensen RE. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem. 2004;279:28298–28303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- 112.Wong ED, et al. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meeusen S, et al. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 114.Mohler WA, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 115.Sapir A, et al. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Podbilewicz B, et al. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 117.Shemer G, et al. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol. 2004;14:1587–1591. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 118.del Campo JJ, et al. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr Biol. 2005;15:413–423. doi: 10.1016/j.cub.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 119.McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elion EA, Trueheart J, Fink GR. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J Cell Biol. 1995;130:1283–1296. doi: 10.1083/jcb.130.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gammie AE, Brizzio V, Rose MD. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jin H, Carlile C, Nolan S, Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sönnichsen B, et al. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jahn R, Scheller R. SNAREs — engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–646. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]


